SUMMARY
The regulation of protein-coding and noncoding RNAs is linked to nuclear processes including chromatin modifications and gene silencing. However, the mechanisms that distinguish RNAs and mediate their functions are poorly understood. We describe a nuclear RNA processing network in fission yeast with a core module comprising the Mtr4-like protein, Mtl1, and the zinc finger protein, Red1. The Mtl1-Red1 core promotes degradation of mRNAs and noncoding RNAs, and associates with different proteins to assemble heterochromatin via distinct mechanisms. Mtl1 also forms Red1-independent interactions with evolutionarily conserved proteins named Nrl1 and Ctr1, which associate with splicing factors. Whereas Nrl1 targets transcripts with cryptic introns to form heterochromatin at developmental genes and retrotransposons, Ctr1 functions in processing intron-containing telomerase RNA. Together with our discovery of widespread cryptic introns, including in noncoding RNAs, these findings reveal unique cellular strategies for recognizing regulatory RNAs and coordinating their functions in response to developmental and environmental cues.
INTRODUCTION
Considerable portions of eukaryotic genomes are transcribed. In addition to mRNAs, genomes encode long noncoding RNAs (lncRNAs) that overlap with protein-coding genes in either sense or antisense orientation, or are derived from intergenic regions (Lee, 2012; Orom and Shiekhattar, 2011). In several cases, expression of lncRNAs is regulated and occurs under specific growth or developmental conditions. lncRNAs are emerging as critical components of epigenetic regulatory mechanisms that direct chromatin modifications (Batista and Chang, 2013; Feng and Jacobsen, 2011; Lee, 2012). However, the mechanisms by which cells recognize these RNAs and mediate their effects are poorly understood.
Cellular RNA levels are tightly controlled at both transcriptional and post-transcriptional levels. RNA processing activities, such as the exosome and RNAi machinery, control the steady-state levels of diverse RNA species (Doma and Parker, 2007; Houseley et al., 2006; Reyes-Turcu and Grewal, 2012; Schmid and Jensen, 2008) in conjunction with 3′ end formation mechanisms that determine the fate of various RNAs (Tuck and Tollervey, 2013). The exosome processes and degrades RNA substrates in the cytoplasm and the nucleus (Houseley et al., 2006). The nuclear exosome contains the 3′ → 5′ exonuclease Rrp6, which processes various RNAs to their mature forms and degrades ncRNA, antisense RNA, and transcripts produced from repeat elements (Houseley et al., 2006; Reyes-Turcu and Grewal, 2012). Cofactors such as TRAMP (Trf4-Air2-Mtr4 polyadenylation), which polyadenylates RNA substrates through its non-canonical poly(A) polymerase Trf4/5 (Cid14 in fission yeast Schizosaccharomyces pombe) and which contains the RNA helicase Mtr4, stimulate exosome activity (Houseley et al., 2006). In S. pombe, TRAMP also activates RNAi to degrade transcripts from repeat elements and antisense RNA (Zhang et al., 2011).
The regulated degradation of RNAs also impacts gene control during differentiation. Meiosis is the most dramatic differentiation program. In S. pombe, meiotic induction is accompanied by upregulation of meiosis-specific genes, which are silenced during vegetative growth (Mata et al., 2002) by an RNA elimination system involving the exosome (Yamamoto, 2010). Polyadenylation of meiotic RNAs facilitates their elimination, and requires the canonical poly(A) polymerase Pla1, which acts together with Mmi1, a protein that binds RNA containing determinant of selective removal (DSR) elements (Harigaya et al., 2006; Sugiyama and Sugioka-Sugiyama, 2011; Yamanaka et al., 2010). TRAMP is dispensable for this process (McPheeters et al., 2009; St-Andre et al., 2010; Yamanaka et al., 2010). Mmi1 recruits the Zn-finger protein Red1, which associates with Pla1 and the exosome to degrade RNAs. The nuclear poly(A) binding protein Pab2, implicated in processing snoRNAs and in turnover of pre-mRNAs by the exosome (Lemay et al., 2010; Lemieux et al., 2011), is also required (St-Andre et al., 2010; Sugiyama and Sugioka-Sugiyama, 2011; Yamanaka et al., 2010).
Transcription and RNA processing promote assembly of heterochromatin, characterized by methylation of histone H3 at lysine 9 and the presence of HP1 proteins (Reyes-Turcu and Grewal, 2012). S. pombe contains three distinct types of heterochromatin, which differ in their dependency on trans-acting factors. The first type corresponds to major H3K9me peaks at centromeres, telomeres and the mating-type locus. RNAi proteins Argonaute (Ago1), Dicer (Dcr1) and RNA-dependent RNA polymerase (Rdp1) target transcripts produced by dg/dh repeats in these regions to generate siRNAs that facilitate loading of the Clr4/Suv39h methyltransferase (Reyes-Turcu and Grewal, 2012). The second type includes small blocks of heterochromatin islands, which include meiotic genes and other loci (Cam et al., 2005; Zofall et al., 2012). Islands are akin to facultative heterochromatin and are dynamically regulated in response to environmental signals (Zofall et al., 2012). The assembly of heterochromatin islands at meiotic genes requires Red1 and Rrp6, but not RNAi (Hiriart et al., 2012; Tashiro et al., 2013; Zofall et al., 2012). The third type of heterochromatin consists of domains called HOODs (heterochromatin domains), which are detected under specific growth conditions and are dynamically regulated (Yamanaka et al., 2013). HOODs preferentially assemble at sexual differentiation genes and retrotransposons, and require RNAi as well as elimination factors (Yamanaka et al., 2013). Red1 and its partners Pla1 and Pab2 are cofactors for both RNAi and the exosome, which act in parallel to degrade transcripts from loci within these domains (Yamanaka et al., 2013).
The mechanisms by which Red1 promotes assembly of RNAi-independent and -dependent heterochromatin, and for the specification of HOOD formation, are not understood. Here we report that Red1 interacts with Mtl1 (Mtr4-like protein 1) to form a core module, MTREC (Mtl1-Red1 Core), which associates with other factors to assemble islands or HOODs. We find that MTREC coordinates degradation of meiotic mRNAs, and localizes to ncRNA loci that regulate gene expression in response to environmental changes. Mtl1 also associates with the putative C. elegans NRDE-2 (Guang et al., 2010) homolog Nrl1, an evolutionarily conserved novel protein named Ctr1 (coiled-coil domain telomerase regulatory protein 1), and splicing factors. We also discover numerous previously unannotated and cryptic introns in the S. pombe genome. Splicing machinery and Nrl1 act on cryptic introns that specify assembly of HOODs, whereas Ctr1 facilitates processing of intron-containing telomerase RNA to maintain telomeres. These results uncover an RNA processing network that targets various RNA substrates and promotes heterochromatin assembly.
RESULTS
Purification of Red1 and Identification of Its Associated Proteins
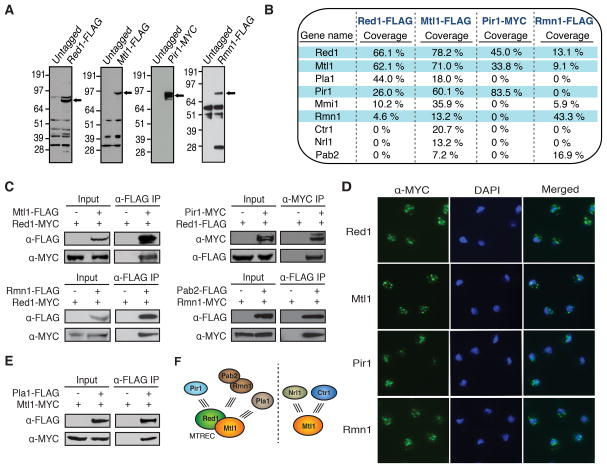
To investigate the role of Red1 in meiotic RNA processing and in RNAi-dependent and –independent heterochromatin formation, we expressed Red1 with 3×FLAG-tag at its C-terminus under its native promoter (Figure 1A). Purification of Red1-FLAG followed by mass spectrometry analysis identified a number of proteins specific to the Red1-FLAG purified fraction, including Red1 and its known interaction partners Mmi1 and Pla1 (Figure 1B) (Sugiyama and Sugioka-Sugiyama, 2011). The purified fraction also contained components of the exosome (Figure S1A), consistent with the role of Red1 in facilitating RNA degradation by the exosome (Sugiyama and Sugioka-Sugiyama, 2011).
Figure 1. Red1 and Mtl1 Form a Common Core that Associates with Other Proteins.
(A) Expression of tagged proteins. Extracts from tagged and untagged strains were analyzed by Western blot. (B) Proteins co-purified with Red1, Mtl1, Pir1, and Rmn1. Proteins highly associated with each other are shaded in blue. (C) Co-IP of associated proteins from strains expressing tagged proteins. (D) Immunofluorescence analysis of MYC-tagged proteins. (E) Co-IP analysis of Pla1 and Mtl1 interaction. (F) Protein interaction network of MTREC and Mtl1. MTREC, comprising Mtl1 and Red1, is the core module that interacts with Pir1, Rmn1-Pab2, or Pla1 to form different functional modules (left). Mtl1 also forms Red1-independent interactions with Nrl1 and Ctr1 (right). See also Figure S1.
We also identified new Red1 interacting partners (Figure 1B and S1A), including the uncharacterized proteins SPAC17H9.02 and SPAC7D4.14c as well as Rmn1, which contains an RNA recognition motif (Cho et al., 2012). SPAC17H9.02 shows significant homology to human hMtr4/SKIV2L2, which belongs to an RNA helicase family related to S. cerevisiae Ski2 (Lubas et al., 2011). The hMTR4 domains, including DEXDc, HELICc, KOW Mtr4, and DSHCT, are conserved in SPAC17H9.02 (Figure S1B). We named this novel protein Mtl1. Notably, Mtl1 is distinct from the previously described Mtr4 component of TRAMP (Buhler et al., 2007; Zhang et al., 2011). We named SPAC7D4.14c, a serine/proline rich protein with no apparent domains or homologs, Pir1 (Protein interacting with Red1-1). Additional reproducibly detected proteins included the zf-C2H2 type zinc finger protein SPBC725.08 (named Pir2, Protein Interacting with Red1-2), RNA polymerase I subunits (Rpa1 and Rpa2), Spt6 involved in chromatin and RNA processing (Kiely et al., 2011), and an assortment of proteins involved in RNA metabolism (Figure S1A). Purification and co-immunoprecipitation (co-IP) experiments also revealed association of Red1 with splicing factors (Figures S1A and S1C).
Red1 and Mtl1 Form a Core Module that Interacts with Different Nuclear Proteins
To explore the interactions among Red1-associated proteins, we appended MYC or FLAG epitope tags to Mtl1, Pir1 and Rmn1 for immuno-affinity purification (Figure 1A). Mass spectrometry analyses of purified fractions revealed specific networks of interactions among Red1-associated proteins. Mtl1 purification yielded all of the major proteins identified in Red1 purifications, as well as additional proteins (Figures 1B and S1A). In particular, Rmn1 and proteins that specifically associated with Mtl1 did not purify with Pir1 (Figures 1B and S1A). Conversely, purification of Rmn1 yielded Red1, Mtl1 and Pab2, but not Pir1 (Figures 1B and S1A). We conclude that Red1 and Mtl1, which were detected in all four purifications, form the common core module of distinct protein assemblies.
To confirm interactions among Red1-associated factors, we performed co-IPs (Figure 1C) and determined their subcellular localization by immunofluorescence (Figure 1D). We found that Pir1 and Rmn1 co-IP with Red1 (Figure 1C). However, Pir1 and Rmn1 did not co-purify (Figures 1B and S1A), consistent with their affiliation with distinct Red1-containing modules. Our analyses also confirmed the Rmn1 and Pab2 interaction (Figure 1C). In addition to the previously established interaction between Pla1 and Red1 (Sugiyama and Sugioka-Sugiyama, 2011), we found that Pla1 and Mtl1 also co-IP (Figure 1E). Mtl1, Rmn1 and Pir1 localized to discrete foci in the nucleus (Figure 1D), similar to Red1 (Sugiyama and Sugioka-Sugiyama, 2011). Furthermore, Mtl1 co-localized with Red1 (Figure S1D), and ChIP-chip analyses revealed shared binding sites across the genome (Figure S1E).
Based on these results, we propose that a core comprising Red1 and Mtl1 (named MTREC) exists in multiple protein assemblies (Figure 1F). One assembly contains Pir1, and another is formed with Rmn1 and Pab2. A third includes Pla1, which is absent from Pir1 and Rmn1 purifications (Figures 1B and S1A). Mtl1 also engages in additional Red1-independent interactions (Fig. 1F; see below).
Red1- and Mtl1-Associated Factors Differentially Affect Heterochromatin Domains
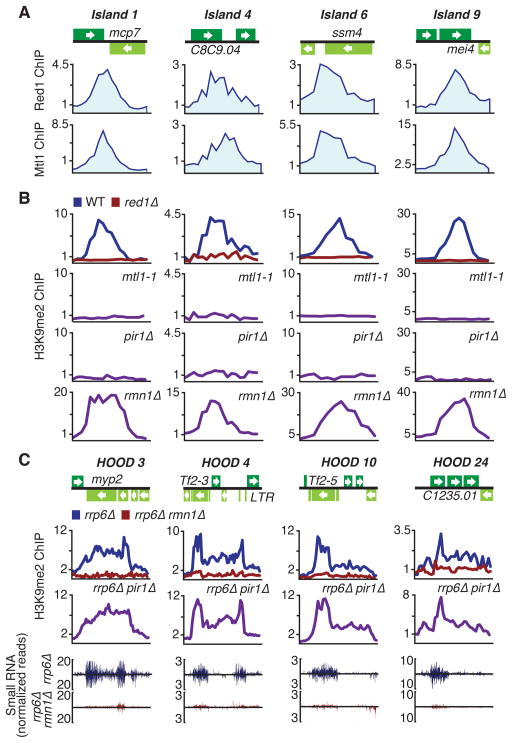
Red1 localizes to meiotic loci and facilitates formation of heterochromatin islands (Tashiro et al., 2013; Zofall et al., 2012). Since Mtl1 is similarly enriched at meiotic islands (Figures 2A and S2A), we examined whether loss of Mtl1 affects heterochromatin islands. Because Mtl1 is an essential protein (Kim et al., 2010), we generated a partial loss of function mutant allele, mtl1-1 (Figure S1B). The mtl1-1 mutant showed severe defects in H3K9me at heterochromatin islands affected in red1Δ (Figures 2B and S2B). Both Red1 and Mtl1 were specifically enriched at islands that require these factors for H3K9me (Figures S2C and S2D), and Red1-dependent islands also showed strong correlation between levels of H3K9me and Mtl1 binding (correlation coefficient 0.90, p < 0.0001). Red1-independent heterochromatin islands that do not show Red1 and Mtl1 enrichment were not affected in mtl1-1 (Figures S2C–E). These results suggest that Mtl1 and Red1 act together as components of MTREC to assemble heterochromatin islands.
Figure 2. Red1- and Mtl1-Associated Factors Differentially Affect Heterochromatin Domains.
(A) ChIP-chip analysis of Red1-MYC and Mtl1-MYC distribution at heterochromatin island loci. (B) ChIP-chip analysis of H3K9me2 distribution at heterochromatin island loci in indicated strains. (C) ChIP-chip analysis of the effect of pir1Δ and rmn1Δ on H3K9me2 distribution at HOODs in rrp6Δ (top). Normalized number of small RNA reads plotted in alignment with HOOD loci (bottom). The signals above and below the line represent small RNAs that map to the top and bottom DNA strands, respectively. See also Figure S2 and Table S1.
We expanded our analyses to include factors associated with MTREC. Interestingly, loss of Pir1, but not Rmn1, led to defects in H3K9me at Red1-dependent heterochromatin islands (Figures 2B, S2B and S2D). A low level of Pir1 enrichment was observed at heterochromatin islands, consistent with its direct involvement, and its loss affected silencing of target loci (Figure S2F). No major changes in H3K9me could be detected in cells carrying a mutation in Pla1, another MTREC-associated factor.
Red1 also affects RNAi-dependent assembly of HOODs (Yamanaka et al., 2013). Pab2 interacts with Rmn1 (Figure 1C), and is required along with Pla1 for H3K9me at all known HOODs (Yamanaka et al., 2013). We investigated the role of MTREC factors in HOOD assembly by creating deletions in cells that lack Rrp6. Rrp6 acts parallel to RNAi to silence loci within HOODs and consequently, rrp6Δ cells show a significant increase in siRNA-dependent heterochromatin modifications under lab growth conditions (Yamanaka et al., 2013). Remarkably, loss of Rmn1, but not Pir1, abolished RNAi-dependent H3K9me at HOODs (Figure 2C and Table S1). Defects in H3K9me correlated with defects in siRNA production at affected loci (Figures 2C and S2G and Table S1). Loss of Red1, Mtl1, Rmn1 or Pir1 did not affect RNAi-mediated heterochromatin assembly at centromeres (Figure S2E). We conclude that MTREC protein assemblies containing Pir1 or Rmn1 differentially affect formation of heterochromatin islands and HOODs.
Mtl1 Regulates Expression of Genes Involved in Sexual Differentiation, Stress Response and Membrane Transport
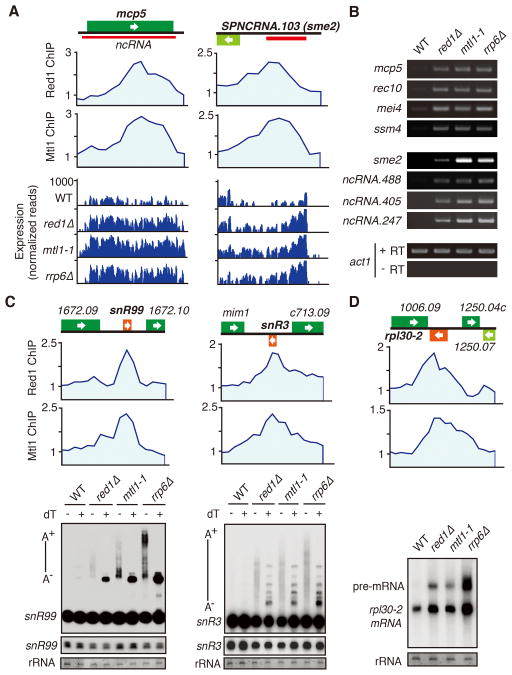
Red1 coordinates exosome-dependent repression of meiotic genes in vegetative cells (Sugiyama and Sugioka-Sugiyama, 2011; Zofall et al., 2012). Mtl1 and Red1 interaction and co-localization at several genomic sites (Figures 1C, S1E and 2A) prompted us to investigate whether they collaborate to regulate gene expression. We detected increased expression of various mRNAs in mtl1-1, as compared to wild type (Table S2). A majority of genes affected in mtl1-1 were also upregulated in red1Δ and rrp6Δ (correlation coefficient 0.63 and 0.65, respectively, p < 0.0001) (Table S2). For example, the mcp5 gene, which is bound by Red1 and Mtl1, was upregulated in mtl1-1 as well as red1Δ and rrp6Δ (Figure 3A). We confirmed that loci affected in red1Δ and rrp6Δ were also upregulated in mtl1-1 by RT-PCR (Figure 3B).
Figure 3. MTREC Regulates Gene Expression and Targets Noncoding RNAs and premRNAs Degraded by the Exosome.
(A) ChIP-chip analysis of Red1-MYC and Mtl1-MYC distribution at indicated loci (top). RNA-Seq analysis of Red1 and Mtl1 bound genes in red1Δ, mtl1-1, and rrp6Δ strains (bottom). (B) RT-PCR analysis of Red1 and Mtl1 bound loci in indicated strains. The transcript level of act1 was used as a control. +RT and −RT indicate presence or absence of reverse transcriptase. (C) ChIP-chip analysis of Red1-MYC and Mtl1-MYC distribution at snoRNAs (top). Northern analysis of snR99 and snR3 processing in mutant strains (bottom). −/+ dT denotes RNase H treatment in the absence or presence of oligo dT. A− /A+ indicates non-polyadenylated and polyadenylated species. Lighter exposures of mature snR99 and snR3 are shown below. rRNA was used as a loading control. (D) Red1-MYC and Mtl1-MYC distribution at rpl30-2 (top). Northern blot analysis of rpl30-2 pre-mRNA processing in the indicated strains (bottom). See also Table S2.
The upregulated loci belong to three distinct groups (Table S2). The first group contains genes involved in meiosis (e.g. crs1, mug1, ssm4 and mei4) or that show an increase in expression during meiotic induction (e.g. mug8, mug9 and meu10). The second group includes genes implicated in cellular stress response (Table S2). The third group consists of genes that encode transmembrane proteins involved in transport of amino acids, ions and nutrients (Table S2). Other loci affected in all three mutants are metabolic genes and pseudogenes. These results indicate that the interaction between Red1 and Mtl1 is important to suppress mRNAs targeted by the exosome.
MTREC Targets Regulatory Noncoding RNA and Pre-mRNA Degraded by the Exosome
The S. pombe genome encodes a large number of ncRNAs (Bitton et al., 2011; Dutrow et al., 2008; Rhind et al., 2011; Wilhelm et al., 2008). Mtl1 and Red1 localized to ribosomal genes and loci encoding noncoding RNAs such as lncRNAs, tRNAs and snoRNAs (Figure 3C). Significantly, mtl1-1, red1Δ and rrp6Δ all showed extensive changes in ncRNA levels (Table S2). A ncRNA produced from the sme2 locus was among a cohort of upregulated transcripts including ~300 previously identified ncRNAs and several unannotated transcripts (Table S2). The sme2 ncRNA accumulates during meiotic induction and promotes pairing of homologous chromosomes (Ding et al., 2012), but is not detected in vegetative cells. Red1 and Mtl1 enrichment at sme2 prevented the accumulation of ncRNA in vegetative cells, similar to Rrp6 (Figure 3A) (Sugiyama and Sugioka-Sugiyama, 2011). We confirmed the increase in sme2 and other ncRNAs in mtl1-1, red1Δ and rrp6Δ mutants by RT-PCR (Figure 3B). Thus, MTREC regulates the abundance of ncRNAs that are substrates of the nuclear exosome.
Red1 and Mtl1 are enriched at majority of snoRNA loci, including snR3 and snR99 (Figure 3C) and ribosomal genes (Figure 3D), the processing of which requires Rrp6 and Pab2 (Lemay et al., 2010; Lemieux et al., 2011). Northern blot analysis using snoRNA probes showed accumulation of a large heterogeneous population of transcripts in rrp6Δ (Figure 3C). The larger species represent hyperadenylated transcripts that appeared as discrete bands upon oligo dT-directed RNase H cleavage (Figure 3C). The mtl1-1 and red1Δ mutants accumulated 3′-extended snoRNAs, but polyadenylation levels in these mutants were lower than in rrp6Δ (Figure 3C). Whereas RNase H cleavage caused 3′-extended snR99 to collapse to a single discrete product, cleavage of snR3 yielded several bands, possibly signifying transcript termination at multiple sites in mtl1-1 and red1Δ (Figure 3C). We propose that Mtl1 and Red1 cooperate with polyadenylation and exosome machinery to process and degrade pre-snoRNAs, and may also play a role in transcription termination.
MTREC was also required for degradation of pre-mRNA from ribosomal gene rpl30-2, which contains a 246 nt intron. Similar to rrp6Δ, mtl1-1 and red1Δ accumulated unspliced pre-mRNAs with a slight increase in spliced rpl30-2 transcript (Figure 3D). Thus, MTREC functions in the turnover of specific intron-containing pre-mRNAs, in addition to meiotic mRNA and ncRNA decay.
Noncoding RNA Regulates Gene Expression in Response to Environmental Changes
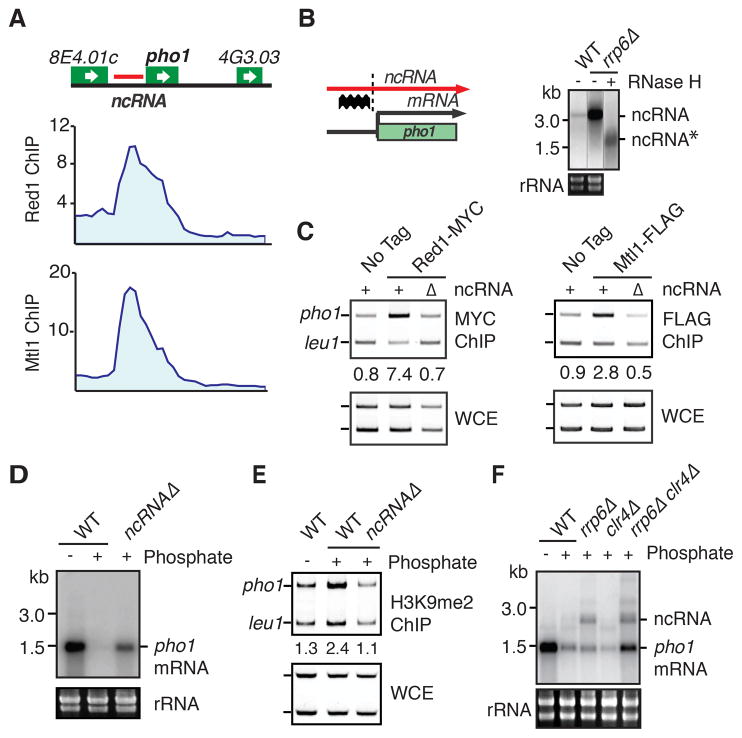
The biological function of widespread ncRNA production is unknown. We noted prominent Red1, Mtl1 and Rrp6 peaks upstream of several protein-coding genes, such as pho1, SPBC1271.09 and SPCC11E10.01 (Figures 4A and S3A–D). These peaks correlate with ncRNAs that accumulate in red1Δ, mtl1-1 and rrp6Δ cells (Figures S3A–C). We hypothesized that upstream ncRNAs recruit RNA processing factors, and might regulate adjacent genes. To test this, we focused on the pho1 locus. We detected upstream ncRNA accumulation in rrp6Δ (Figures 4B and S3B). Results from oligo-directed RNase H cleavage were consistent with upstream initiation and continuation of transcription through pho1 in rrp6Δ cells (Figure 4B).
Figure 4. ncRNA Regulates Gene Expression in Response to Growth Conditions.
(A) ChIPchip analysis of Red1-MYC and Mtl1-MYC distribution at the pho1 locus. (B) Schematic depicting transcription of ncRNA and mRNA at the pho1 locus (left). The location of the Northern blot probe is indicated (thick wavy line). The dotted line indicates the site cut by RNase H. Northern blot of ncRNA in wild-type and rrp6Δ strains (right). The asterisk denotes ncRNA cleaved by oligo-directed RNase H treatment. Lanes shown are from the same Northern blot with additional control lanes removed. (C) ChIP analysis of Red1-MYC and Mtl1-FLAG enrichment at the pho1 locus. Numbers shown below ChIP lanes represent fold enrichments. (D) Northern blot analysis of pho1 mRNA expression in ncRNAΔ. (E) ChIP analysis of H3K9me2 enrichment at pho1 in wild-type and ncRNAΔ. Numbers shown below ChIP lanes represent fold enrichments. (F) Northern blot analysis of ncRNA and pho1 mRNA expression in the indicated strains. See also Figure S3.
Expression of pho1 occurs only in phosphate-limiting conditions. To explore the effect of the ncRNA on pho1, we replaced the −400 to −1200 bp region upstream of pho1 with ura4+ to generate ncRNAΔ. Deletion of ncRNA abolished Red1 and Mtl1 recruitment to pho1 (Figure 4C), and de-repressed pho1 mRNA in the presence of phosphate (Figure 4D). Moreover, in the presence of phosphate H3K9me could be detected at pho1 in wild-type, but not in ncRNAΔ cells (Figure 4E). Loss of the sole H3K9 methyltransferase, Clr4, had no major impact on pho1 expression (Figure 4F). However, when clr4Δ was combined with rrp6Δ, synergistic accumulation of pho1 mRNA was observed (Figure 4F). Together, these results implicate ncRNA in the regulation of gene expression. To perform this function, ncRNA utilizes not only heterochromatin factor Clr4, but also RNA processing factors to degrade gene transcripts.
Mtl1 Associates with Nrl1 and Ctr1 without Red1
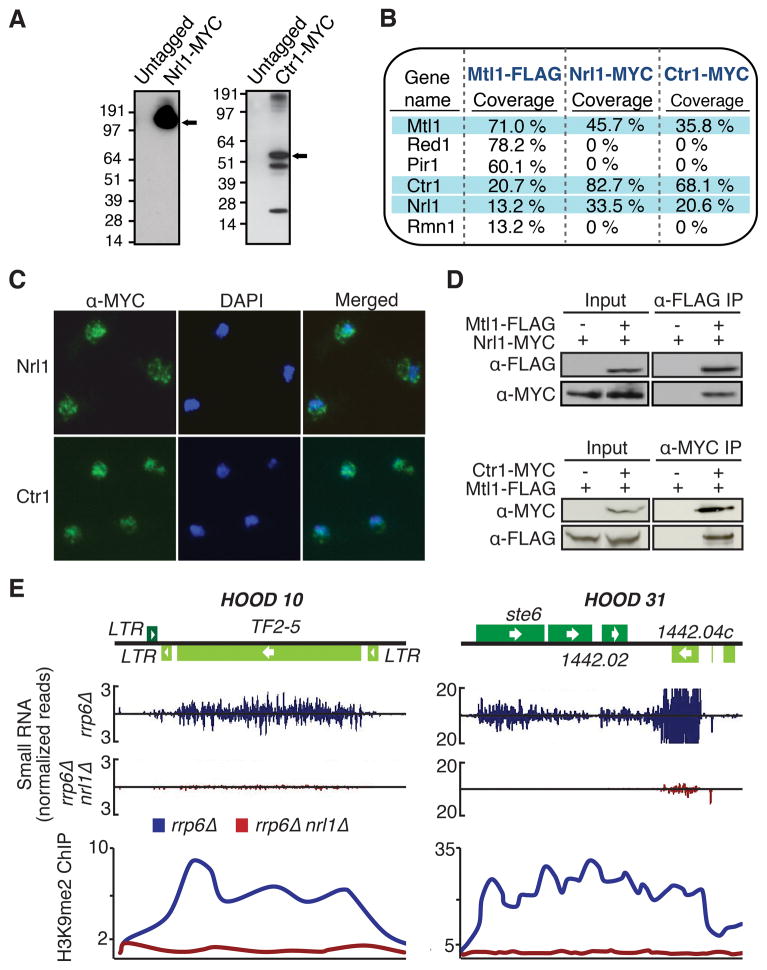
Among the proteins specific to the Mtl1 purification were the putative homolog of C. elegans NRDE-2, Nrl1 (SPBC20F10.05), and the coiled-coil and DUF4078-domain-containing protein SPAC140.04, which we named Ctr1 (Figures 1B and 1F). Ctr1 is evolutionarily conserved and shares homology with human CCDC174 protein of unknown function (Figure S1B). We tagged Nrl1 and Ctr1 with the MYC epitope (Figure 5A). Nrl1-MYC purification identified Mtl1 and Ctr1 (Figure 5B), as well as several splicing factors (Figure S4A, see below). Red1, Pir1, Rmn1 and the exosome were not present (Figures 5B and S4A). Ctr1-MYC purification also identified Nrl1, Mtl1 and splicing factors, as well as the core spliceosomal Sm proteins and the Tgs1 RNA methyltransferase (Figure S4A), suggesting a possible role in splicing related processes. Nrl1 and Ctr1 localized to the nucleus, similar to Mtl1 (Figure 5C). Interactions between these factors were confirmed by co-IP (Figure 5D).
Figure 5. Mtl1 Forms Red1-independent Interactions with Nrl1 and Ctr1.
(A) Expression of Nrl1-MYC and Ctr1-MYC. The arrow indicates MYC-tagged Nrl1 or Ctr1. (B) Proteins associated with Mtl1, Nrl1 or Ctr1. Proteins highly associated with each other are shaded in blue. (C) Immunofluorescence analysis of MYC-tagged Nrl1 and Ctr1. (D) Co-IP analysis of Mtl1 with Nrl1 and Ctr1. (E) Normalized number of small RNA reads plotted in alignment with HOOD 10 and HOOD 31 loci in rrp6Δ and rrp6Δ nrl1Δ strains (top). ChIP-chip analysis of H3K9me2 at HOODs (bottom). See also Figure S4 and Table S1.
Nrl1 Promotes Assembly of HOODs at Genes and Retrotransposons
C. elegans NRDE-2 is involved in the nuclear RNAi pathway and mediates changes in chromatin structure (Guang et al., 2010). Production of small RNAs and heterochromatin formation at HOODs are severely affected in nrl1Δ rrp6Δ cells (Figures 5E and S4B; Table S1). However, nrl1Δ had no major impact on siRNAs and H3K9me at centromeres (data not shown) or assembly of heterochromatin islands formed by RNAi-independent mechanisms (Figure S4C). Thus, Nrl1 selectively affects RNAi-dependent assembly of heterochromatin at retrotransposons and developmental genes, and is dispensable for heterochromatin formation at centromeres and meiotic islands.
Nrl1 Interacts with Splicing Factors That Assemble HOODs via Cryptic Introns
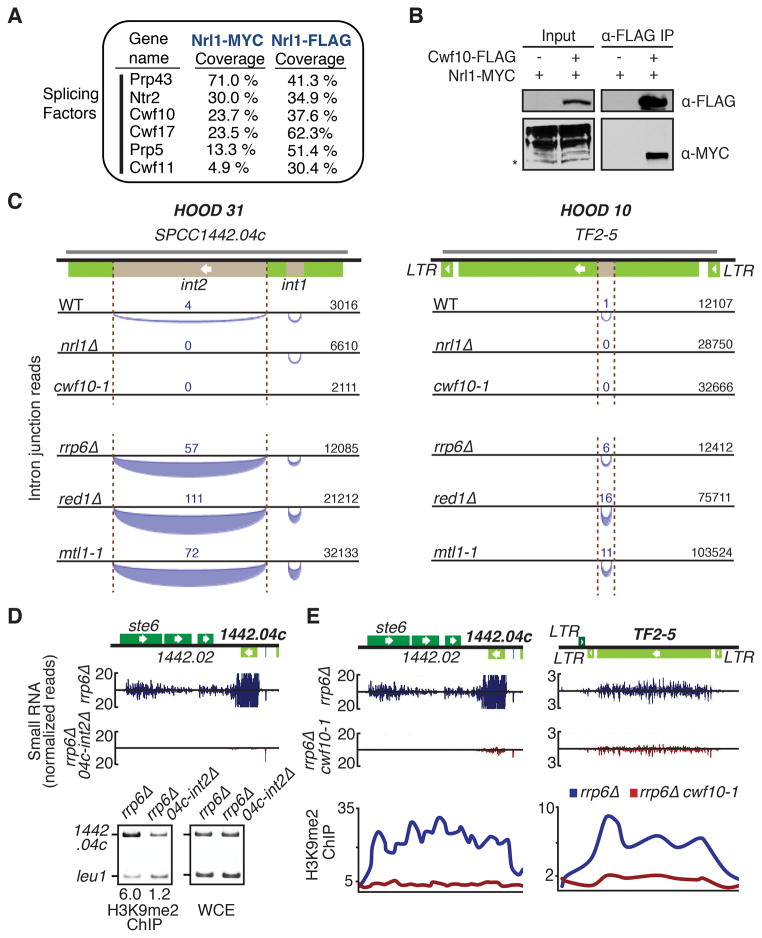
As described above, Nrl1 co-purified with factors involved in pre-mRNA splicing. Independent purifications of Nrl1-MYC and Nrl1-FLAG contained components of U2 and U5 small nuclear ribonucleic particles (snRNPs), and other splicing factors (Figure 6A). Among these proteins was Cwf10, a homolog of S. cerevisiae U5 snRNP Snu114 and of human EFTUD2 (Jurica and Moore, 2003), which has been shown to interact with Cid12, a subunit of the RNA-dependent RNA polymerase involved in RNAi (Bayne et al., 2008). Co-IP analysis confirmed Nrl1 interaction with Cwf10 (Figure 6B).
Figure 6. Nrl1 Interacts with Splicing Factors That Promote HOOD Assembly at Loci Encoding Cryptic Introns.
(A) Splicing factors identified in Nrl1 purifications. (B) Co-IP analysis of the interaction between Cwf10 and Nrl1. Asterisk indicates the correct size of Nrl1. Multiple Nrl1 bands may indicate protein modifications. (C) Schematic of cryptic introns detected at HOODs by RNA-Seq in indicated strains. Tf2 and SPCC1442.04c are shown with cryptic introns (brown). The arcs below the line represent intron junction reads that map to the bottom DNA strands. The thickness of the arc corresponds to the number of reads detected. The total unnormalized read count for each locus, which is indicative of sequencing depth, is shown. The grey line denotes antisense RNA. (D) Normalized number of small RNA reads at HOOD 31 in rrp6Δ and rrp6Δ 04c-int2Δ (top). ChIP analysis of H3K9me2 enrichment (bottom). Numbers shown below ChIP lanes represent fold enrichments. (E) Normalized number of small RNA reads at HOOD 31 and HOOD 10 in rrp6Δ and rrp6Δ cwf10-1 (top). ChIP-chip analysis of H3K9me2 (bottom). See also Figures S5–7 and Tables S1 and S3.
We hypothesized that Nrl1 might cooperate with splicing factors to promote siRNA production at retrotransposons and genes. Surprisingly, analyses of RNA-Seq data from wild-type and mutants revealed unannotated cryptic introns in Tf2 and SPCC1442.04c loci showing siRNA clusters (Figures 6C and S5A). Unlike known introns, splicing of cryptic introns in Tf2 and SPCC1442.04c is detected in a small fraction of total reads. The red1Δ, mtl1-1 and rrp6Δ mutants that were defective in silencing of SPCC1442.04c and Tf2 showed higher levels of spliced reads, as compared to wild-type. However, splicing was not detected in nrl1Δ, even amongst a greater number of Tf2 and SPCC1442.04c sequencing reads as compared to wild type (Figure 6C).
We asked whether the introns in RNAi-targeted loci are involved in generation of small RNAs and H3K9me. Deletion of an intron in SPCC1442.04c severely affected production of siRNAs and H3K9me at this locus (Figure 6D), and also caused defects in production of siRNAs at surrounding loci including ste6, which is involved in sexual differentiation (Figure 6D). siRNA production at centromeric repeats and other HOODs was not affected. These results are consistent with our previous finding that deletion of RNAi-dependent heterochromatin nucleation sites affects expression of neighboring loci within HOODs (Yamanaka et al., 2013). Our results suggest that a cryptic intron, which is differentially spliced in the absence of Nrl1, serves to recruit RNAi and target H3K9me.
We next explored the role of splicing machinery in the formation of HOODs. Analysis of rrp6Δ and rrp6Δ cwf10-1 strains grown at permissive temperature (26 °C) revealed cwf10-1 caused major reductions in siRNAs and H3K9me at a majority of HOODs (Table S1 and Figures S6A–B), including Tf2 and SPCC1442.04c loci (Figure 6E). Importantly, higher levels of siRNA and H3K9me could be detected at HOOD 12, which contains mcp3 (Figure S6C), where the RNA binding protein Mmi1 directs RNAi (Yamanaka et al., 2013). Thus, the observed changes were not due to general defects in splicing of genes involved in RNAi. Indeed, whereas splicing of cryptic introns, such as in Tf2 and SPCC1442.04c, was not detected in cwf10-1 (Figure 6C), splicing of annotated introns was not affected (Figure S6D). These results suggest that factors involved in splicing of introns embedded in HOOD loci direct RNAi to assemble heterochromatin domains.
Noncoding RNAs and Read-through Transcripts Contain Introns
We expanded our analyses of the RNA-Seq data to search for additional introns, and discovered 3,218 unannotated introns. Some introns were detected only in red1Δ, mtl1-1, rrp6Δ and/or nrl1Δ mutants, while others were also found in wild-type (Table S3). These introns showed a broader length distribution, and a small fraction of them contained variant splice junctions (Figure S7A). An intron in the centromeric dg repeat, but not the dh repeat, was previously reported (Chinen et al., 2010). We detected spliced RNA products from both dg and dh repeats in ago1Δ cells, and found that these introns mapped to regions containing siRNA clusters (Figure S5B). Interestingly, a substantial portion of the new introns was located within ncRNAs, or in regions that contain no annotated features (Table S3 and Figure S7B). In many instances, intron-containing transcripts were upregulated in red1Δ, mtl1-1 and/or rrp6Δ (Table S2). Moreover, meiotic mRNAs that are normally spliced specifically during meiosis were spliced during vegetative growth in mutants (Figure S7C).
We also found introns located within UTRs of protein-coding genes. Many transcripts containing introns in their 3′ UTR are produced from convergent genes while in other cases, introns are within unannotated transcripts near the 5′ or 3′ region of genes (Figures S7D–E). We observed elevated levels of read-through transcripts, normally suppressed by Rrp6 (Zhang et al., 2011; Zofall et al., 2009), at several loci in cells defective in Mtl1 or its associated factors (Figure S7D). Cells lacking Nrl1 or Red1 showed changes in splice site utilization at several sites across the genome (Figure S7F and Table S3), indicating that these factors affect splicing of pre-mRNAs.
Mtl1 and Ctr1 Promote Telomerase RNA Biogenesis and Telomere Maintenance
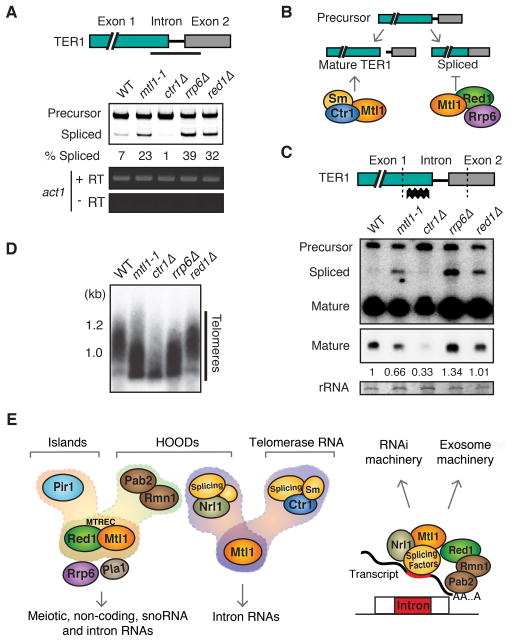
Spliceosome Sm proteins and the Tgs1 RNA methyltransferase were detected in the Ctr1 purification (Figure S4A), and are required for processing of intron-containing precursor telomerase RNA (TER1) into a mature form (Box et al., 2008; Tang et al., 2012). Core spliceosomal Sm proteins generate the mature 3′ end of TER1, releasing the active form of RNA without exon ligation (Box et al., 2008). Blocking the first step or permitting the completion of splicing generates inactive forms of TER1 (Box et al., 2008).
We tested whether Mtl1-associated factors affect TER1 processing. Spliced product accumulated in mtl1-1, red1Δ and rrp6Δ. However, ctr1Δ caused defective splicing of TER1 (Figure 7A). One explanation is that Red1- and Ctr1-containing protein assemblies carry out distinct functions. Mtl1-Red1 and Rrp6 could prevent splicing or promote degradation of spliced product, while Mtl1-Ctr1 could cooperate with the spliceosome to generate mature TER1 (Figure 7B). In this case, Mtl1 would perform dual functions by preventing accumulation of spliced product and generating mature TER1. Supporting this idea, mtl1-1 or ctr1Δ reduced the level of mature TER1, although a more severe phenotype was observed in ctr1Δ compared to the partial loss of function mtl1-1 mutant (Figure 7C). No major reduction in mature TER1 levels was observed in red1Δ and rrp6Δ, which caused accumulation of spliced product (Figure 7C). These results extend previous studies and suggest that Ctr1 and Mtl1, which associate with splicing factors, are required for production of mature TER1. Loss of Nrl1, which did not co-purify with Sm proteins, had no major effect on processing of TER1 (data not shown).
Figure 7. Ctr1 Mediates Telomere Maintenance by Regulating Telomerase RNA Processing.
(A) Schematic of TER1 RNA showing the RT-PCR amplification region (top). RT-PCR analysis of splicing in indicated strains (bottom). (B) Proposed roles for different factors in TER1 RNA processing. The Sm and Ctr1-Mtl1 proteins process TER1 to the mature form. Red1-Mtl1 targets spliced TER1 for degradation by the exosome but might also inhibit complete splicing. (C) Schematic of TER1 RNA showing the probe used for Northern analysis (top). Dotted lines indicate regions cut by oligo-directed RNase H. Northern blot analysis of TER1 (bottom). A lighter exposure of mature TER1 is shown below. Relative quantitation of mature TER1 to wild-type is indicated. (D) Southern blot analysis of telomere length. (E) Schematic illustrating the role of MTREC and Mtl1-Nrl1 protein assemblies in degradation of various RNA species and formation of heterochromatin at islands or HOODs. Mtl1 in association with Ctr1 also promotes processing of telomerase RNA. Intron-containing RNA species can be targeted by the Mtl1-Nrl1 complex in association with splicing factors, or by MTREC bound to Pab2-Rmn1 and Pla1 (not shown), to degrade RNAs and assemble HOODs through recruitment of RNAi machinery, or promote RNA decay by the exosome. See also Figure S4A.
A reduction in mature TER1 is expected to cause shortening of telomeres. As expected, both mtl1-1 and ctr1Δ caused significant reduction in telomere length, with ctr1Δ producing a stronger phenotype, whereas red1Δ did not cause reduction (Figure 7D). Interestingly, telomere shortening was evident in rrp6Δ cells, despite normal levels of mature TER1. The exact cause remains to be investigated, but it is possible that the exosome is required for maturation of TER1 or indirectly interferes with telomere maintenance.
DISCUSSION
The diverse RNA species produced by eukaryotic genomes provides the basis for biological diversity and differentiation into various cell types (Licatalosi and Darnell, 2010; Sharp, 2009). RNAs are not only messengers of genetic information for protein synthesis but also regulatory hubs that govern expression of genetic information. These RNA functions are enacted through associated proteins, but how regulatory RNAs are recognized remains largely unknown. We utilized the S. pombe system, which contains conserved RNAi and heterochromatin assembly mechanisms, and discovered a nuclear RNA processing network that silences genes and retrotransposons, and is required for adaptive regulation of the genome.
Mtl1 is a conserved protein related to human Mtr4, and is a component of multiple protein assemblies involved in regulation of various RNAs including mRNAs, snoRNA and telomerase RNA. We also detected unannotated and cryptic introns in ncRNA and gene transcripts. Mtl1 associates with conserved proteins and splicing factors that act through these introns to promote RNAi-dependent assembly of heterochromatin at developmental genes and retrotransposons. Below we discuss potential advantages of ncRNA- and intron-based heterochromatin assembly pathways for reprogramming the genome under different growth conditions and during differentiation.
Mtl1 and Red1 Exist in Multiple Protein Assemblies With Distinct Functions
Red1 was identified as a factor that promotes degradation of meiotic mRNAs via mechanisms that require Pla1, Pab2 and Rrp6 (Sugiyama and Sugioka-Sugiyama, 2011). It was later reported that Red1, but not Pab2 or Pla1, is involved in the assembly of heterochromatin islands (Hiriart et al., 2012; Tashiro et al., 2013; Zofall et al., 2012). In contrast, Red1 as well as Pla1 and Pab2 are required for RNAi-dependent assembly of HOODs (Yamanaka et al., 2013). The molecular basis of these distinct functions of Red1 was not known.
We show that Red1 functions with Mtl1 as part of the MTREC module (Figure 7E). Similar to Red1, Mtl1 is required for meiotic gene silencing, and regulates stress response genes, genes encoding membrane transporters, and ncRNAs. In addition, both Red1 and Mtl1 affect polyadenylation and degradation of pre-mRNAs and snoRNAs, the processing of which also requires Pab2 and Rrp6 (Lemay et al., 2010; Lemieux et al., 2011; St-Andre et al., 2010). MTREC associated factors coordinate the distinct functions attributed to Red1 (Figure 7E). For example, Pir1 assembles heterochromatin islands, whereas Rmn1 directs HOOD formation with Pab2 and Pla1 (Figure 2) (Yamanaka et al., 2013). Current evidence suggests two modes for MTREC involvement in RNA degradation and heterochromatin formation. First, MTREC could act as a scaffold that directly engages ribonucleolytic activities, including the exosome that interacts with Red1 (Sugiyama and Sugioka-Sugiyama, 2011) or its associated factors. Second, the core likely promotes polyadenylation of RNA substrates by Pla1, which in turn recruits Pab2 and its associated proteins to deliver RNA to the exosome and/or trigger RNAi to assemble HOODs. The latter function resembles roles performed by TRAMP (Houseley et al., 2006). The putative RNA helicase Mtl1 might unwind RNA substrates to aid their processing.
The Mtl1-Red1 interaction network is reminiscent of the human NEXT complex, which includes hMtr4 among other factors (Lubas et al., 2011). hMtr4 couples the exosome to its various cofactors including ZFC3H1, the Zn-knuckle and Ser/Pro-rich protein that appears to be related to Red1 and Pir1, which contain Zn finger and Ser/Pro-rich domains, respectively. However, whether hMtr4 also interacts with factors that trigger RNAi or guide epigenetic chromatin modifications directed by lncRNAs is unknown.
Noncoding RNAs and Environmental Gene Control
Many conditionally expressed ncRNAs, whose roles are not understood, are derived from regions containing genes and intergenic regions (Dutrow et al., 2008; Rhind et al., 2011; Wilhelm et al., 2008). We demonstrate that ncRNA targeted by the nuclear exosome regulates pho1 expression in response to environmental cues. Deletion of ncRNA decreases H3K9me at pho1 and causes its de-repression in the presence of phosphate. Loss of heterochromatin machinery alone does not de-repress pho1. We find that heterochromatin and the exosome act in parallel to fully repress pho1, as clr4Δ rrp6Δ shows a synergistic increase in pho1 mRNA expression (Figure 4F).
How ncRNA facilitates heterochromatin nucleation and RNA processing is an important question. ncRNA could recruit Red1 and Mtl1, which are cofactors for the exosome and participate in RNAi-dependent and –independent assembly of heterochromatin. In this case, ncRNA would provide high affinity binding sites for RNA processing factors that regulate gene expression through mRNA turnover and heterochromatin formation. Environmental changes could modulate RNA processing factors, which regulate levels of ncRNA and/or their entry into pathways that nucleate heterochromatin. Transcription of upstream RNA could potentially also affect gene expression by interfering with binding of activators to the gene promoter, as observed in S. cerevisiae (Martens et al., 2004).
The regulation of gene expression by ncRNA in response to the environment is a conserved feature in eukaryotes. In S. cerevisiae, which lacks RNAi, ncRNAs modulate gene expression in response to growth conditions by targeting a histone deacetylase (Camblong et al., 2007; Kim et al., 2012; van Werven et al., 2012). Similarly, the antisense RNA COOLAIR regulates flowering time by inducing chromatin modifications in Arabidopsis (Ietswaart et al., 2012). Further analyses may uncover conceptual parallels and highlight the roles of RNA processing factors in directing chromatin changes in response to developmental and environmental signals. We note that an elaborate array of ncRNAs is produced during sexual differentiation in S. pombe, which can impact gene function (Bitton et al., 2011). It is therefore conceivable that targeting of ncRNAs by MTREC is a fundamental component of gene expression reprogramming.
Introns, Splicing and Epigenetic Genome Control
Mmi1 binds specific RNAs and directs RNAi to assemble HOODs at several locations (Hiriart et al., 2012; Yamanaka et al., 2013). However, the specificity for assembly of HOODs at other loci has remained unclear. We show that within certain HOODs, RNAi targets contain cryptic introns. Deletion of such an intron in the SPCC1442.04c locus abolished siRNA production and H3K9me across the entire heterochromatin domain. Moreover, cwf10-1 severely affects siRNA production and H3K9me at a majority of HOODs. We also find that Nrl1 associates with splicing factors and facilitates formation of HOODs. Together, these analyses significantly extend previous work and reveal that cryptic introns and the spliceosome, which acts co-transcriptionally, play an important role in defining the targets of RNAi-mediated heterochromatin assembly at various loci, including developmental genes and Tf2 retrotransposons (Figure 7E).
How might the spliceosome and Nrl1 trigger RNAi? Nrl1 could associate with RNAi machinery, as C. elegans NRDE-2 does (Guang et al., 2010). However, no RNAi factors were identified in our Nrl1 purification. Moreover, cwf10-1 has broad effects on HOOD assembly, including loci not affected by Nrl1 (Table S1). Therefore, it is possible that the spliceosome itself recruits RNAi proteins, and Nrl1 helps in this process by engaging the spliceosome to cryptic introns. Indeed, Nrl1 affects both the splicing efficiency and alternative splicing of various RNA substrates, including RNAi targets (Figures 6C and S7; Table S3). Supporting a direct role in triggering RNAi, splicing machinery interacts with components of the RNA-dependent RNA polymerase complex (Bayne et al., 2008), which is involved in production of siRNA and H3K9me at HOODs (Yamanaka et al., 2013). In Cryptococcus neoformans, stalling of the spliceosome is required for synthesis of siRNAs by a spliceosome-associated RNAi complex, although it is not known if this process facilitates formation of heterochromatin (Dumesic et al., 2013). In addition to engaging RNAi factors, the spliceosome may prevent the release of target transcripts from chromatin and make them available for double strand RNA production through hybridization with opposite strand RNA. Indeed, transcription from the opposite strand occurs at most loci associated with siRNA clusters, including SPCC1442.04c and Tf2 (Yamanaka et al., 2013) (Figure 6C).
The fact that cryptic introns and the spliceosome affect HOOD assembly is a highly significant finding. Splicing factors have been identified in screens for RNAi components in other systems (Ausin et al., 2012; Tabach et al., 2013; Zhang et al., 2013) and human lncRNAs contain inefficiently spliced introns (Tilgner et al., 2012), suggesting this mechanism is likely conserved in higher eukaryotes. Since splicing can be affected by environmental and developmental signals (Averbeck et al., 2005; Keren et al., 2010; McPheeters et al., 2009; Pleiss et al., 2007), a regulatory cascade involving cryptic introns might be responsible for the assembly/disassembly of HOODs observed under specific growth conditions (Yamanaka et al., 2013). Introns may serve as “sensors” that can reprogram gene expression by degrading mRNAs and forming heterochromatin.
An intron-based mechanism might also facilitate RNAi-independent degradation of RNAs, including certain meiotic genes and read-through transcripts targeted by the exosome (Harigaya et al., 2006; Zhang et al., 2011; Zofall et al., 2009). This is supported by the observation that RNA elimination factor Red1 also associates with splicing machinery (Figure 7E), and that various RNAs containing cryptic introns accumulate in cells lacking Red1 and Mtl1, as well as Rrp6 (Tables S2–3 and Figure S7D). In these cases, the degradation of RNA by factors such as MTREC and Rrp6 might be functionally coupled to regulation of their splicing. Notable in this regard, Mmi1 is involved in both meiotic gene silencing and regulation of intron splicing (Chen et al., 2011; McPheeters et al., 2009). Moreover, splicing of conserved introns in the 3′ UTR of transcripts regulates gene expression in mammals by targeting mRNA for degradation (McGlincy and Smith, 2008).
Processing of TER1 precursor RNA, which undergoes maturation by splicing-related mechanisms (Box et al., 2008), by Mtl1 protein assemblies uncovers an important aspect of proper telomere maintenance. Whereas MTREC and Rrp6 prevent formation or accumulation of inactive spliced product, Mtl1-Ctr1 and splicing factors generate mature TER1. The spliceosome may function as part of a complex in which splicing factors join Ctr1 and Mtl1 to process TER1. The exact roles of Ctr1 and Mtl1 are not clear, but they could be involved in spliceosomal cleavage, 3′-end formation and/or protection of mature TER1 from degradation by ribonucleases. Ctr1 and Mtl1 are highly conserved, and could be required for telomere maintenance in higher eukaryotes.
EXPERIMENTAL PROCEDURES
Strains
Strains used in this study are listed in Table S4. Strain constructions are described in Extended Experimental Procedures.
Purification and Mass Spectrometry
Purifications of FLAG- or MYC-tagged proteins were performed by standard methods, using anti-FLAG M2 affinity gel (Sigma) or anti-MYC agarose affinity gel (Sigma), respectively. Purified proteins were analyzed by Western blot or mass spectrometry. Details are described in Extended Experimental Procedures.
Chromatin Immunoprecipitation (ChIP) and ChIP-chip
ChIP and ChIP-chip are described in Extended Experimental Procedures. ChIPs were performed with antibody against H3K9me2 (Abcam), anti-FLAG M2 affinity gel (Sigma), or anti-MYC (Covance). DNA isolated from immunoprecipitated and whole cell crude extract (WCE) fractions was analyzed by multiplex PCR or used for microarray-based ChIP-chip analysis by hybridization to a custom 4X44K oligonucleotide array.
Northern Analysis
RNA was purified using the MasterPure™ Yeast RNA Purification Kit (Epicentre) according to the manufacturer’s instructions. Northern blot analysis was performed as described in the Extended Experimental Procedures.
Small-RNA and RNA-Seq
Libraries were sequenced on the Illumina MiSeq platform. Library preparation and analysis are described in Extended Experimental Procedures.
Supplementary Material
HIGHLIGHTS.
Mtl1 (Mtr4-like) forms an RNA processing network targeting mRNA, ncRNA, and introns
MTREC (Mtl1-Red1 core) and its associated factors assemble heterochromatin domains
Cryptic introns and Mtl1-associated Nrl1 target RNAi to genes and retrotransposons
Mtl1 and Ctr1 mediate telomerase TER1 RNA processing to maintain telomeres
Acknowledgments
We thank P. FitzGerald for intron analysis, R. Allshire for the cwf10-1 mutant, J. Barrowman for her valuable help in editing the manuscript, and M. Lichten for comments. This research was supported by the Intramural Research Program of the National Institutes of Health (NIH), National Cancer Institute, and utilized the Helix Systems and the Biowulf Linux cluster at the NIH.
Footnotes
ACCESSION NUMBERS
Small RNA, RNA-sequencing, and ChIP-chip data are deposited in the European Nucleotide Archive under accession numbers PRJEB4733 and E-MTAB-1962.
Supplemental Information includes Extended Experimental Procedures, seven figures, four tables and can be found with this article online.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ausin I, Greenberg MV, Li CF, Jacobsen SE. The splicing factor SR45 affects the RNA-directed DNA methylation pathway in Arabidopsis. Epigenetics. 2012;7:29–33. doi: 10.4161/epi.7.1.18782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Averbeck N, Sunder S, Sample N, Wise JA, Leatherwood J. Negative control contributes to an extensive program of meiotic splicing in fission yeast. Mol Cell. 2005;18:491–498. doi: 10.1016/j.molcel.2005.04.007. [DOI] [PubMed] [Google Scholar]
- Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayne EH, Portoso M, Kagansky A, Kos-Braun IC, Urano T, Ekwall K, Alves F, Rappsilber J, Allshire RC. Splicing factors facilitate RNAi-directed silencing in fission yeast. Science. 2008;322:602–606. doi: 10.1126/science.1164029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitton DA, Grallert A, Scutt PJ, Yates T, Li Y, Bradford JR, Hey Y, Pepper SD, Hagan IM, Miller CJ. Programmed fluctuations in sense/antisense transcript ratios drive sexual differentiation in S. pombe. Mol Syst Biol. 2011;7:559. doi: 10.1038/msb.2011.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Box JA, Bunch JT, Tang W, Baumann P. Spliceosomal cleavage generates the 3′ end of telomerase RNA. Nature. 2008;456:910–914. doi: 10.1038/nature07584. [DOI] [PubMed] [Google Scholar]
- Buhler M, Haas W, Gygi SP, Moazed D. RNAi-dependent and -independent RNA turnover mechanisms contribute to heterochromatic gene silencing. Cell. 2007;129:707–721. doi: 10.1016/j.cell.2007.03.038. [DOI] [PubMed] [Google Scholar]
- Cam HP, Sugiyama T, Chen ES, Chen X, FitzGerald PC, Grewal SI. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat Genet. 2005;37:809–819. doi: 10.1038/ng1602. [DOI] [PubMed] [Google Scholar]
- Camblong J, Iglesias N, Fickentscher C, Dieppois G, Stutz F. Antisense RNA stabilization induces transcriptional gene silencing via histone deacetylation in S. cerevisiae. Cell. 2007;131:706–717. doi: 10.1016/j.cell.2007.09.014. [DOI] [PubMed] [Google Scholar]
- Chen HM, Futcher B, Leatherwood J. The fission yeast RNA binding protein Mmi1 regulates meiotic genes by controlling intron specific splicing and polyadenylation coupled RNA turnover. PloS One. 2011;6:e26804. doi: 10.1371/journal.pone.0026804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinen M, Morita M, Fukumura K, Tani T. Involvement of the spliceosomal U4 small nuclear RNA in heterochromatic gene silencing at fission yeast centromeres. J Biol Chem. 2010;285:5630–5638. doi: 10.1074/jbc.M109.074393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YS, Jang S, Yoon JH. Isolation of a novel rmn1 gene genetically linked to spnab2 with respect to mRNA export in fission yeast. Mol Cells. 2012;34:315–321. doi: 10.1007/s10059-012-0157-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding DQ, Okamasa K, Yamane M, Tsutsumi C, Haraguchi T, Yamamoto M, Hiraoka Y. Meiosis-specific noncoding RNA mediates robust pairing of homologous chromosomes in meiosis. Science. 2012;336:732–736. doi: 10.1126/science.1219518. [DOI] [PubMed] [Google Scholar]
- Doma MK, Parker R. RNA quality control in eukaryotes. Cell. 2007;131:660–668. doi: 10.1016/j.cell.2007.10.041. [DOI] [PubMed] [Google Scholar]
- Dumesic PA, Natarajan P, Chen C, Drinnenberg IA, Schiller BJ, Thompson J, Moresco JJ, Yates JR, 3rd, Bartel DP, Madhani HD. Stalled spliceosomes are a signal for RNAi-mediated genome defense. Cell. 2013;152:957–968. doi: 10.1016/j.cell.2013.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutrow N, Nix DA, Holt D, Milash B, Dalley B, Westbroek E, Parnell TJ, Cairns BR. Dynamic transcriptome of Schizosaccharomyces pombe shown by RNA-DNA hybrid mapping. Nat Genet. 2008;40:977–986. doi: 10.1038/ng.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Jacobsen SE. Epigenetic modifications in plants: an evolutionary perspective. Curr Opin Plant Biol. 2011;14:179–186. doi: 10.1016/j.pbi.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guang S, Bochner AF, Burkhart KB, Burton N, Pavelec DM, Kennedy S. Small regulatory RNAs inhibit RNA polymerase II during the elongation phase of transcription. Nature. 2010;465:1097–1101. doi: 10.1038/nature09095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harigaya Y, Tanaka H, Yamanaka S, Tanaka K, Watanabe Y, Tsutsumi C, Chikashige Y, Hiraoka Y, Yamashita A, Yamamoto M. Selective elimination of messenger RNA prevents an incidence of untimely meiosis. Nature. 2006;442:45–50. doi: 10.1038/nature04881. [DOI] [PubMed] [Google Scholar]
- Hiriart E, Vavasseur A, Touat-Todeschini L, Yamashita A, Gilquin B, Lambert E, Perot J, Shichino Y, Nazaret N, Boyault C, et al. Mmi1 RNA surveillance machinery directs RNAi complex RITS to specific meiotic genes in fission yeast. EMBO J. 2012;31:2296–2308. doi: 10.1038/emboj.2012.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, LaCava J, Tollervey D. RNA-quality control by the exosome. Nat Rev Mol Cell Biol. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- Ietswaart R, Wu Z, Dean C. Flowering time control: another window to the connection between antisense RNA and chromatin. Trends Genet. 2012;28:445–453. doi: 10.1016/j.tig.2012.06.002. [DOI] [PubMed] [Google Scholar]
- Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- Keren H, Lev-Maor G, Ast G. Alternative splicing and evolution: diversification, exon definition and function. Nat Rev Genet. 2010;11:345–355. doi: 10.1038/nrg2776. [DOI] [PubMed] [Google Scholar]
- Kiely CM, Marguerat S, Garcia JF, Madhani HD, Bahler J, Winston F. Spt6 is required for heterochromatic silencing in the fission yeast Schizosaccharomyces pombe. Mol Cell Biol. 2011;31:4193–4204. doi: 10.1128/MCB.05568-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DU, Hayles J, Kim D, Wood V, Park HO, Won M, Yoo HS, Duhig T, Nam M, Palmer G, et al. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol. 2010;28:617–623. doi: 10.1038/nbt.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T, Xu Z, Clauder-Munster S, Steinmetz LM, Buratowski S. Set3 HDAC mediates effects of overlapping noncoding transcription on gene induction kinetics. Cell. 2012;150:1158–1169. doi: 10.1016/j.cell.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- Lemay JF, D’Amours A, Lemieux C, Lackner DH, St-Sauveur VG, Bahler J, Bachand F. The nuclear poly(A)-binding protein interacts with the exosome to promote synthesis of noncoding small nucleolar RNAs. Mol Cell. 2010;37:34–45. doi: 10.1016/j.molcel.2009.12.019. [DOI] [PubMed] [Google Scholar]
- Lemieux C, Marguerat S, Lafontaine J, Barbezier N, Bahler J, Bachand F. A Pre-mRNA degradation pathway that selectively targets intron-containing genes requires the nuclear poly(A)-binding protein. Mol Cell. 2011;44:108–119. doi: 10.1016/j.molcel.2011.06.035. [DOI] [PubMed] [Google Scholar]
- Licatalosi DD, Darnell RB. RNA processing and its regulation: global insights into biological networks. Nat Rev Genet. 2010;11:75–87. doi: 10.1038/nrg2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubas M, Christensen MS, Kristiansen MS, Domanski M, Falkenby LG, Lykke-Andersen S, Andersen JS, Dziembowski A, Jensen TH. Interaction profiling identifies the human nuclear exosome targeting complex. Mol Cell. 2011;43:624–637. doi: 10.1016/j.molcel.2011.06.028. [DOI] [PubMed] [Google Scholar]
- Martens JA, Laprade L, Winston F. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature. 2004;429:571–574. doi: 10.1038/nature02538. [DOI] [PubMed] [Google Scholar]
- Mata J, Lyne R, Burns G, Bahler J. The transcriptional program of meiosis and sporulation in fission yeast. Nat Genet. 2002;32:143–147. doi: 10.1038/ng951. [DOI] [PubMed] [Google Scholar]
- McGlincy NJ, Smith CW. Alternative splicing resulting in nonsense-mediated mRNA decay: what is the meaning of nonsense? Trend Biochem Sci. 2008;33:385–393. doi: 10.1016/j.tibs.2008.06.001. [DOI] [PubMed] [Google Scholar]
- McPheeters DS, Cremona N, Sunder S, Chen HM, Averbeck N, Leatherwood J, Wise JA. A complex gene regulatory mechanism that operates at the nexus of multiple RNA processing decisions. Nat Struct Mol Biol. 2009;16:255–264. doi: 10.1038/nsmb.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orom UA, Shiekhattar R. Long non-coding RNAs and enhancers. Curr Opin Genet Dev. 2011;21:194–198. doi: 10.1016/j.gde.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleiss JA, Whitworth GB, Bergkessel M, Guthrie C. Rapid, transcript-specific changes in splicing in response to environmental stress. Mol Cell. 2007;27:928–937. doi: 10.1016/j.molcel.2007.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Grewal SI. Different means, same end-heterochromatin formation by RNAi and RNAi-independent RNA processing factors in fission yeast. Curr Opin Genet Dev. 2012;22:156–163. doi: 10.1016/j.gde.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhind N, Chen Z, Yassour M, Thompson DA, Haas BJ, Habib N, Wapinski I, Roy S, Lin MF, Heiman DI, et al. Comparative functional genomics of the fission yeasts. Science. 2011;332:930–936. doi: 10.1126/science.1203357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Jensen TH. The exosome: a multipurpose RNA-decay machine. Trend Biochem Sci. 2008;33:501–510. doi: 10.1016/j.tibs.2008.07.003. [DOI] [PubMed] [Google Scholar]
- Sharp PA. The centrality of RNA. Cell. 2009;136:577–580. doi: 10.1016/j.cell.2009.02.007. [DOI] [PubMed] [Google Scholar]
- St-Andre O, Lemieux C, Perreault A, Lackner DH, Bahler J, Bachand F. Negative regulation of meiotic gene expression by the nuclear poly(a)-binding protein in fission yeast. J Biol Chem. 2010;285:27859–27868. doi: 10.1074/jbc.M110.150748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, Sugioka-Sugiyama R. Red1 promotes the elimination of meiosis-specific mRNAs in vegetatively growing fission yeast. EMBO J. 2011;30:1027–1039. doi: 10.1038/emboj.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabach Y, Billi AC, Hayes GD, Newman MA, Zuk O, Gabel H, Kamath R, Yacoby K, Chapman B, Garcia SM, et al. Identification of small RNA pathway genes using patterns of phylogenetic conservation and divergence. Nature. 2013;493:694–698. doi: 10.1038/nature11779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Kannan R, Blanchette M, Baumann P. Telomerase RNA biogenesis involves sequential binding by Sm and Lsm complexes. Nature. 2012;484:260–264. doi: 10.1038/nature10924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro S, Asano T, Kanoh J, Ishikawa F. Transcription-induced chromatin association of RNA surveillance factors mediates facultative heterochromatin formation in fission yeast. Genes Cells. 2013;18:327–339. doi: 10.1111/gtc.12038. [DOI] [PubMed] [Google Scholar]
- Tilgner H, Knowles DG, Johnson R, Davis CA, Chakrabortty S, Djebali S, Curado J, Snyder M, Gingeras TR, Guigo R. Deep sequencing of subcellular RNA fractions shows splicing to be predominantly co-transcriptional in the human genome but inefficient for lncRNAs. Genome Res. 2012;22:1616–1625. doi: 10.1101/gr.134445.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuck AC, Tollervey D. A Transcriptome-wide atlas of RNP composition reveals diverse classes of mRNAs and lncRNAs. Cell. 2013;154:996–1009. doi: 10.1016/j.cell.2013.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Werven FJ, Neuert G, Hendrick N, Lardenois A, Buratowski S, van Oudenaarden A, Primig M, Amon A. Transcription of two long noncoding RNAs mediates mating-type control of gametogenesis in budding yeast. Cell. 2012;150:1170–1181. doi: 10.1016/j.cell.2012.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm BT, Marguerat S, Watt S, Schubert F, Wood V, Goodhead I, Penkett CJ, Rogers J, Bahler J. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature. 2008;453:1239–1243. doi: 10.1038/nature07002. [DOI] [PubMed] [Google Scholar]
- Yamamoto M. The selective elimination of messenger RNA underlies the mitosis-meiosis switch in fission yeast. Proc Jpn Acad Ser B Phys Biol Sci. 2010;86:788–797. doi: 10.2183/pjab.86.788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka S, Mehta S, Reyes-Turcu FE, Zhuang F, Fuchs RT, Rong Y, Robb GB, Grewal SI. RNAi triggered by specialized machinery silences developmental genes and retrotransposons. Nature. 2013;493:557–560. doi: 10.1038/nature11716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka S, Yamashita A, Harigaya Y, Iwata R, Yamamoto M. Importance of polyadenylation in the selective elimination of meiotic mRNAs in growing S. pombe cells. EMBO J. 2010;29:2173–2181. doi: 10.1038/emboj.2010.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CJ, Zhou JX, Liu J, Ma ZY, Zhang SW, Dou K, Huang HW, Cai T, Liu R, Zhu JK, et al. The splicing machinery promotes RNA-directed DNA methylation and transcriptional silencing in Arabidopsis. EMBO J. 2013;32:1128–1140. doi: 10.1038/emboj.2013.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Fischer T, Porter RL, Dhakshnamoorthy J, Zofall M, Zhou M, Veenstra T, Grewal SI. Clr4/Suv39 and RNA quality control factors cooperate to trigger RNAi and suppress antisense RNA. Science. 2011;331:1624–1627. doi: 10.1126/science.1198712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zofall M, Fischer T, Zhang K, Zhou M, Cui B, Veenstra TD, Grewal SI. Histone H2A.Z cooperates with RNAi and heterochromatin factors to suppress antisense RNAs. Nature. 2009;461:419–422. doi: 10.1038/nature08321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zofall M, Yamanaka S, Reyes-Turcu FE, Zhang K, Rubin C, Grewal SI. RNA elimination machinery targeting meiotic mRNAs promotes facultative heterochromatin formation. Science. 2012;335:96–100. doi: 10.1126/science.1211651. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.