Summary
One of the key questions in understanding the biology of an organism is how to correlate cellular fate and function with gene expression patterns. This is particularly relevant for pathogenic organisms, like the parasitic protozoa Trypanosoma brucei, who often cycle between different hosts, thereby encountering vastly different environments. Survival in and adaptation to new surroundings requires activation of specific gene networks, which is most often achieved by regulatory mechanisms embedded in the transcriptional machinery. However, in T. brucei and related trypanosomatids these responses appear to be accomplished mainly by post-transcriptional mechanisms. Although an understanding of how this parasite modulates gene regulatory networks is in the early stages, RNA-binding proteins (RBPs) are beginning to take center stage. Here, we discuss recent progress in the identification of RBPs with crucial roles in different stages of the T. brucei life cycle, and in elucidating targets of RBPs.
Introduction
The protozoan parasite Trypanosoma brucei, the causative agent of African trypanosomiasis in humans (HAT) and nagana in animals, is transmitted by the tsetse fly (Glossina spp.), the blood-feeding dipteran vector. Like many parasites, T. brucei undergoes remarkable transformations during its life cycle, each adapting to distinct surroundings in the mammalian host or the insect vector (Vickerman et al., 1988). Within the bloodstream of the mammal, the parasites exist as proliferative slender forms, which establish parasitaemia. The differentiation of slender to quiescent, stumpy bloodstream forms occurs in response to cell density (Vassella et al., 1997) and proceeds gradually through transitional forms, referred to as intermediates. Stumpy trypanosomes are arrested in their cell cycle and are primed to respond to the environmental changes associated with the uptake by the tsetse fly (Matthews, 2011). In the insect midgut, stumpy forms differentiate into procyclic forms that are no longer infectious to mammals (Dyer et al., 2013). Reacquisition of infectivity is achieved through a complex developmental program that culminates in the tsetse salivary glands with the generation of metacyclic forms, which again are non-dividing forms ready to cope with a change in their environment when they are transmitted to the mammalian host. Trypanosomes rapidly respond to transmission between hosts by a complex cellular differentiation that includes radical changes in surface protein expression, metabolism, organelle function, and cytoskeletal architecture. For example, throughout the life cycle, the trypanosome plasma membrane is covered by densely packed surface coats consisting of GPI-anchored proteins. In the bloodstream the variant surface glycoprotein (VSG) coat is the paradigm for antigenic variation (Horn et al., 2010), in the tsetse midgut, trypanosomes express procyclins, a family of EP (Glu/Pro repeat-containing) and GPEET (Gly/Pro/Glu/Glu/Thr repeat-containing) proteins (Roditi et al., 1987) and epimastigote forms express a family of proteins known as brucei alanine-rich proteins or BARPs (Urwyler et al., 2007).
Whereas the T. brucei life cycle was first described early in the 20th century, we are just beginning to get insights into regulatory mechanisms operating at different life cycle stages. Traditionally, an analysis of circuits controlling developmental processes mainly concentrated on the transcriptional machinery. However, in recent years it has become increasingly evident that post-transcriptional processes play equally important roles in the output of gene products and in some organisms, like the trypanosomatids (including the genera Trypanosoma and Leishmania), the regulation of gene expression appears to have moved away almost entirely from transcriptional control to post-transcriptional regulatory mechanisms. The conceptual shift in trypanosomatids started with the realization that protein coding genes in these organisms are organized into long unidirectional clusters, where multiple genes are co-transcribed by RNA polymerase II (Pol II). In general, there is no apparent functional connection between neighboring genes or genes in an array. Polycistronic pre-mRNAs are processed to mature mRNAs by trans-splicing of the spliced leader (SL) at the 5′ end and 3′ end formation/polyadenylation (Preusser et al., 2012). Current evidence is consistent with Pol II transcribing all protein coding genes at comparable rates, although this is inferred from limited experimental data. However, the final output of mature mRNAs and proteins can vary tremendously between genes, and even between adjacent genes. In addition, expression levels of the same gene differ dramatically between various developmental stages. This conundrum elevated post-transcriptional mechanisms to major players in the regulation of gene expression. These include, but are not limited to, differential pre-mRNA processing (efficiency of trans-splicing and polyadenylation, alternative trans-splicing and polyadenylation), mRNA transport and subcellular localization (efficiency of export from the nucleus, sequestration in a variety of cytoplasmic mRNP bodies/granules), mRNA stability, mRNA translation, protein stability and post-translational modifications. In this scenario, RNA-binding proteins (RBPs) are emerging as key factors. The evolving picture is that, as in other eukaryotes, trypanosome RBPs interact through cis-acting elements with any part of the pre-mRNA or the mature mRNA, untranslated regions (3′UTR or 5′UTR) or coding sequence (CDS), and are suspected to have dominant roles in controlling gene expression in trypanosomatids at all steps of mRNA metabolism. Only recently evidence has emerged to support the view that RBPs can drive profound changes in the gene expression profile in these parasites, and as a result, influence trypanosome development. Here we briefly review the T. brucei RBPs playing a role in gene expression and life-cycle progression. Our overview does not cover general factors involved in RNA degradation, pre-mRNA splicing, polyadenylation, export and translation, and for a more complete view on RNA metabolism in trypanosomes, the reader is referred to additional excellent recent reviews on the subject covered here (Kramer et al., 2011, Clayton, 2013), mRNA splicing in trypanosomes (Gunzl, 2010, Michaeli, 2011, Preusser et al., 2012), mRNA degradation and translation initiation in trypanosomes and leishmanias (Clayton et al., 2007) and mitochondrial RNA processing in trypanosomes (Aphasizhev et al., 2011).
Repertoire of RNA-binding proteins
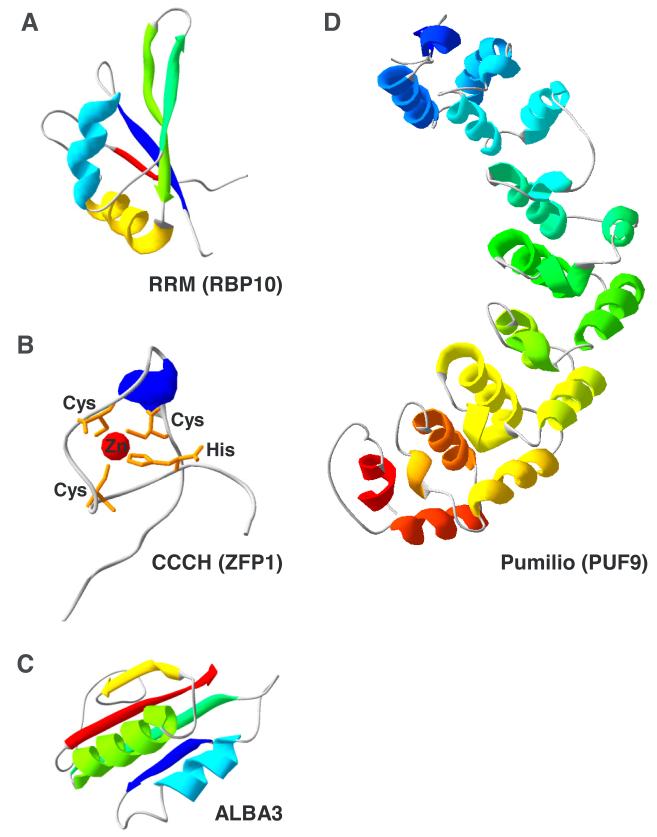
The RNA-Recognition Motif (RRM) domain is one of the most commonly found protein domains in nature. The structure of this versatile module of ~90 amino acids (Fig. 1A) consists of a four-stranded β-sheet packed against two α-helices (Nagai et al., 1990). T. brucei, like all other eukaryotes, contains a large number of RRM-containing RBPs, around 70 (De Gaudenzi et al., 2005, Wurst et al., 2009). RRMs most often are involved in sequence-specific interaction with single-stranded RNA, but can also interact with DNA, as well as other proteins, including themselves.
Fig. 1.
Tertiary structure models using the alignment interface of SWISS-MODEL (Bordoli et al., 2009) for the RRM domain of RBP10 (A), the Cys3His zinc finger of ZFP1 (B), the nucleic acid binding domain of ALBA3 (C), and the Pumilio domain of PUF9 (D).
The Cys3His Zinc finger or CCCH domain represents a subclass of the large zinc finger family with a characteristic disk-like structure (Amann et al., 2003)(Fig. 1B) and an apparent preference for binding to AU-rich RNA elements. The T. brucei genome encodes 48 proteins with this signature (Kramer et al., 2010).
There are four T. brucei proteins with a recognizable ALBA (acetylation lowers binding affinity) domain. In addition, these proteins contain a C-terminal stretch of multiple RGG repeats. The monomer structure (Fig. 1C) consists of four stranded β-sheet and two α-helices (Wardleworth et al., 2002). Proteins of this family have both DNA and RNA binding activity.
Pumilio domain RBPs
PUF proteins (Drosophila Pumilio and C. elegans Fem-3-binding factor BF are founding members of this family) were initially characterized in other eukaryotes as RBPs that can regulate the stability and the translation of their targets with binding sites in the 3′ UTR of the affected mRNAs. Some members have functions in pre-rRNA processing (Thomson et al., 2007). The core of the pumilio RNA-binding domain (Fig. 1D) consists of α-helical tandem repeats (Edwards et al., 2001, Wang et al., 2001), typically eight, each interacting with one base of the RNA sequence recognized by the protein (Wang et al., 2002). The RNA elements bound by PUF proteins usually contain the trinucleotide UGU (Wang et al., 2002). T. brucei harbors 11 PUF protein genes (Luu et al., 2006).
RBPs involved in the differentiation from bloodstream to procyclic forms
ZFP1 (Tb927.6.3490) and ZFP2 (Tb927.11.14950), two CCCH-family proteins, were the first trypanosome RBPs to be implicated in development and its regulation (Hendriks et al., 2001). ZFP1 was transiently up-regulated during the differentiation from bloodstream to procyclic forms and also expressed at higher levels in established procyclic cells. ZFP2 was present at equal levels in bloodstream and procyclic cells. RNAi knockdown of ZFP2 inhibited differentiation to procyclic forms, as judged by reduced EP procyclin expression, impaired kinetoplast repositioning and inhibited morphological restructuring (Hendriks et al., 2001). Additionally, ectopic expression of ZFP2 in procyclic forms resulted in gross morphological changes termed “nozzle” phenotype, the result of polar extension of the trypanosome cytoskeleton in cells which are early in the cell cycle. ZFP1 null bloodstream form cell lines were shown to be compromised in their ability to differentiate to procyclics, specifically in kinetoplast repositioning (Hendriks et al., 2005). ZFP3 (Tb927.3.720) was identified by means of its similarity in primary structure to ZFP1 and ZFP2 (Paterou et al., 2006). Ectopic ZFP3 expression in bloodstream T. brucei was shown to enhance the differentiation to procyclics. Expression in procyclics induced the “nozzle” phenotype previously observed for ZFP2, supporting the conclusion that these RBPs function in the same differentiation pathway. Protein-protein interactions ZFP1-ZFP2 and ZFP1-ZFP3 were demonstrated with a yeast two-hybrid system and all three proteins could be co-immunoprecipitated from cell extracts (Paterou et al., 2006). ZFP3 was also shown to be enriched on polysomes in procyclic cells, but not in bloodstream cells, and this enrichment correlated with the “nozzle” phenotype and enhancement of differentiation. Later studies (Walrad et al., 2012) demonstrated that ZFP3 associates with a subset of mRNAs that was enriched in stumpy bloodstream form trypanosomes, the abundance of these transcripts was modulated by ZFP3 levels and was mediated by sequences in the 3′UTRs of the mRNA targets.
ZC3H18 (Tb927.7.2140), a CCCH-family protein with two characteristic domains, was shown to be required for differentiation of bloodstream trypanosomes to procyclic forms (Benz et al., 2011), but its mode of action needs to be investigated.
RBP10 modulates mRNAs in bloodstream-form trypanosomes
RBP10 (Tb927.8.2780), a small RRM-containing protein was the first example of an RBP with profound life-cycle stage-specific effects on the global trypanosome transcriptome, and its developmental regulation (Wurst et al., 2012). RBP10 was found to localize primarily in the cytoplasm and its expression was shown to be up-regulated in bloodstream-form trypanosomes. The protein was also detected to be differentially phosphorylated (Urbaniak et al., 2012) with the modification found mostly in the bloodstream form. RNAi knockdown of RBP10 in bloodstream trypanosomes resulted in the down-regulation of a large number of mRNAs normally found elevated in bloodstream forms, and conversely, overexpression of the protein in procyclics led to an increase of many bloodstream-form specific mRNAs (Wurst et al., 2012). Thus, RBP10 was able to promote a bloodstream-form transcriptome pattern in T. brucei. Genes involved in sugar transport and metabolism were among the most affected by RBP10 and flagellum and cytoskeleton mRNAs were also modulated broadly. Finally, forced expression of RBP10 in bloodstream forms inhibited their differentiation to procyclics. At present it is unclear whether RBP10 interacts with mRNAs, however the RBP10 effects on mRNA metabolism were clearly shown to depend on the 3′UTRs of the affected transcripts (Wurst et al., 2012).
RBPs during trypanosome development in the tsetse fly
A second example of an RRM protein with a remarkable role in regulating trypanosome development is RBP6 (Tb927.3.2930). RBP6 was found to be highly enriched at the mRNA level in trypanosomes inhabiting the proventriculus of infected flies, when compared to procyclics from the midgut (Kolev et al., 2012). Inducible overexpression of RBP6 in procyclics, in an effort to mimic events in the proventriculus, resulted in progression through the developmental program of the parasite. Epimastigotes and, within a week of RBP6 expression, metacyclic trypomastigotes appeared in culture in the absence of any other developmental triggers. The metacyclic cells produced in vitro by inducible RBP6 expression were shown to express the VSG coat and importantly, the activated VSG genes possessed a metacyclic-type VSG Pol I promoter and were found in monocistronic transcription units (Kolev et al., 2012). More recently, it was suggested that RBP6 is a remote ELAV-like T. brucei homolog that binds in vitro to an AU-rich element (AUUUAUU) present in the 3′UTRs of mRNAs (Najafabadi et al., 2013). Interestingly, expression of RBP6 in procyclic cells was found to down-regulate mRNAs containing the identified sequence signature mRNAs (Najafabadi et al., 2013), an opposite effect in comparison to available data for ELAV family proteins from other species. Identifying the precise in vivo RNA-binding sites for RBP6 will undoubtedly clarify the repertoire of RBP6 targets and shed more light on its mode of action. The combined examples of RBP10 and RBP6 as potent triggers of developmental changes are a strong indication for the critical role of RBPs as post-transcriptional regulators of gene expression in T. brucei and other trypanosomatids.
Recently, the four T. brucei ALBA proteins were characterized (Mani et al., 2011, Subota et al., 2011). ALBA3 and ALBA4 (Tb927.4.2040/Tb927.4.2030) are primarily cytoplasmic proteins found to be expressed throughout the T. brucei life cycle, except in developmental stages found in the proventriculus of tsetse representing the transition from procyclic to epimastigote forms (Subota et al., 2011). Nutritional stress, a likely condition in the fly proventriculus, caused ALBA3/4 to localize to stress granules together with the helicase DHH1. Knockdown by RNAi of ALBA3/4 in procyclic trypanosomes grown in culture, mimicking the down-regulation of the proteins in the proventriculus, resulted in cellular morphology resembling the development in the fly proventriculus, specifically, elongation of the cell body and repositioning of the nucleus and the kinetoplast in a epimastigote configuration. Overexpression of these proteins in trypanosomes in the fly impaired the normal differentiation taking place in the proventriculus. These results represent yet another striking example of perturbation of T. brucei development by modulating the expression of proteins with RNA-binding potential (Subota et al., 2011). ALBA1/ALBA2 (Tb927.11.4460/Tb927.11.4450) are also primarily cytoplasmic (Mani et al., 2011). They were identified as proteins interacting with regulatory elements in the GPEET procyclin mRNA 3′UTR, and shown to form complexes with ALBA3 and ALBA4, suggestive of multiple homodimers/heterodimers possibilities between the T. brucei ALBA family members. Like ALBA3/4, ALBA1/2 are also recruited to stress mRNP granules upon starvation, a likely indication that they may be involved in translational control, supported by their partial co-migration with polysomes during density gradient centrifugation and co-purification with translation initiation factors and poly(A)-binding proteins (Mani et al., 2011).
Differential regulation of mRNA metabolism
The T. brucei homolog of hnRNP F/H (Tb927.2.3880) is a member of the RRM superfamily and was shown to play a major role in the differential regulation of mRNA metabolism (Gupta et al., 2013). This RBP is highly up-regulated in bloodstream-form trypanosomes and shows primarily nuclear localization, but is also present in the cytoplasm. Just like in other eukaryotes, where hnRNP F/H proteins regulate alternative splicing, the trypanosome homolog regulates the efficiency of trans-splicing and acts as a repressor. Additionally, the protein has a role in controlling differential mRNA stability in the bloodstream and procyclic life-cycle stages. The protein preferentially contacts its RNA substrates at an AAGAA sequence motif, found enriched in both the 3′UTR (for mRNA stability control) and upstream of the CDS (for effects on trans-splicing). HnRNP F/H is the first example of a differentially expressed splicing regulator in trypanosomes (Gupta et al., 2013).
RBPs with housekeeping functions
An RRM protein contacting its targets in the CDS is RBP42 (Tb927.6.4440). By using the powerful HITS-CLIP approach, it was shown that in vivo RBP42 binding sites cluster in the CDS of mRNAs, many of them coding for proteins involved in cellular energy metabolism (Das et al., 2012). RBP42 is a cytoplasmic protein that is associated with polysomes.
ZC3H11 (Tb927.5.810), a CCCH-family protein, is a phosphorylated cytoplasmic protein up-regulated upon heat shock and was shown to preferentially bind and stabilize mRNAs for a group of chaperones required for protein refolding, thus regulating the heat shock response in T. brucei (Droll et al., 2013). The interaction of ZC3H11 and mRNA was shown to be mediated by AUU repeats in the 3′UTRs, and the differential regulation of these mRNAs is dependent on these motifs.
Cell cycle and rRNA processing
PUF9 (Tb927.1.2600) was shown to be up-regulated in the S-phase of the cell cycle and inducible PUF9 RNAi knockdown resulted in accumulation of cells in G2/M (Archer et al., 2009). The protein was found localized primarily to the cytoplasm. Four specific mRNA targets were identified by PUF9 co-immunoprecipitation and it was demonstrated that PUF9 stabilizes 3 of these messages, leading to their up-regulation in S-phase of the cell cycle. This stimulatory effect of a trypanosome PUF protein on the expression of its targets is a deviation from the more typical repressive role assigned to these proteins. Nevertheless, the sequence motif 5′-UUGUACC-3′, containing the signature UGU recognized by many PUF proteins was found overrepresented in the 3′ UTRs of the identified targets. Additionally, a correlation between the presence of the motif, expression of PUF9 and cell-cycle changes in abundance of the target mRNAs strongly supported the case for stimulatory role of PUF9 in mRNA metabolism, likely by modulating RNA stability (Archer et al., 2009).
Another cytoplasmically localized protein of this family, PUF1 (Tb927.10.4430), was shown to interact with the expression site associated gene 8 (ESAG8) protein, which is enriched in the nucleolus, and to positively affect the levels of both ESAG8 mRNA and ESAG8 protein (Hoek et al., 2002). Overexpression of a HA-tagged PUF1 in bloodstream-form trypanosomes dramatically reduced their infectivity in mice. PUF1 has also been reported to associate with transcripts from repeat elements INGI and DIRE, and ribosomal RNA (Luu et al., 2006). PUF7 (Tb927.11.14960) localizes to the nucleolus, associates with a protein termed nuclear cyclophilin 1, and has an effect on pre-rRNA processing (Droll et al., 2010). A remarkable connection between pre-rRNA processing and regulation of the developmentally regulated Pol I-transcribed GPEET mRNAs was recently demonstrated, and two Pumilo domain proteins, PUF7 and PUF10 were implicated in this regulatory process (Schumann Burkard et al., 2013). By screening a genomic RNAi library for negative regulators of GPEET expression, a protein termed NRG1 was identified, and its binding partners included PUF7 and PUF10. Knockdown of each of these genes resulted in higher propensity for GPEET expression, lower levels of 5.8S rRNA and higher levels of rRNA precursors. The links between Pol I transcription, pre-rRNA processing and Pol I-transcribed mRNAs metabolism will likely continue to surface in the future, since shared protein factors and close proximity of nuclear compartments, where rRNA and surface coat mRNAs are produced, possibly dictate close connections in regulatory mechanisms. It will be of great interest to elucidate how changes in rRNA production rates during differentiation from dividing to quiescent cells prior to a new cycle of host invasion affect the production and stability of Pol I-transcribed mRNAs coding for surface proteins.
RBPs awaiting functional assignment
UBP1 (Tb927.11.500) and UBP2 (Tb927.11.510) are two closely related abundant RRM proteins (Hartmann et al., 2007). They were shown to localize primarily to the cytoplasm, but were not excluded from the nucleus. Modulating the expression levels of UBP1/2 affected the abundance of CFB1 mRNAs via an element in the 3′UTRs of these messages (Hartmann et al., 2007). An interesting feature of the UBP1 and UBP2 genes is the presence of a very long 3′UTR in the corresponding mRNAs (discussed later).
PUF5 (Tb927.7.4730) is localized primarily in the cytoplasm and makes contacts with cellular RNA of unknown nature (Jha et al., 2013). ZC3H12 (Tb927.5.1570) and ZC3H13 (Tb927.5.1580) are examples of two phosphorylated cytoplasmic zinc finger proteins, but their function is unclear (Ouna et al., 2012). ZC3H20 (Tb927.7.2660) was demonstrated to bind to the 3′UTRs and stabilize at least two developmentally regulated mRNAs (Ling et al., 2011).
Conclusions
RBPs have been universally shown to have a combinatorial mode of action, i.e. many RBPs are bound to the same mRNA and some may have opposing roles in RNA metabolism. Additionally, most RBPs have multiple mRNA targets. The wide-spread, overlapping and often different effects of RBPs on the transcriptome present a great challenge in delineating the function of trypanosome RBPs in regulating gene expression and development. Future priorities should include precise cataloging of RNA targets and definition of preferred in vivo binding sites and distinguishing between direct and indirect effects on target RNA metabolism. Most importantly, we should strive to identify the molecular mechanisms driving the differential expression or differential action of RBPs in particular life-cycle stages of parasite development. There is a distinct possibility that expression of RBPs themselves is controlled by RBPs, and illuminating the cellular processes and signals that mediate sensing changes in the parasite environment (host identity and available nutrients) and signal transduction to the level of RBP expression holds the key to understanding the network of regulatory events that determine the gene expression program in different developmental stages. Interestingly, the 3′UTRs of several of the RBPs discussed here (Fig. 2) are well above the median length of 388-400 nucleotides determined in transcriptome-wide studies (Kolev et al., 2010, Siegel et al., 2010). If we consider 3′UTRs as the most commonly utilized platform for RNP regulatory complexes assembly, then the size of the 3′UTR could be an indication for the level of intricacy in the post-transcriptional control of expression for a particular gene. In longer 3′UTRs many more sequence and/or structure elements are available for binding to different RBPs and the number of possible RBP combinations associated with an mRNA increases. Thus, there is the potential for a larger number of signaling pathways involving RNA-binding components (proteins, regulatory RNAs, and even metabolites) to exert effect(s) on the expression of an RBP. Many other predicted RBPs in the trypanosome genome contain much longer than average untranslated regions in their mRNAs, suggesting that we have only scratched the surface of the RBP toolbox that T. brucei uses to modulate and fine-tune gene expression post-transcriptionally.
Fig. 2.
Schematic representation of the mRNAs coding for RNA-binding proteins discussed in this review. The drawings are according to the predicted sizes of the 3′UTRs based on high throughput sequencing data (Kolev et al., 2010, Siegel et al., 2010).The numbers listed are rounded to the closest 0.5 kb.
Acknowledgements
Work in the authors laboratory is supported by Public Health Service grants AI28798 and AI56333 to E.U. and AI043594 to C.T. from the National Institute of Allergy and Infectious Diseases. The funding agency had no role in the writing and the decision to publish the article. We apologize to all those colleagues whose relevant work could not be cited due to space limitations.
References
- Amann BT, Worthington MT, Berg JM. A Cys3His zinc-binding domain from Nup475/tristetraprolin: a novel fold with a disklike structure. Biochemistry. 2003;42:217–221. doi: 10.1021/bi026988m. [DOI] [PubMed] [Google Scholar]
- Aphasizhev R, Aphasizheva I. Mitochondrial RNA processing in trypanosomes. Res Microbiol. 2011;162:655–663. doi: 10.1016/j.resmic.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer SK, Luu VD, de Queiroz RA, Brems S, Clayton C. Trypanosoma brucei PUF9 regulates mRNAs for proteins involved in replicative processes over the cell cycle. PLoS Pathog. 2009;5:e1000565. doi: 10.1371/journal.ppat.1000565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benz C, Mulindwa J, Ouna B, Clayton C. The Trypanosoma brucei zinc finger protein ZC3H18 is involved in differentiation. Mol Biochem Parasitol. 2011;177:148–151. doi: 10.1016/j.molbiopara.2011.02.007. [DOI] [PubMed] [Google Scholar]
- Bordoli L, Kiefer F, Arnold K, Benkert P, Battey J, Schwede T. Protein structure homology modeling using SWISS-MODEL workspace. Nat Protoc. 2009;4:1–13. doi: 10.1038/nprot.2008.197. [DOI] [PubMed] [Google Scholar]
- Clayton C. The Regulation of Trypanosome Gene Expression by RNA-Binding Proteins. PLoS Pathog. 2013;9:e1003680. doi: 10.1371/journal.ppat.1003680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton C, Shapira M. Post-transcriptional regulation of gene expression in trypanosomes and leishmanias. Mol Biochem Parasitol. 2007;156:93–101. doi: 10.1016/j.molbiopara.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Das A, Morales R, Banday M, Garcia S, Hao L, Cross GA, et al. The essential polysome-associated RNA-binding protein RBP42 targets mRNAs involved in Trypanosoma brucei energy metabolism. Rna. 2012;18:1968–1983. doi: 10.1261/rna.033829.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gaudenzi J, Frasch AC, Clayton C. RNA-binding domain proteins in Kinetoplastids: a comparative analysis. Eukaryot Cell. 2005;4:2106–2114. doi: 10.1128/EC.4.12.2106-2114.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droll D, Archer S, Fenn K, Delhi P, Matthews K, Clayton C. The trypanosome Pumilio-domain protein PUF7 associates with a nuclear cyclophilin and is involved in ribosomal RNA maturation. FEBS Lett. 2010;584:1156–1162. doi: 10.1016/j.febslet.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droll D, Minia I, Fadda A, Singh A, Stewart M, Queiroz R, Clayton C. Post-transcriptional regulation of the trypanosome heat shock response by a zinc finger protein. PLoS Pathog. 2013;9:e1003286. doi: 10.1371/journal.ppat.1003286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer NA, Rose C, Ejeh NO, Acosta-Serrano A. Flying tryps: survival and maturation of trypanosomes in tsetse flies. Trends Parasitol. 2013;29:188–196. doi: 10.1016/j.pt.2013.02.003. [DOI] [PubMed] [Google Scholar]
- Edwards TA, Pyle SE, Wharton RP, Aggarwal AK. Structure of Pumilio reveals similarity between RNA and peptide binding motifs. Cell. 2001;105:281–289. doi: 10.1016/s0092-8674(01)00318-x. [DOI] [PubMed] [Google Scholar]
- Gunzl A. The pre-mRNA splicing machinery of trypanosomes: complex or simplified? Eukaryot Cell. 2010;9:1159–1170. doi: 10.1128/EC.00113-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta SK, Kosti I, Plaut G, Pivko A, Tkacz ID, Cohen-Chalamish S, et al. The hnRNP F/H homologue of Trypanosoma brucei is differentially expressed in the two life cycle stages of the parasite and regulates splicing and mRNA stability. Nucleic Acids Res. 2013;41:6577–6594. doi: 10.1093/nar/gkt369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann C, Benz C, Brems S, Ellis L, Luu VD, Stewart M, et al. Small trypanosome RNA-binding proteins TbUBP1 and TbUBP2 influence expression of F-box protein mRNAs in bloodstream trypanosomes. Eukaryot Cell. 2007;6:1964–1978. doi: 10.1128/EC.00279-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks EF, Matthews KR. Disruption of the developmental programme of Trypanosoma brucei by genetic ablation of TbZFP1, a differentiation-enriched CCCH protein. Mol Microbiol. 2005;57:706–716. doi: 10.1111/j.1365-2958.2005.04679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks EF, Robinson DR, Hinkins M, Matthews KR. A novel CCCH protein which modulates differentiation of Trypanosoma brucei to its procyclic form. Embo J. 2001;20:6700–6711. doi: 10.1093/emboj/20.23.6700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoek M, Zanders T, Cross GA. Trypanosoma brucei expression-site-associated-gene-8 protein interacts with a Pumilio family protein. Mol Biochem Parasitol. 2002;120:269–283. doi: 10.1016/s0166-6851(02)00009-9. [DOI] [PubMed] [Google Scholar]
- Horn D, McCulloch R. Molecular mechanisms underlying the control of antigenic variation in African trypanosomes. Curr Opin Microbiol. 2010;13:700–705. doi: 10.1016/j.mib.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha BA, Archer SK, Clayton CE. The Trypanosome Pumilio Domain Protein PUF5. PLoS One. 2013;8:e77371. doi: 10.1371/journal.pone.0077371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolev NG, Franklin JB, Carmi S, Shi H, Michaeli S, Tschudi C. The transcriptome of the human pathogen Trypanosoma brucei at single-nucleotide resolution. PLoS Pathog. 2010;6(5):e1001090. doi: 10.1371/journal.ppat.1001090. doi:1001010.1001371/journal.ppat.1001090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolev NG, Ramey-Butler K, Cross GA, Ullu E, Tschudi C. Developmental progression to infectivity in Trypanosoma brucei triggered by an RNA-binding protein. Science. 2012;338:1352–1353. doi: 10.1126/science.1229641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer S, Carrington M. Trans-acting proteins regulating mRNA maturation, stability and translation in trypanosomatids. Trends Parasitol. 2011;27:23–30. doi: 10.1016/j.pt.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer S, Kimblin NC, Carrington M. Genome-wide in silico screen for CCCH-type zinc finger proteins of Trypanosoma brucei, Trypanosoma cruzi and Leishmania major. BMC Genomics. 2010;11:283. doi: 10.1186/1471-2164-11-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling AS, Trotter JR, Hendriks EF. A zinc finger protein, TbZC3H20, stabilizes two developmentally regulated mRNAs in trypanosomes. J Biol Chem. 2011;286:20152–20162. doi: 10.1074/jbc.M110.139261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luu VD, Brems S, Hoheisel JD, Burchmore R, Guilbride DL, Clayton C. Functional analysis of Trypanosoma brucei PUF1. Mol Biochem Parasitol. 2006;150:340–349. doi: 10.1016/j.molbiopara.2006.09.007. [DOI] [PubMed] [Google Scholar]
- Mani J, Guttinger A, Schimanski B, Heller M, Acosta-Serrano A, Pescher P, et al. Alba-domain proteins of Trypanosoma brucei are cytoplasmic RNA-binding proteins that interact with the translation machinery. PLoS One. 2011;6:e22463. doi: 10.1371/journal.pone.0022463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews KR. Controlling and coordinating development in vector-transmitted parasites. Science. 2011;331:1149–1153. doi: 10.1126/science.1198077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaeli S. Trans-splicing in trypanosomes: machinery and its impact on the parasite transcriptome. Future Microbiol. 2011;6:459–474. doi: 10.2217/fmb.11.20. [DOI] [PubMed] [Google Scholar]
- Nagai K, Oubridge C, Jessen TH, Li J, Evans PR. Crystal structure of the RNA-binding domain of the U1 small nuclear ribonucleoprotein A. Nature. 1990;348:515–520. doi: 10.1038/348515a0. [DOI] [PubMed] [Google Scholar]
- Najafabadi HS, Lu Z, MacPherson C, Mehta V, Adoue V, Pastinen T, Salavati R. Global identification of conserved post-transcriptional regulatory programs in trypanosomatids. Nucleic Acids Res. 2013;41:8591–8600. doi: 10.1093/nar/gkt647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouna BA, Stewart M, Helbig C, Clayton C. The Trypanosoma brucei CCCH zinc finger proteins ZC3H12 and ZC3H13. Mol Biochem Parasitol. 2012;183:184–188. doi: 10.1016/j.molbiopara.2012.02.006. [DOI] [PubMed] [Google Scholar]
- Paterou A, Walrad P, Craddy P, Fenn K, Matthews K. Identification and stage-specific association with the translational apparatus of TbZFP3, a CCCH protein that promotes trypanosome life-cycle development. J Biol Chem. 2006;281:39002–39013. doi: 10.1074/jbc.M604280200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preusser C, Jae N, Bindereif A. mRNA splicing in trypanosomes. Int J Med Microbiol. 2012;302:221–224. doi: 10.1016/j.ijmm.2012.07.004. [DOI] [PubMed] [Google Scholar]
- Roditi I, Carrington M, Turner M. Expression of a polypeptide containing a dipeptide repeat is confined to the insect stage of Trypanosoma brucei. Nature. 1987;325:272–274. doi: 10.1038/325272a0. [DOI] [PubMed] [Google Scholar]
- Schumann Burkard G, Kaser S, de Araujo PR, Schimanski B, Naguleswaran A, Knusel S, et al. Nucleolar proteins regulate stage-specific gene expression and ribosomal RNA maturation in Trypanosoma brucei. Mol Microbiol. 2013;88:827–840. doi: 10.1111/mmi.12227. [DOI] [PubMed] [Google Scholar]
- Siegel TN, Hekstra DR, Wang X, Dewell S, Cross GA. Genome-wide analysis of mRNA abundance in two life-cycle stages of Trypanosoma brucei and identification of splicing and polyadenylation sites. Nucleic Acids Res. 2010;38:4946–4957. doi: 10.1093/nar/gkq237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subota I, Rotureau B, Blisnick T, Ngwabyt S, Durand-Dubief M, Engstler M, Bastin P. ALBA proteins are stage regulated during trypanosome development in the tsetse fly and participate in differentiation. Mol Biol Cell. 2011;22:4205–4219. doi: 10.1091/mbc.E11-06-0511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson E, Rappsilber J, Tollervey D. Nop9 is an RNA binding protein present in pre-40S ribosomes and required for 18S rRNA synthesis in yeast. Rna. 2007;13:2165–2174. doi: 10.1261/rna.747607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbaniak MD, Guther ML, Ferguson MA. Comparative SILAC proteomic analysis of Trypanosoma brucei bloodstream and procyclic lifecycle stages. PLoS One. 2012;7:e36619. doi: 10.1371/journal.pone.0036619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urwyler S, Studer E, Renggli CK, Roditi I. A family of stage-specific alanine-rich proteins on the surface of epimastigote forms of Trypanosoma brucei. Mol Microbiol. 2007;63:218–228. doi: 10.1111/j.1365-2958.2006.05492.x. [DOI] [PubMed] [Google Scholar]
- Vassella E, Reuner B, Yutzy B, Boshart M. Differentiation of African trypanosomes is controlled by a density sensing mechanism which signals cell cycle arrest via the cAMP pathway. J Cell Sci. 1997;110(Pt 21):2661–2671. doi: 10.1242/jcs.110.21.2661. [DOI] [PubMed] [Google Scholar]
- Vickerman K, Tetley L, Hendry KA, Turner CM. Biology of African trypanosomes in the tsetse fly. Biol Cell. 1988;64:109–119. doi: 10.1016/0248-4900(88)90070-6. [DOI] [PubMed] [Google Scholar]
- Walrad PB, Capewell P, Fenn K, Matthews KR. The post-transcriptional trans-acting regulator, TbZFP3, co-ordinates transmission-stage enriched mRNAs in Trypanosoma brucei. Nucleic Acids Res. 2012;40:2869–2883. doi: 10.1093/nar/gkr1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, McLachlan J, Zamore PD, Hall TM. Modular recognition of RNA by a human pumilio-homology domain. Cell. 2002;110:501–512. doi: 10.1016/s0092-8674(02)00873-5. [DOI] [PubMed] [Google Scholar]
- Wang X, Zamore PD, Hall TM. Crystal structure of a Pumilio homology domain. Mol Cell. 2001;7:855–865. doi: 10.1016/s1097-2765(01)00229-5. [DOI] [PubMed] [Google Scholar]
- Wardleworth BN, Russell RJ, Bell SD, Taylor GL, White MF. Structure of Alba: an archaeal chromatin protein modulated by acetylation. Embo J. 2002;21:4654–4662. doi: 10.1093/emboj/cdf465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurst M, Robles A, Po J, Luu VD, Brems S, Marentije M, et al. An RNAi screen of the RRM-domain proteins of Trypanosoma brucei. Mol Biochem Parasitol. 2009;163:61–65. doi: 10.1016/j.molbiopara.2008.09.001. [DOI] [PubMed] [Google Scholar]
- Wurst M, Seliger B, Jha BA, Klein C, Queiroz R, Clayton C. Expression of the RNA recognition motif protein RBP10 promotes a bloodstream-form transcript pattern in Trypanosoma brucei. Mol Microbiol. 2012;83:1048–1063. doi: 10.1111/j.1365-2958.2012.07988.x. [DOI] [PubMed] [Google Scholar]