Abstract
We present the complete 2,843,201-bp genome sequence of Treponema denticola (ATCC 35405) an oral spirochete associated with periodontal disease. Analysis of the T. denticola genome reveals factors mediating coaggregation, cell signaling, stress protection, and other competitive and cooperative measures, consistent with its pathogenic nature and lifestyle within the mixed-species environment of subgingival dental plaque. Comparisons with previously sequenced spirochete genomes revealed specific factors contributing to differences and similarities in spirochete physiology as well as pathogenic potential. The T. denticola genome is considerably larger in size than the genome of the related syphilis-causing spirochete Treponema pallidum. The differences in gene content appear to be attributable to a combination of three phenomena: genome reduction, lineage-specific expansions, and horizontal gene transfer. Genes lost due to reductive evolution appear to be largely involved in metabolism and transport, whereas some of the genes that have arisen due to lineage-specific expansions are implicated in various pathogenic interactions, and genes acquired via horizontal gene transfer are largely phage-related or of unknown function.
The Gram-negative oral spirochete Treponema denticola is predominantly associated with the incidence and severity of human periodontal disease (1). This polymicrobial infection and inflammation of the gingiva occurs in 80% of the adult population at some time in their lives and can evolve to severe forms including refractory periodontitis and acute necrotizing gingivitis, resulting in bone resorption and tooth loss. Treatment regimens to combat periodontitis are difficult and costly involving extensive antibiotic treatment and intricate surgery. T. denticola dwells in a complex and diverse microbial community within the oral cavity, and as such is highly specialized to survive within this milieu. This aerotolerant anaerobe (2) is related to the syphilis-causing obligate human pathogen, Treponema pallidum subsp. pallidum. T. denticola is one of ≈60 treponemal species or uncharacterized phylotypes found in dental plaque (3). Spirochetes comprise a monophyletic phylum that exhibits overall structural similarity and rRNA relatedness but great variability in habitat, physiologic properties, and genome size and organization (Table 1).
Table 1. General genome features.
| T. denticola | T. pallidum* | B. burgdorferi* | L. interrogans† | |
|---|---|---|---|---|
| Size, bp | 2,843,201 | 1,138,012 | 910,725 | 4,691,184 |
| G+C content, % | 37.9 | 52.8 | 28.6 | 36.0 |
| Protein-coding genes | ||||
| No. with assigned function | 1,223 | 542 | 487 | 2,060 |
| No. of unknown function‡ | 352 | 35 | 22 | 146 |
| No. of conserved hypotheticals§ | 477 | 175 | 102 | 569 |
| No. with no database match¶ | 734 | 288 | 242 | 1,952 |
| Total | 2,786∥ | 1,040 | 853 | 4,727 |
| Average CDS size, bp | 939 | 1,017 | 992 | 778 |
| Coding, % | 92.1 | 93.0 | 93.5 | 78.4 |
| rRNA | 6 | 6 | 5 | 4 |
| tRNA | 44 | 45 | 34 | 37 |
The distribution of CDSs in the T. pallidum and B. burgdorferi chromosomes are derived from the original annotation. These numbers, particularly hypothetical and conserved hypothetical proteins, may be significantly different with updated blast searches and annotation.
The genome information for L. interrogans represents combined data from both chromosomes.
Unknown function, significant sequence similarity to a named protein for which no function is currently attributed.
Conserved hypothetical protein, sequence similarity to a translation of another ORF, however no experimental evidence for protein expression exists.
Hypothetical protein, no significant similarity to any other sequenced protein.
Twenty-five of the total number of CDSs in T. denticola possess one or more authentic frameshifts, point mutations, or are truncated.
Comparative analysis with previously sequenced spirochetes (T. pallidum, Borrelia burgdorferi, and Leptospira interrogans) (4–6) yielded insights into the basis for differences in their lifestyle and disease manifestations. The genome of T. denticola clearly reflects its adaptations for colonization and survival within the biofilm environment of subgingival dental plaque. Compared to other spirochetes, T. denticola is relatively easy to cultivate and manipulate genetically, making this an excellent model for spirochete research.
Methods
Bacteria. T. denticola strain 35405 was initially isolated and designated as the type strain by Chan et al. (7). Bacteria used in this study were obtained from the American Type Culture Collection.
Sequencing and Gene Identification. The complete genome of T. denticola strain 35405 was sequenced by using the random shotgun method described for genomes sequenced by The Institute for Genomic Research (5). ORFs likely to encode proteins (CDSs) were predicted by glimmer (8). All predicted proteins >30 aa were analyzed for sequence similarity against a nonredundant protein database. Two sets of hidden Markov models were used to determine CDS membership in families and superfamilies: pfam v5.5 (9) and tigrfams (10). Domain-based paralogous families were built by performing all-versus-all searches on the remaining protein sequences. The 5′ regions of each CDS were inspected to define initiation codons by using homologies, position of ribosomal binding sites, and transcriptional terminators. Sequences containing frameshifts and point mutations were reexamined and corrected where appropriate. Protein membrane-spanning domains were identified by toppred (11). Putative signal peptides were identified with signalp (12).
Trinucleotide Composition. Distribution of all 64 trinucleotides (3-mers) for each chromosome was determined, and the 3-mer distribution in 2,000-bp windows that overlapped by half their length (1,000 bp) across the genome was computed. For each window, we computed the χ2 statistic on the difference between its 3-mer content and that of the whole chromosome. A large value for χ2 indicates the 3-mer composition in this window is different from the rest of the chromosome. Probability values for this analysis are based on assumptions that the DNA composition is relatively uniform throughout the genome, and that 3-mer composition is independent. Because these assumptions may be incorrect, we prefer to interpret high χ2 values as indicators of regions on the chromosome that appear unusual and demand further scrutiny.
Comparative Genomics. The T. pallidum and T. denticola genomes were compared at the nucleotide level by suffix tree analysis using mummer, and their ORF sets were compared by using blast. Additionally, all T. denticola CDSs were compared by blastp against the complete set of CDSs from T. pallidum, L. interrogans, and B. burgdorferi using an E value cutoff of 10–5.
Results and Discussion
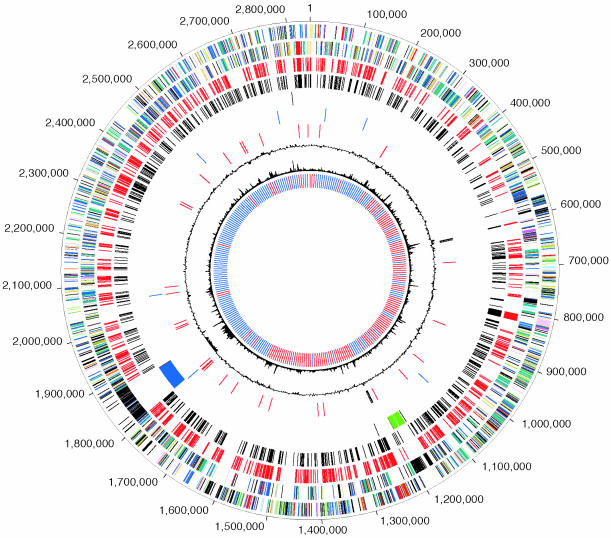
General Genome Features. The 2,843,201-bp chromosome of T. denticola (ATCC 35405) is predicted to encode 2,786 CDSs, of which 26.5% (734 CDSs) are unique. Although the T. denticola and T. pallidum genome sizes are significantly different, the number of stable RNAs present in each is near identical (Table 1 and Fig. 1). The organization of the T. denticola chromosomal origin of replication is disparate from the organization (dnaA-dnaN-recF-gyrB) typical for bacteria, including other spirochetes (Fig. 4, which is published as supporting information on the PNAS web site). Base pair 1 was assigned in an intergenic region between dnaA and dnaE based on previous analyses (13). However the transition in GC skew (G – C/G + C) typically associated with the origin of replication (14) does not correspond with the assigned base pair 1. Instead, a GC inversion occurs near a distally located recF, with an adjacent dnaN, and a gene encoding an ATPase domain protein that may serve as a gyrase. The relative disruption in the organization of replication genes and the GC skew analysis suggests that there may have been a lineage-specific genome rearrangement in the region of the replication origin. Annotated genome sequence and analysis for T. denticola is available at www.tigr.org/tigr-scripts/CMR2/GenomePage3.spl?database=gtd.
Fig. 1.
Circular representation of the T. denticola (ATCC 35405) overall genome structure. The outer scale designates coordinates in base pairs. The first circle shows predicted coding regions on the plus strand color-coded by role categories: violet, amino acid biosynthesis; light blue, biosynthesis of cofactors, prosthetic groups, and carriers; light green, cell envelope; red, cellular processes; brown, central intermediary metabolism; yellow, DNA metabolism; light gray, energy metabolism; magenta, fatty acid and phospholipid metabolism; pink, protein synthesis and fate; orange, purines, pyrimidines, nucleosides, and nucleotides; olive, regulatory functions and signal transduction; dark green, transcription; teal, transport and binding proteins; gray, unknown function; salmon, other categories; blue, hypothetical proteins. The second circle shows predicted coding regions on the minus strand color-coded by role categories. The third circle shows the core set of CDSs conserved in all other sequenced spirochete genomes. The fourth circle shows CDSs with best matches to predicted CDSs in T. pallidum. The fifth circle shows putative phage regions and isolated phage genes. The sixth circle shows IS elements in black. The seventh circle shows rRNA genes in black and tRNA genes in red. The eighth circle shows trinucleotide composition in black. The ninth circle shows percentage G + C in relation to the mean G + C in a 2,000-bp window. The 10th circle shows GC-skew curve in red (positive residues) and blue (negative residues).
Comparative Genomics. It has been proposed that the 1.14 Mb T. pallidum genome was derived from the genome of T. denticola or other host-associated treponemes through deletion and divergence (15). Comparisons reveal that the T. pallidum genome shares limited nucleotide identity with the T. denticola genome. About one-fourth of T. denticola genes (728 CDSs) have their best matches to CDSs in the T. pallidum genome (representing 68% of its genome), and on average, these share only 53% amino acid identity (70.6% similarity). Essentially no synteny (conservation of gene order) exists between the T. denticola and T. pallidum genomes [with the exception of highly conserved operons encoding ribosomal (TDE0766-TDE0792) and flagellar proteins (TDE1198-TDE1219)]. This result, as well as differences in G + C content (Table 1) and rRNA sequences, indicates that divergence of T. denticola and T. pallidum from a common ancestor was an ancient event relative to the recent divergence of many bacterial groups (for which complete genome sequences are available), such as the Brucella and Rickettsia genera and some members of the Enterobacteriaceae.
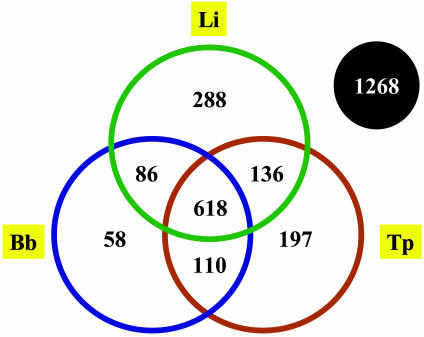
Comparisons of the total set of T. denticola CDSs with that of the other sequenced spirochetes by using blastp reveals a core set of 618 CDSs found conserved in the other three spirochete genomes (Fig. 2 and Table 2, which is published as supporting information on the PNAS web site). Of these, ≈90% have an assigned role, most of which are housekeeping functions (DNA replication, repair, cell division, transcription, translation, energy metabolism, etc.); others include ATP-binding cassette (ABC) transporters (≈120 CDS), and flagellum and chemotaxis genes (≈55 CDS). A total of 1,268 T. denticola CDSs have no matches in any of the other spirochete genomes. Over half of these are hypothetical genes, whereas the remainder have their best matches in various sequenced Gram-positive species like Clostridium spp., Streptococcus spp., as well as Fusobacterium nucleatum, a primary colonizer during dental plaque formation, found in conjunction with T. denticola and Porphyromonas gingivalis, and thought to be required for their colonization (16). These include various membrane proteins, ABC transporters, transcriptional regulators, and enzymes involved in amino acid metabolism and glycogen synthesis.
Fig. 2.
Venn diagram showing the number of T. denticola predicted proteins with significant homology (E < 10–5) with the predicted products of the pathogenic spirochetes T. pallidum, B. burgdorferi, and L. interrogans. The number outside the circles (1,268) represents the number of T. denticola proteins that do not have significant homologs in any of the spirochete species examined.
A small subset of the 618 predicted gene products conserved in all spirochetes did not have significant matches in other phyla; these may represent spirochete-specific proteins. These include characterized flagellar components, FliL (TDE2764), FliE (TDE1214) (17), putative outer layer protein FlaA (TDE1408) (18) and its distant paralogs TDE1712 and TDE1409, that perhaps account for the unique periplasmic flagellar location and motility properties of spirochetes. The other spirochete-associated genes (TDE2598, TDE2751, TDE1300, and TDE1334) have no predicted functions, and two have regions of similarity to fibronectin (TDE0446), and tetratricopeptide repeat (TPR)-domain proteins (TDE2696). Overall, this group may be responsible for spirochete-specific properties, including morphology and host–pathogen interactions.
A total of 1,700 (≈60%) T. denticola CDSs have no matches in T. pallidum (Table 3, which is published as supporting information on the PNAS web site), and 162 T. pallidum CDSs have no matches in T. denticola (Table 4, which is published as supporting information on the PNAS web site). There are several possible reasons for this disparity in genome size and encoded CDSs, including (i) elimination of genes in one genome (reductive evolution), (ii) acquisition of genes via lateral transfer, and (iii) lineage-specific expansions, all subsequent to the divergence of the two species. Our comparisons reveal that all of these factors contribute in varying degrees to the differences in gene content (Fig. 2). About one-sixth of these 1,700 CDSs have their best matches to other CDSs within T. denticola and may represent recent duplication events or expansions within the T. denticola lineage (Table 5, which is published as supporting information on the PNAS web site). Over half of these putative duplicated genes are situated adjacent to each other, suggesting that they arose via tandem duplication events. Many of these may be implicated in virulence functions, for example, YD-repeat proteins, internalin-related proteins, and bacteriocin ABC transporters.
Of the 1,700 T. denticola CDSs that lack T. pallidum homologs, 129 have homologs in B. burgdorferi and 313 have homologs in L. interrogans, suggesting that these may have been lost by gene reduction in the T. pallidum lineage. Of the 162 T. pallidum CDSs with no significant match in T. denticola (Table 4), 106 are hypothetical proteins. The remainder includes transporters, components of a V-type ATPase, some proteases and enzymes involved in various metabolic activities including proline and asparagine synthesis, and the oxidative branch of the pentose phosphate pathway. In the case of the 106 hypothetical CDSs, one-third are <100 aa in length and may not necessarily represent expressed genes.
The biofilm nature of dental plaque allows for population diversity and coexistence of various aerobes, anaerobes, and microaerophiles. Exchange of genetic information is probable in such a milieu; for example, plasmid transfer from T. denticola to oral bacteria such as Streptococcus gordonii has been demonstrated (19). A number of genomic regions with unusual trinucleotide composition (see Methods) were identified that may signify laterally acquired DNA. For example, a 65-kb region (coordinates ≈1,807,000–1,872,000, TDE1756–TDE1843) encodes 66 hypothetical proteins and 15 conserved hypothetical proteins (all with the same transcriptional orientation). Genes for a single tRNA and a site-specific recombinase (TDE1844) flank this region, suggesting a phage-mediated integration event. A second 13-kb intergenic region (coordinates 367,000–370,000) possesses 54 copies of a 36-bp “clustered regularly interspaced short palindromic repeat” (CRISPR) (20) with four adjacently located CRISPR-associated cas genes (TDE0327–TDE0330). These direct repeats are interspaced by a nonrepetitive sequence of 30 bp. Although no function is assigned to the CRISPR locus, it has been hypothesized to be a mobile element. A third region, a single cryptic phage, is seen at coordinates 1,164,840–1,202,400 encoding a cluster including eight phage-related genes, 28 hypothetical genes, and a ParB nuclease domain protein. In addition, miscellaneous phage remnants and seven assorted insertion sequence transposases are also found scattered in the genome. At least three operons encoding type -I and -II restriction-modification (RM) systems are present. These are speculated to serve as a barrier against interspecies gene transfer (21) and may be important for survival in the biofilm. Homologs of these RM components are absent in the other spirochetes, with the exception of a single type I RM system seen in L. interrogans. Three predicted type IV RM systems have also been described in B. burgdorferi (22). In contrast, T. pallidum, which survives predominantly in the relatively secluded environment of human tissue, has no recognized RM systems, insertion sequence elements, or bacteriophage.
Overall, genes lost due to reductive evolution appear to be largely involved in basic metabolic functions and transport, whereas some of the genes that have arisen due to lineage-specific expansions are implicated in pathogenesis, and genes acquired via horizontal gene transfer are phage-related or of unknown function. These analyses emphasize that differences in genome sizes of T. pallidum and T. denticola are not solely due to reductive evolution of T. pallidum. Like many other obligate parasites, T. pallidum had undergone some reduction in metabolic capabilities indicating an increased dependence on host (and oral microbial community) for nutritional purposes. Lineage-specific expansions and lateral gene transfer, on the other hand, may reflect niche-specific adaptations and differences in their pathogenic potential.
Host–Pathogen Interactions. T. denticola has been shown to adhere to various cell types and basement membranes via binding to fibronectin, collagen, laminin, fibrinogen, and other substrates (23, 24). Additionally, because T. denticola is a late colonizer during plaque biofilm formation, adhesion to other oral bacteria is critical; binding to Fusobacterium, B. forsythus, and P. gingivalis has been demonstrated (25, 26). A whole-genome survey for potential surface-exposed proteins revealed an array of adhesins and other factors that may play a role in binding to host-cell components and/or coaggregation (Table 6, which is published as supporting information on the PNAS web site). These genes include a four-member family of YD repeat proteins (carbohydrate-binding), 11 tetratricopeptide (TPR) repeat-containing proteins (protein–protein interactions), peptidases, and proteases among others. Tracts of iterative DNA located either in the promoter region or the N terminus of some of these genes may suggest a potential for undergoing antigenic or phase variation.
The T. pallidum genome encodes a 12-member family (TprA-L) of putative membrane proteins (4). This T. pallidum gene family exhibits heterogeneity among different subspecies and strains, indicative of recombination events among the different tpr alleles (27). In addition, protective immune responses and generation of opsonic activity against Tpr proteins has been reported (28) but disputed (29). T. denticola possesses only `a single member (TDE0405, major outer sheath protein) related to this gene family, suggesting that this family has been specifically expanded in the T. pallidum lineage. The outer sheath of spirochetes is reported to be unusual in its composition and properties compared to the outer membrane of typical Gram-negative bacteria. Previous studies with T. denticola lipopoly-saccharide (LPS) are contradictory, ranging from the detection of LPS-like material (30) to the absence of any LPS structural components (31). TDE1419–TDE1441 encode various glycosyl hydrolases and RfbAB that may be involved in synthesis of surface polysaccharides; however, other enzymes for synthesis of Lipid A, KDO, or other LPS components are absent. However, two forms of LPS have been isolated from Treponema pectinovorum, indicating that at least some of the oral treponemes are capable of synthesizing LPS (32).
Proteinases and peptidases may aid in degradation of host tissue and bacterial components to fulfill nutritional requirements, and may contribute to cytotoxicity of periodontal infections (33–35). Over 25 assorted peptidases, hydrolases, and other putative degradative enzymes were identified in the genome. Because T. denticola is known to disrupt epithelial barriers and perturb actin and actin-regulating pathways in host cells (36), a three-member family of internalin-related proteins (two of which have signal peptides) may facilitate this process (Table 6).
Stress Responses. The dental biofilm environment transitions continually between feast and famine, aerobic and anaerobic conditions, and high and low pH. T. denticola possesses various mechanisms to counteract oxidative, osmotic, and other stresses. It has been reported to have NADH oxidase, NADH peroxidase, and superoxide dismutase (SOD)/reductase activities (37). These activities may be fulfilled by Nox (TDE0096), alkyl hydroperoxide reductase/peroxiredoxin (AhpC, TDE0011) in combination with Nox, and desulfoferrodoxin/neelaredoxin (TDE1754) (38), respectively. In addition, a single rubrerythrin (TDE0434), implicated in oxidative stress protection via catalytic reduction of intracellular H2O2, may account for the observed SOD activity (39). Homologs of Bacteroides fragilis aerotolerance-associated genes, batAB (TDE1250, TDE1252) (40), may be implicated in aerotolerance of T. denticola as well.
Prokaryotic selenoproteins catalyze redox reactions and formation of selenoethers in (stress-induced) metabolism and energy production. Previous assessment of the nutritional requirements of T. denticola revealed a strict growth dependence on selenium (41). Genome sequence reveals the complete set of genes for selenoprotein synthesis: selA, (TDE2477), selD (TDE2461), and selB (TDE1963). In E. coli, the selenium is incorporated into formate dehydrogenase (fdh) subunits; although no fdh is identified in the T. denticola genome, a thioredoxin (TDE0238), glycine reductase complex selenoproteins (grdA, TDE0745, grdB, TDE2119, TDE0078), and glutathione peroxidase (TDE1729), all contain predicted selenocysteine sites. Given the potential role of these selenoproteins in antioxidant functions, deficiency of selenium may have detrimental effects on T. denticola's ability to withstand oxidative stress. Other stress–response mechanisms include an arginine deiminase (TDE0451) to counter acidification by producing ammonia from arginine, and betaine aldehyde dehydrogenase (TDE0080) and a possible transporter (TDE1261) for synthesis or uptake of betaine, an osmoprotectant. T. denticola possesses enzymes for glycogen synthesis (unlike the other spirochetes), which may be a specific adaptation for survival during starvation conditions. T. denticola has a proportionately greater number of regulatory factors (relative to genome size) compared to T. pallidum, including seven sigma factor homologs (TDE0070, TDE0091, TDE0937, TDE1346, TDE2320, TDE2404, and TDE2683), nine CDSs encoding two-component signal transduction systems, and 28 miscellaneous transcriptional regulators.
Transport and Metabolism. Consistent with the known proteolytic abilities of T. denticola, analysis of the transporter content of the genome reveals an array of peptide and amino acid uptake systems, including eight ABC-type peptide uptake systems. By contrast, there are very limited capabilities for uptake of other organic nutrients, including a glucose/galactose ABC transporter (TDE2215–TDE2217), and probable transporters for lactose, gluconate, and carboxylates. Like T. pallidum, T. denticola possesses HPr, enzyme I and enzyme IIA of the phospho-transferase system (PTS), but no PTS transporter complex, suggesting that these proteins play a purely regulatory role. Iron acquisition appears to be an important priority for T. denticola with the identification of eight probable ABC-type uptake systems for iron chelates or siderophores.
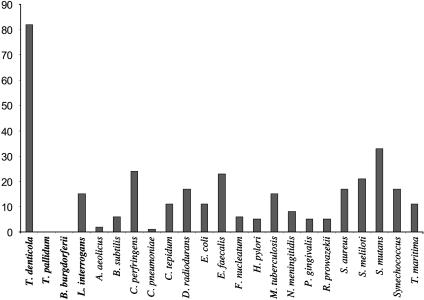
Unexpectedly, T. denticola encodes a very large complement of predicted ABC-type drug efflux systems. Eighty-three predicted ABC family drug efflux proteins were identified, comprising at least 47 different ABC efflux systems, significantly more than any sequenced spirochete or other prokaryotes (Fig. 3). Three of these systems (TDE0719–TDE0720; TDE0425–TDE0426; TDE2431–TDE2430) are probable bacteriocin secretion systems, based on the presence of three proximally encoded bacteriocin-type signal domain-containing CDSs (TDE0416, TDE0422, TDE0424). The secreted bacteriocins may be bacteriostatic for competition against other organisms, or may be for signaling within the oral biofilm. Homologs of the lux genes are not present in the T. denticola genome in contrast to other dental microbes that produce autoinducer (AI-2) signaling peptides. Speculatively, the remainder of the ABC efflux systems may play a role in secretion of effectors mediating host interactions or in providing resistance against antagonistic factors (toxins, secondary metabolites, antibiotics) produced by other microbes in the dental biofilm. T. denticola also possesses nine probable sodiumion driven MATE family drug efflux systems as well as other predicted drug efflux systems.
Fig. 3.
Number of predicted ABC efflux transporter genes present in various microbial species.
Comparative analysis of metabolic profiles across all four sequenced spirochete genomes reveals some distinctions of T. denticola metabolism. T. pallidum and B. burgdorferi have limited biosynthetic abilities in accordance with their reduced genome sizes, whereas L. interrogans and T. denticola have somewhat greater capabilities. Genes encoding enzymes for glycolysis, gluconeogenesis, and a pentose phosphate pathway are present (Table 7, which is published as supporting information on the PNAS web site). Unlike T. pallidum and B. burgdorferi, the pentose phosphate pathway in T. denticola (and L. interrogans) lacks the oxidative branch. The existence of glycolysis and absence of a tricarboxylic acid (TCA) cycle in T. denticola suggests that ATP is generated by sugar fermentation as in the case of B. burgdorferi and T. pallidum, whereas L. interrogans possesses both a TCA cycle and an electron transport chain (ETC). The lack of cytochromes and quinone biosynthesis genes in T. denticola indicates that it does not possess an ETC for energy production.
In T. pallidum and B. burgdorferi, the membrane potential essential for transport, motility and other cellular functions is apparently established by V1V0 ATPases; these V-type complexes typically consume ATP to translocate H+ or Na+ ions across the membrane. T. pallidum has two V1V0-type ATPase operons, with different subunit compositions, each speculated to possess different specificities for Na+ or H+. The single V-type ATPase in T. denticola shares similarity with H+-specific ATPase systems; however, the operon has undergone considerable rearrangement (genes encoding the subunits are ordered K-I-D-B-A, with a subunit E gene located 300 kb upstream), and the subunit I gene contains a frameshift, which might render the ATPase nonfunctional. Another component that may also contribute to the proton gradient is a proton-translocating pyrophosphatase (TDE0651), although this gene also has a frameshift. TDE0834–TDE0838 have sequence similarity to the Na+-translocating NADH/quinone reductase complex (nqrABCDE) of other organisms. It has been speculated that this complex in T. denticola and other organisms maintains a Na+ gradient used by several transport systems (42). However, T. denticola and many other host-associated organisms (including Chlamydia, Porphyromonas, and Clostridium species) that encode NQR complexes do not have recognizable biosynthetic pathways for ubiquinone, a required cofactor for the NQR systems characterized to date. An interesting possibility is that these bacteria may scavenge quinone compounds from the surrounding micro-environment. This concept is not unprecedented, in that Escherichia coli and Salmonella typhimurium are not capable of synthesizing the o-quinone pyrroloquinolone quinone and must import it as a cofactor to express glucose dehydrogenase activity (43). However, the T. denticola genome does not encode a recognizable quinone oxidase. Therefore, it is likely that the T. denticola system utilizes another electron donor/receptor, such as ferridoxin or rubredoxin, to fulfill the electron exchange part of this reaction. Finally, Na+/H+ antiporter family proteins TDE2203 and TDE2204 may permit exchange between the Na+ and H+ gradients.
The T. denticola genome also contains all five required genes of a glycine reductase complex (grdA–grdE), including two divergent copies each of grdB and grdE. This complex acts in consort with thioredoxin and thioredoxin reductase to deaminate and phosphorylate the substrate, thus contributing to the pool of phosphorylated compounds (44). Alternate substrates in Clostridium and Eubacterium species include betaine, sarcosine, and N-methyl derivatives of glycine; the polypeptide subunits derived from the multiple grdB and grdE genes in T. denticola may affect substrate specificity and permit the utilization of amino acids and other compounds for substrate phosphorylation and ATP production. This possibility is consistent with the growth of T. denticola in peptone/yeast extract/serum medium in the absence of glucose with conversion of amino acids to ammonia and acetic, lactic, succinic, and formic acids (45).
Unlike T. pallidum and B. burgdorferi, de novo synthesis of fatty acids, cofactors, and nucleotides is possible. T. denticola may also use a number of sugars like glucose, galactose, glycerol, melibiose, fucose, and sorbitol. In contrast to the other spirochetes, enzymes for synthesis of glycogen (TDE2035 and TDE1582) as well as degradation to maltodextrin (TDE2411 and additional α-amylases) are apparent. Although amino acid biosynthesis is deficient, numerous genes for uptake, interconversion, and catabolism are present.
Conclusions
The analysis of the T. denticola genome and comparisons with other spirochetes has revealed insights into the evolution and adaptive responses within the spirochete phylum. T. denticola is primarily restricted to the subgingival plaque and does not cause the systemic infections and manifestations characteristic of T. pallidum, Borrelia, and Leptospira. However, a number of factors that may contribute to periodontal disease were identified. The genome sequence together with recently developed genetic techniques (46, 47) will permit analysis of genes involved in the colonization, survival, growth, and pathobiology of T. denticola in this unique polymicrobial environment.
Supplementary Material
Acknowledgments
We thank S. Salzberg, O. White, M. Heaney, S. Lo, M. Holmes, M. Covarrubias, J. Sitz, A. Resnick, J. Zhao, M. Zhurkin, R. Deal, R. Karamchedu, and V. Sapiro for informatics, database, and software support. This work was supported by the National Institute of Dental and Craniofacial Research Grant RO1-DE12488.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: CDS, protein-coding sequences.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AE017226).
References
- 1.Loesche, W. J. & Grossman, N. S. (2001) Clin. Microbiol. Rev. 14, 727–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Syed, S. A., Makinen, K. K., Makinen, P. L., Chen, C. Y. & Muhammad, Z. (1993) Res. Microbiol. 144, 317–326. [DOI] [PubMed] [Google Scholar]
- 3.Paster, B. J., Boches, S. K., Galvin, J. L., Ericson, R. E., Lau, C. N., Levanos, V. A., Sahasrabudhe, A. & Dewhirst, F. E. (2001) J. Bacteriol. 183, 3770–3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fraser, C. M., Norris, S. J., Weinstock, G. M., White, O., Sutton, G. G., Dodson, R., Gwinn, M., Hickey, E. K., Clayton, R., Ketchum, K. A., et al. (1998) Science 281, 375–388. [DOI] [PubMed] [Google Scholar]
- 5.Fraser, C. M., Casjens, S., Huang, W. M., Sutton, G. G., Clayton, R., Lathigra, R., White, O., Ketchum, K. A., Dodson, R., Hickey, E. K., et al. (1997) Nature 390, 580–586. [DOI] [PubMed] [Google Scholar]
- 6.Ren, S. X., Fu, G., Jiang, X. G., Zeng, R., Miao, Y. G., Xu, H., Zhang, Y. X., Xiong, H., Lu, G., Lu, L. F., et al. (2003) Nature 422, 888–893. [DOI] [PubMed] [Google Scholar]
- 7.Chan, E. C., Siboo, R., Keng, T., Psarra, N., Hurley, R., Cheng, S. L. & Iugovaz, I. (1993) Int. J. Syst. Bacteriol. 43, 196–203. [DOI] [PubMed] [Google Scholar]
- 8.Delcher, A. L., Harmon, D., Kasif, S., White, O. & Salzberg, S. L. (1999) Nucleic Acids Res. 27, 4636–4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bateman, A., Birney, E., Durbin, R., Eddy, S. R., Howe, K. L. & Sonnhammer, E. L. (2000) Nucleic Acids Res. 28, 263–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haft, D. H., Loftus, B. J., Richardson, D. L., Yang, F., Eisen, J. A., Paulsen, I. T. & White, O. (2001) Nucleic Acids Res. 29, 41–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Claros, M. G. & von Heijne, G. (1994) Comput. Appl. Biosci. 10, 685–686. [DOI] [PubMed] [Google Scholar]
- 12.Nielsen, H., Engelbrecht, J., Brunak, S. & von Heijne, G. (1997) Protein Eng. 10, 1–6. [DOI] [PubMed] [Google Scholar]
- 13.Greene, S. R. & Stamm, L. V. (2000) Gene 253, 259–269. [DOI] [PubMed] [Google Scholar]
- 14.Lobry, J. R. (1996) Mol. Biol. Evol. 13, 660–665. [DOI] [PubMed] [Google Scholar]
- 15.MacDougall, J. & Saint Girons, I. (1995) J. Bacteriol. 177, 1805–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Socransky, S. S., Haffajee, A. D., Cugini, M. A., Smith, C. & Kent, R. L., Jr. (1998) J. Clin. Periodontol. 25, 134–144. [DOI] [PubMed] [Google Scholar]
- 17.Stamm, L. V. & Bergen, H. L. (1999) FEMS Microbiol. Lett. 179, 31–36. [DOI] [PubMed] [Google Scholar]
- 18.Champion, C. I., Blanco, D. R., Exner, M. M., Erdjument-Bromage, H., Hancock, R. E., Tempst, P., Miller, J. N. & Lovett, M. A. (1997) J. Bacteriol. 179, 1230–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang, B. Y., Chi, B. & Kuramitsu, H. K. (2002) Oral Microbiol. Immunol. 17, 108–112. [DOI] [PubMed] [Google Scholar]
- 20.Jansen, R., Embden, J. D., Gaastra, W. & Schouls, L. M. (2002) Mol. Microbiol. 43, 1565–1575. [DOI] [PubMed] [Google Scholar]
- 21.Bourniquel, A. A. & Bickle, T. A. (2002) Biochimie 84, 1047–1059. [DOI] [PubMed] [Google Scholar]
- 22.Lawrenz, M. B., Kawabata, H., Purser, J. E. & Norris, S. J. (2002) Infect. Immun. 70, 4798–4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thomas, D. D. (1996) Crit. Rev. Oral Biol. Med. 7, 4–11. [DOI] [PubMed] [Google Scholar]
- 24.Ellen, R. P., Lepine, G. & Nghiem, P. M. (1997) Adv. Dent. Res. 11, 33–42. [DOI] [PubMed] [Google Scholar]
- 25.Yao, E. S., Lamont, R. J., Leu, S. P. & Weinberg, A. (1996) Oral Microbiol. Immunol. 11, 35–41. [DOI] [PubMed] [Google Scholar]
- 26.Kolenbrander, P. E., Andersen, R. N., Blehert, D. S., Egland, P. G., Foster, J. S. & Palmer, R. J., Jr. (2002) Microbiol. Mol. Biol. Rev. 66, 486–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Centurion-Lara, A., Sun, E. S., Barrett, L. K., Castro, C., Lukehart, S. A. & Van Voorhis, W. C. (2000) J. Bacteriol. 182, 2332–2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Centurion-Lara, A., Castro, C., Barrett, L., Cameron, C., Mostowfi, M., Van Voorhis, W. C. & Lukehart, S. A. (1999) J. Exp. Med. 189, 647–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hazlett, K. R., Sellati, T. J., Nguyen, T. T., Cox, D. L., Clawson, M. L., Caimano, M. J. & Radolf, J. D. (2001) J. Exp. Med. 193, 1015–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dahle, U. R., Tronstad, L. & Olsen, I. (1996) Endod. Dent. Traumatol. 12, 202–205. [DOI] [PubMed] [Google Scholar]
- 31.Schultz, C. P., Wolf, V., Lange, R., Mertens, E., Wecke, J., Naumann, D. & Zahringer, U. (1998) J. Biol. Chem. 273, 15661–15666. [DOI] [PubMed] [Google Scholar]
- 32.Walker, S. G., Xu, X., Altman, E., Davis, K. J., Ebersole, J. L. & Holt, S. C. (1999) Oral Microbiol. Immunol. 14, 304–308. [DOI] [PubMed] [Google Scholar]
- 33.Makinen, K. K. & Makinen, P. L. (1996) Med. Microbiol. Immunol. 185, 1–10. [DOI] [PubMed] [Google Scholar]
- 34.Sela, M. N. (2001) Crit. Rev. Oral Biol. Med. 12, 399–413. [DOI] [PubMed] [Google Scholar]
- 35.Chi, B., Qi, M. & Kuramitsu, H. K. (2003) Res. Microbiol. 154, 637–643. [DOI] [PubMed] [Google Scholar]
- 36.Baehni, P. C., Song, M., McCulloch, C. A. & Ellen, R. P. (1992) Infect. Immun. 60, 3360–3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caldwell, C. E. & Marquis, R. E. (1999) Oral Microbiol. Immunol. 14, 66–72. [DOI] [PubMed] [Google Scholar]
- 38.Jovanovic, T., Ascenso, C., Hazlett, K. R., Sikkink, R., Krebs, C., Litwiller, R., Benson, L. M., Moura, I., Moura, J. J., Radolf, J. D., et al. (2000) J. Biol. Chem. 275, 28439–28448. [DOI] [PubMed] [Google Scholar]
- 39.Lehmann, Y., Meile, L. & Teuber, M. (1996) J. Bacteriol. 178, 7152–7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang, Y. P., Dallas, M. M. & Malamy, M. H. (1999) Mol. Microbiol. 32, 139–149. [DOI] [PubMed] [Google Scholar]
- 41.Rother, M., Bock, A. & Wyss, C. (2001) Arch. Microbiol. 177, 113–116. [DOI] [PubMed] [Google Scholar]
- 42.Hase, C. C., Fedorova, N. D., Galperin, M. Y. & Dibrov, P. A. (2001) Microbiol. Mol. Biol. Rev. 65, 353–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stites, T. E., Mitchell, A. E. & Rucker, R. B. (2000) J. Nutr. 130, 719–727. [DOI] [PubMed] [Google Scholar]
- 44.Andreesen, J. R. (1994) Antonie Leeuwenhoek 66, 223–237. [DOI] [PubMed] [Google Scholar]
- 45.Smibert, R. M. (1984) in Bergey's Manual of Systematic Bacteriology, eds. Kreig, N. R. & Holt, J. G. (Williams & Wilkins, Baltimore), Vol. 1, pp. 49–57. [Google Scholar]
- 46.Girons, I. S., Chi, B. & Kuramitsu, H. (2000) J. Mol. Microbiol. Biotechnol. 2, 443–445. [PubMed] [Google Scholar]
- 47.Chi, B., Limberger, R. J. & Kuramitsu, H. K. (2002) Infect. Immun. 70, 2233–2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.