Abstract
Background
Differentiating surveillance from non-surveillance colonoscopy for colorectal cancer in patients with inflammatory bowel disease (IBD) using electronic medical records (EMR) is important for practice improvement and research purposes, but diagnosis code algorithms are lacking. The automated retrieval console (ARC) is natural language processing (NLP)-based software that allows text-based document-level classification.
Aims
The purpose of this study was to test the feasibility and accuracy of ARC in identifying surveillance and non-surveillance colonoscopy in IBD using EMR.
Methods
We performed a split validation study of electronic reports of colonoscopy pathology for patients with IBD from the Michael E. DeBakey VA Medical Center. A gastroenterologist manually classified pathology reports as either derived from surveillance or non-surveillance colonoscopy. Pathology reports were randomly split into two sets: 70 % for algorithm derivation and 30 % for validation. An ARC generated classification model was applied to the validation set of pathology reports. The performance of the model was compared with manual classification for surveillance and non-surveillance colonoscopy.
Results
A total of 575 colonoscopy pathology reports were available on 195 IBD patients, of which 400 reports were designated as training and 175 as testing sets. Within the testing set, a total of 69 pathology reports were classified as surveillance by manual review, whereas the ARC model classified 66 reports as surveillance for a recall of 0.77, precision of 0.80, and specificity of 0.88.
Conclusions
ARC was able to identify surveillance colonoscopy for IBD without customized software programming. NLP-based document-level classification may be used to differentiate surveillance from non-surveillance colonoscopy in IBD.
Keywords: Crohn’s disease, Ulcerative colitis, Machine learning, Automated retrieval console
Introduction
Some groups of patients with inflammatory bowel disease (IBD) are at an increased risk of developing colorectal cancer (CRC), and therefore practice guidelines recommend that these patients undergo colonoscopy surveillance to detect dysplasia or early cancer [1, 2]. Two prior studies reported rates of CRC surveillance among IBD patients that varied between 25 and 90 % [3, 4]. Identifying surveillance colonoscopy and estimating surveillance rates in IBD patients have important implications both for measuring and improving quality of care, as well as for research purposes. However, the methods employed in ascertaining surveillance colonoscopy by prior studies are limited. One of these studies differentiated surveillance from non-surveillance colonoscopy using a retrospective chart review, while the second study used a text retrieval strategy based on manual pathology review and the number of pathology specimens. Retrospective chart review is impractical in large or multisite cohorts, and classification based on the number of pathology specimens may not be transferrable in all practice settings.
The availability of disease registries and administrative databases provides the possibility of using automated data retrieval on very large numbers of patients with IBD. For example, we recently identified over 45,000 unique patients with IBD in the national veterans affairs (VA) outpatient and inpatient databases. However, the absence of specific procedure codes for surveillance and non-surveillance colonoscopy limits the use of administrative data for researching surveillance practices and outcomes and quality of care measurements and improvement. Research utilizing databases is currently limited to less specific administrative-level data or requires time-intensive chart review. However, an additional resource of extensive databases with text-based medical information has recently become available with the advent of electronic medical records (EMR). Efficient methods of retrieving this data for clinical or research purposes should be explored.
Natural language processing (NLP) is a sub-discipline of computer science and linguistics focused on developing methods to automatically add structure to otherwise unstructured free text. Automated document-level classification can be used to help classify patient conditions or treatments in which standard diagnostic or procedural codes are not available or inaccurate. The Automated Retrieval Console (ARC) is NLP-based software that allows investigators without programming expertise to design and perform NLP assisted document-level classification. ARC works by combining features derived from NLP pipelines with supervised machine learning classification algorithms [5]. ARC has been used successfully in a number of studies to facilitate EMR based research including studies to classify diagnostic versus therapeutic pathology reports for breast and prostate cancer and to identify psychotherapy treatment types for patients with post-traumatic stress disorder [6, 7]. In addition to advancing opportunities and accuracy of database research, NLP has potential for clinical applications when embedded into EMR to allow real-time clinical feedback of clinical performance.
Our aim was to test the feasibility and accuracy of ARC in document-level classification of surveillance and non-surveillance colonoscopy for IBD.
Materials and Methods
This study was approved by the Institutional Review Boards of Baylor College of Medicine and the Michael E. DeBakey VA Medical Center in Houston, Texas.
Manual Classification
Subjects with IBD diagnoses at the Michael E. DeBakey VA Medical Center in Houston, TX were identified by International Classification of Diseases, 9th revision (ICD-9) code and confirmed by manual review of the EMR. All available colonoscopy and related pathology reports were extracted from the computerized patient record system (CPRS) in text format. Colonoscopy related pathology reports were randomly divided into a 70 % training set for algorithm derivation and a 30 % testing set for validation. Pathology reports related to colonoscopy procedures were manually classified as either surveillance or non-surveillance for dysplasia by a gastroenterologist (JKH). Pathology reports were classified as surveillance using a priori predefined criteria. Reports were classified as surveillance if the indication for “surveillance” was explicitly described or if the pattern of biopsies was consistent with surveillance biopsies, i.e. multiple biopsies (>1) were obtained from each segment of the colon or four quadrant biopsies every 10 cm, if the indication was not explicitly described. Reports in which the procedure was performed for symptoms, i.e. “rectal bleeding” or “diarrhea,” without explicit description of surveillance were classified as non-surveillance. Pathology reports were chosen for this study because of the relative uniformity in reporting and documentation as compared to endoscopic reports.
NLP with ARC
ARC classifies text documents using NLP pipelines to breakdown documents into structured fragments of text based on parts of speech, negated terms and a library of medical and non-medical terms. ARC uses the clinical text analysis and knowledge extraction system (cTAKES), an unstructured information management architecture (UIMA)-based analysis pipeline designed specifically to parse medical documents [8]. Document classification is performed in ARC using the conditional random fields (CRF) implementation from machine learning for language toolkit (MALLET), a text-based information retrieval tool [9]. To deliver acceptable classification performance across domains with no custom software development, ARC automatically attempts combinations of features for a given classification problem, calculating the performance of several models against the training set using tenfold cross validation. Users of ARC are shown all models attempted and the recall, precision, and harmonic mean (F-measure) of each. Selected models can be deployed on a larger cohort of documents.
For this study, documents in the training set were judged in ARC models as derived from surveillance or non-surveillance colonoscopy as determined by manual classification. A classification algorithm was generated to identify pathology documents from surveillance colonoscopy using a case insensitive CRF algorithm with binary vectors and overlapping features. The best performing algorithm was selected based on the F-measure, the harmonic mean of recall (estimate of sensitivity) and precision (estimate of positive predictive value [PPV]) as estimated by tenfold validation. The best performing model was then applied to the testing cohort, and its performance in terms of recall and precision was compared with manual classification of pathology reports by a gastroenterologist.
Data Analysis
Performance of ARC classification was reported as precision (estimate of PPV) = {relevant documents} ⋂ {retrieved documents}/{retrieved documents}, recall (estimate of sensitivity) = {relevant documents} ⋂ {retrieved documents}/{relevant documents}, and specificity = 1 – {non-relevant documents} ⋂ {retrieved documents}/{non-relevant documents}. Agreement was calculated as the number of identified surveillance documents based on ARC classification of pathology records that were also classified as surveillance based manual review of colonoscopy reports divided by the total number of manually classified surveillance documents. Likewise, agreement for non-surveillance was calculated as the number of documents classified as non-surveillance based on ARC retrieved divided by the total number of manually classified non-surveillance documents.
A sensitivity analysis was performed using a classification algorithm using a number of pathology specimen jars of ≥4 or ≥5 to define surveillance colonoscopy status; these cutoff numbers were based on a prior study [3]. The number of pathology specimen jars was determined by manual review from each pathology report. Sensitivity was calculated as the number of pathology reports correctly classified as derived from surveillance colonoscopy by both specimen jar number and manual classification divided by the total number of pathology reports classified as surveillance by manual classification. Specificity was calculated as the number of pathology reports classified as non-surveillance by specimen jar number and manual classification divided by the total number of pathology reports classified as non-surveillance by manual classification.
Categorical variables were compared using χ2 tests. Continuous variables were compared using t-tests. Statistical analysis was performed using Stata version 11 software.
Results
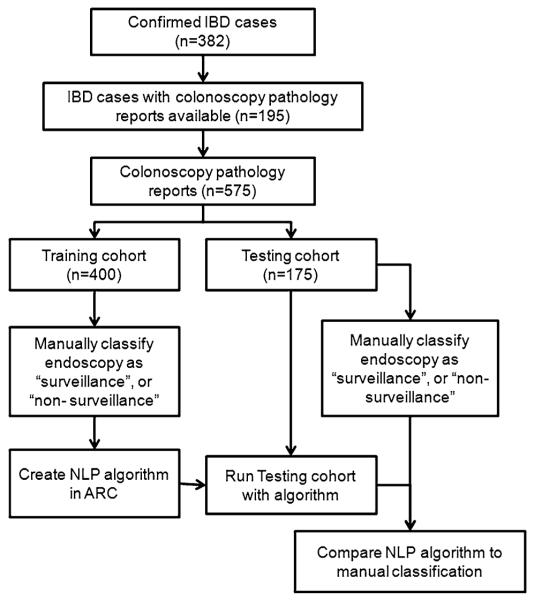
A total of 382 unique IBD patients were identified by medical chart review during 1998–2009 (Fig. 1); 92 % were men, 73 % white, 43 % Crohn’s disease (CD), 56 % ulcerative colitis (UC), 1 % indeterminate colitis (IC); mean age was 63 years old (SD 16.2). A total of 575 colonoscopy pathology reports were available on 195 IBD patients, of which 400 pathology reports were designated as the training cohort and 175 as the testing cohort. There were no significant demographic differences between the patients who had colonoscopy reports available and those who did not in mean age (62 vs. 65 years, p = 0.10), IBD type (40 % CD, 58 % UC, 1 % IC vs. 47 % CD, 52 % UC, 1 % IC, p = 0.37) or race (73 % white vs. 79 % white, p = 0.17).
Fig. 1.
Split validation workflow for natural language processing (NLP) and manual colonoscopy classification
Manual Review of Reports
All 575 colonoscopy pathology reports were manually reviewed and classified as derived from surveillance or non-surveillance colonoscopy. Of the 575 colonoscopy pathology reports reviewed, 251 (44 %) were manually classified as surveillance and 324 (56 %) were classified as non-surveillance.
Training Set
In the training set, 182 pathology reports (46 %) were manually classified as derived from a surveillance colonoscopy procedure. The best performing ARC model had an F-measure of 0.79, recall of 0.80, and precision of 0.80 by tenfold cross-validation. Cross validation is the process in which the training set is divided into ten equal groups. A classification algorithm is created based on the first nine of ten groups and is validated on the tenth group. The classification algorithm is similarly applied to the other nine groups and the performance parameters are averaged. Recall of 0.80 means that 80 % of all pathology reports from surveillance colonoscopy were correctly identified as surveillance by NLP. Precision of 0.80 means that 80 % of the reports classified as surveillance by NLP were also manually classified as surveillance colonoscopy.
Testing Set
In the testing set, 69 pathology reports were classified as derived from surveillance colonoscopy by manual review, whereas the ARC model (derived in the steps described above) classified 66 reports as surveillance, with a 77 % agreement for surveillance reports and an 88 % agreement for non-surveillance reports compared to manual classification (Table 1). ARC classification performance had a recall (estimate of sensitivity) of 0.77 (95 % confidence interval [CI] 0.66, 0.85), precision (estimate of positive predictive value) of 0.80 (95 % CI 0.72, 0.90), and specificity of 0.88 (95 % CI 0.80, 0.93) compared with manual classification.
Table 1.
Performance of ARC-derived algorithm for classification of surveillance and non-surveillance colonoscopy pathology reports and manual review
| Reports | Manual classification (n) |
NLP classification (n) |
Agreement (n) | Agreement, % (95 % CI) |
|---|---|---|---|---|
| Surveillance | 69 | 66 | 53 | 77 (67, 87) |
| Non-surveillance | 106 | 109 | 93 | 88 (82, 94) |
NLP natural language processing, ARC automated retrieval console
Sensitivity Analysis
We analyzed in a traditional (non-NLP) test characteristics analysis the sensitivity and specificity of different cutoff values of number of pathology jars in predicting surveillance or non-surveillance studies.
A total of 2,425 specimen jars were obtained in the 575 pathology reports reviewed, with a mean of 4.3 (standard deviation 2.6) jars per report. A total of 324 pathology reports had ≥ 4 specimen jars, with a sensitivity of 0.86 (95 % CI 0.80, 0.89), and specificity of 0.65 (95 % CI 0.60, 0.70) of predicting a surveillance study compared to manual classification. A total of 260 pathology reports had ≥ 5 specimen jars, with a sensitivity of 0.74 (95 % CI 0.69, 0.79) and specificity of 0.76 (95 % CI 0.72, 0.81) of predicting a surveillance colonoscopy compared to manual classification.
Discussion
We performed a split validation study of NLP-based pathology document classification of surveillance and non surveillance colonoscopy for IBD patients using ARC software. ARC derived models were able to retrieve pathology reports that were derived from a surveillance colonoscopy with a recall (sensitivity) of 0.77 and specificity of 0.88 compared with manual review of the pathology reports.
Much of traditional database research has been dependent upon the availability and accuracy of administrative databases which were designed for billing, not research purposes. For some questions, database research provides an efficient way to study the epidemiology and practice patterns of certain disease. IBD database research has been limited by the absence of administrative codes for key disease related variables such as intent of colonoscopy, disease extent, and disease severity.
Text-based data have been accumulating in databases since the advent of EMRs and may complement available administrative data for IBD research purposes. As an example, access for research purposes to text-based data from the VA is being developed through the Veterans’ Informatics, Information and Computing Infrastructure (VINCI). We have identified over 45,000 VA users with IBD (unpublished data), and the development of accurate information retrieval and NLP-based tools where administrative data is lacking may create opportunities to answer epidemiological and health services research questions such as quality of care and utilization using existing data.
This pilot study shows that ARC can reliably perform document classification of surveillance and non-surveillance endoscopy. All data acquisition and NLP analyses for this study were performed using ARC by a gastroenterologist without training in computer programming. The performance of classification was adequate with a specificity of 88 % and recall (sensitivity) of 77 %. Based on experience in other domains we believe the performance of ARC will improve with larger datasets as the NLP algorithms improve with increasing training set size [6, 7].
Velayos et al. [3] used a computerized algorithm that searched for text phrases related to surveillance or number of pathology specimens to distinguish surveillance from non-surveillance pathology reports, and reported high performance with a PPV of 97 % and negative predictive value (NPV) of 80 %. Our sensitivity analysis using only the number of pathology jars did not perform as well as previously described. We hypothesize the performance of an algorithm using only number of pathology jars was lower in our study than that reported by Velayos et al. due to facility differences in biopsy collection patterns, interpretation of pathology reports, and the additional use of the word “surveillance” in their algorithm. In order to achieve high level of accuracy, text searches such as derivatives of the word “surveillance” must be enumerative of all patterns of interest and programmed to account for negative references (e.g., “no surveillance”). In contrast, ARC’s combination of features from grammatical NLP pipelines and supervised machine learning has several advantages. First, the use of robust feature sets accounts for negated descriptions (e.g., no, absence of, rule out). Second, rather than making assumptions as to which features are most relevant, ARC “learns” the most relevant features iteratively. The incorporation of machine learning algorithms implies that performance is likely to improve as the number of examples increase. Finally, ARC’s approach can easily be applied to other domains and tasks with no custom software development.
This was a pilot study to determine the feasibility and accuracy of ARC in using automated NLP to classify surveillance and non-surveillance colonoscopy in IBD patients. Text data were manually extracted from only one VA facility, and additional training and validation will need to be performed in a larger cohort from multiple facilities to account for variation in pathology reporting and endoscopy biopsy practice patterns. The proportions of IBD patients with surveillance colonoscopy reported here should not be interpreted as an audit of surveillance rates as subjects were included based only on a confirmed diagnosis of IBD and clinical factors such as disease extent and duration were not taken into account in the analyses. Furthermore, there may be inter- and intra-observer variability in the manual interpretation of pathology reports for surveillance and non-surveillance. Although predefined criteria for surveillance were used, there may be subjectivity in surveillance classification by a single reviewer. Pathology reports were specifically chosen because of perceived relative uniformity in reporting and documentation as compared to endoscopic documentation. In addition, there are multiple formats of colonoscopy reports that may make text-based searching more difficult in large datasets.
In summary, NLP based EMR document classification using ARC software is feasible and accurate in retrieving surveillance colonoscopy in IBD patients when compared to manual classification of EMR. Although our study did not address surveillance practice patterns, we have demonstrated ARC may be used to examine this question in larger datasets for future studies. NLP based applications may open new opportunities in database research in the field of IBD by allowing EMR-based determination of variables, such as the reason for endoscopy, that are not available by using administrative data alone. Further studies of NLP applications embedded into EMR may provide real-time feedback of clinical performance and practice patterns.
Acknowledgments
The research reported here was supported by the American College of Gastroenterology Junior Faculty Development Award to J.K. Hou, a pilot grant from the Houston VA HSR&D Center of Excellence (HFP90-020) to J.K. Hou, the Department of Veterans Affairs, Veterans Health Administration, Health Services Research and Development Service, grant MRP05-305 to J.R. Kramer, and the National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Disease Center grant P30 DK56338, K24 DK078154-05.
Abbreviations
- IBD
Inflammatory bowel disease
- ARC
Automated retrieval console
- NLP
Natural language processing
- CRC
Colorectal cancer
- VA
Veterans affairs
- ICD-9
International classification of diseases, 9th revision
- EMR
Electronic medical records
- CPRS
Computerized patient record system
- cTAKES
Clinical text analysis and knowledge extraction system
- UIMA
Unstructured information management architecture
- CRF
Conditional random fields
- MALLET
Machine learning for language toolkit
- PPV
Positive predictive value
- CD
Crohn’s disease
- UC
Ulcerative colitis
- IC
Indeterminate colitis
- CI
Confidence interval
- NPV
Negative predictive value
Footnotes
Conflict of interest None.
Contributor Information
Jason K. Hou, Houston VA HSR&D Center of Excellence, Michael E. DeBakey Veterans Affairs Medical Center, Houston, TX, USA; Department of Medicine, Baylor College of Medicine, Houston, TX, USA; Section of Gastroenterology and Hepatology, Department of Medicine, Baylor College of Medicine, Houston, TX, USA; 1709 Dryden Road, Suite 8.40, Houston, TX 77030, USA
Mimi Chang, Houston VA HSR&D Center of Excellence, Michael E. DeBakey Veterans Affairs Medical Center, Houston, TX, USA; Department of Medicine, Baylor College of Medicine, Houston, TX, USA.
Thien Nguyen, Massachusetts Veterans Epidemiology Research and Information Center (MAVERIC) Cooperative Studies Coordinating Center, VA Boston Healthcare System, Jamaica Plain, MA, USA.
Jennifer R. Kramer, Houston VA HSR&D Center of Excellence, Michael E. DeBakey Veterans Affairs Medical Center, Houston, TX, USA; Department of Medicine, Baylor College of Medicine, Houston, TX, USA
Peter Richardson, Houston VA HSR&D Center of Excellence, Michael E. DeBakey Veterans Affairs Medical Center, Houston, TX, USA; Department of Medicine, Baylor College of Medicine, Houston, TX, USA.
Shubhada Sansgiry, Houston VA HSR&D Center of Excellence, Michael E. DeBakey Veterans Affairs Medical Center, Houston, TX, USA; Department of Medicine, Baylor College of Medicine, Houston, TX, USA.
Leonard W. D’Avolio, Massachusetts Veterans Epidemiology Research and Information Center (MAVERIC) Cooperative Studies Coordinating Center, VA Boston Healthcare System, Jamaica Plain, MA, USA; Division of Aging, Harvard Medical School, Boston, MA, USA
Hashem B. El-Serag, Houston VA HSR&D Center of Excellence, Michael E. DeBakey Veterans Affairs Medical Center, Houston, TX, USA; Department of Medicine, Baylor College of Medicine, Houston, TX, USA; Section of Gastroenterology and Hepatology, Department of Medicine, Baylor College of Medicine, Houston, TX, USA
References
- 1.Kornbluth A, Sachar DB. Practice parameters committee of the American College of Gastroenterology. Ulcerative colitis practice guidelines in adults: American College of Gastroenterology, practice parameters committee. Am J Gastroenterol. 2010;105:501–523. doi: 10.1038/ajg.2009.727. [DOI] [PubMed] [Google Scholar]
- 2.Farraye FA, Odze RD, Eaden J, et al. AGA technical review on the diagnosis and management of colorectal neoplasia in inflammatory bowel disease. Gastroenterology. 2010;138:746–774. doi: 10.1053/j.gastro.2009.12.035. [DOI] [PubMed] [Google Scholar]
- 3.Velayos FS, Liu L, Lewis JD, et al. Prevalence of colorectal cancer surveillance for ulcerative colitis in an integrated health care delivery system. Gastroenterology. 2010;139:1511–1518. doi: 10.1053/j.gastro.2010.07.039. [DOI] [PubMed] [Google Scholar]
- 4.Kottachchi D, Yung D, Marshall JK. Adherence to guidelines for surveillance colonoscopy in patients with ulcerative colitis at a Canadian quaternary care hospital. Can J Gastroenterol. 2009;23:613–617. doi: 10.1155/2009/691850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical text analysis and knowledge extraction system (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc. 2010;17:507–513. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.D’Avolio LW, Nguyen TM, Farwell WR, et al. Evaluation of a generalizable approach to clinical information retrieval using the automated retrieval console (ARC) J Am Med Inform Assoc. 2010;17:375–382. doi: 10.1136/jamia.2009.001412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shiner B, D’Avolio LW, Nguyen TM, et al. Automated classification of psychotherapy note text: implications for quality assessment in PTSD care. J Eval Clin Pract. 2012;18:698–701. doi: 10.1111/j.1365-2753.2011.01634.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farwell WR, D’Avolio LW, Scranton RE, et al. Statins and prostate cancer diagnosis and grade in a veterans population. J Natl Cancer Inst. 2011;103:885–892. doi: 10.1093/jnci/djr108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCallum AK. MALLET: A machine learning for language toolkit. 2012 http://mallet.cs.umass.edu.