Abstract
Mechanisms generating diverse cell types from multipotent progenitors are crucial for normal development. Neural crest cells (NCCs) are multipotent stem cells that give rise to numerous cell-types, including pigment cells. Medaka has four types of NCC-derived pigment cells (xanthophores, leucophores, melanophores and iridophores), making medaka pigment cell development an excellent model for studying the mechanisms controlling specification of distinct cell types from a multipotent progenitor. Medaka many leucophores-3 (ml-3) mutant embryos exhibit a unique phenotype characterized by excessive formation of leucophores and absence of xanthophores. We show that ml-3 encodes sox5, which is expressed in premigratory NCCs and differentiating xanthophores. Cell transplantation studies reveal a cell-autonomous role of sox5 in the xanthophore lineage. pax7a is expressed in NCCs and required for both xanthophore and leucophore lineages; we demonstrate that Sox5 functions downstream of Pax7a. We propose a model in which multipotent NCCs first give rise to pax7a-positive partially fate-restricted intermediate progenitors for xanthophores and leucophores; some of these progenitors then express sox5, and as a result of Sox5 action develop into xanthophores. Our results provide the first demonstration that Sox5 can function as a molecular switch driving specification of a specific cell-fate (xanthophore) from a partially-restricted, but still multipotent, progenitor (the shared xanthophore-leucophore progenitor).
Author Summary
How individual cell fates are specified from multipotent progenitor cells is a fundamental question in developmental and stem cell biology. Accumulating evidence indicates that stem cells develop into each of their final, diverse cell-types after progression through one or more partially-restricted intermediates, but the molecular mechanisms underlying final fate choice are largely unknown. Neural crest cells (NCCs) give rise to diverse cell-types including multiple pigment cells and thus are a favored model for understanding the mechanism of fate specification. We have investigated how a specific fate choice is made from partially-restricted pigment cell progenitors in medaka. We show that Sry-related transcription factor Sox5 is required for fate determination between yellow xanthophore and white leucophore, and its loss causes excessive formation of leucophores and absence of xanthophores. We demonstrate that Sox5 functions cell-autonomously in the xanthophore lineage in medaka. Furthermore, pax7a is expressed in the partially-restricted progenitor cells shared with xanthophore and leucophore lineages, and Sox5 acts in some of these cells to promote xanthophore lineage. Our work reveals the role of Sox5 as a molecular switch determining xanthophore versus leucophore fate choice from the shared progenitor, and identifies an important mechanism regulating pigment cell fate choice from NCCs.
Introduction
Elucidation of mechanisms that control cell fate specification from multipotent progenitors is one of the most important topics in developmental biology. The neural crest is a vertebrate-specific multipotent cell population that arises at the border of the neural plate and prospective epidermis. The neural crest cells (NCCs) migrate to various destinations along stereotyped pathways and thereby give rise to a diversity of different cell types, including sensory, enteric and autonomic neurons, glia of the peripheral nervous system, skeletogenic fates such as craniofacial cartilage, and pigment cells [1], [2]. Their stem cell-like characteristics, and potential therapeutic uses in regenerative medicine, make NCCs attractive as a model for studying fate specification of multipotent progenitor cells in stem cell biology.
Neural crest-derived pigment cells in vertebrates are classified into diverse cell types and each can be easily identified by its natural coloration. Whereas mammals and birds have a single type of pigment cell, melanocytes, fish have up to six [3]. Zebrafish has three distinct types of pigment cells, melanophores, yellow xanthophores and iridescent iridophores [4], [5]. In medaka and some other fish species, there is a fourth pigment cell type, the white leucophore [6]–[8]. The white coloration and auto-fluorescence of leucophores depend on light reflection from intracellular organellar crystals of uric acid, which is a purine-related substance similar to guanine and hypoxanthine in iridophores [9]. In addition, leucophores in medaka embryos and larvae appear to be orange, due to production of a pteridine pigment, drosopterin, during embryonic/larval stages [10]. Xanthophores contain a different pteridine, yellow sepiapterin. Biochemical studies suggest that both drosopterin and sepiapterin are generated via a common synthesis pathway for H4biopterine from GTP [11], indicating a possible close evolutionary relationship between xanthophores and leucophores, although the embryological/genetic relationship between these two cell-types has not been studied.
Previous studies by in vitro clonal analysis showed that multipotent NCCs become gradually restricted in their potential to generate certain derivatives, forming partially-restricted intermediate progenitors before eventually becoming specified to an individual fate [12]–[14]. However, the presence, identity and diversity of these intermediate progenitors in vivo remains unclear. Furthermore, the molecular mechanisms regulating fate specification from the intermediate progenitors are also incompletely understood. Previous genetic studies identified a few key transcription factors required for fate specification within NCCs [15]. Some of the best-known examples are transcription factors controlling development of melanocytes (functionally and genetically equivalent to melanophores in fish). Sox10 is required to drive transcription of Mitf, a master gene for melanocyte lineage in multipotent NCCs [16]–[21]. Pax3 acts synergistically with Sox10 to regulate the mouse Mitf promoter [22]–[25]. Whereas the molecular mechanisms driving melanocyte differentiation are relatively well understood, the equivalent mechanisms for other pigment cell-types remain largely mysterious, as do the interactions between these factors that control the balance of pigment cell fates. One exception comes from the demonstration in zebrafish and chick that Foxd3 functions to inhibit differentiation of melanocytes from NCCs by repressing Mitf function; in zebrafish, FoxD3 also promotes iridophore development [26]–[29].
The mutant collections in medaka provide models for investigating the genetic basis of fate choice in NCCs. A series of loci affecting both xanthophore and leucophore development in medaka offer novel insight into how these two pigment cell types are specified [30], [31]. Among them, leucophore free and leucophore free-2 (lf and lf-2) mutants have no discernible leucophores nor xanthophores in the embryo/larva, although lf-2 mutants have occasional escaper leucophores [31]. Our previous studies revealed that the lf-2 locus encodes pax7a, and lf-2 mutants failed to develop xanthophores and leucophores, suggesting a role of pax7a in fate specification of a shared, partially-restricted progenitor of the xanthophore and leucophore lineages (Kimura et al, unpublished data). We also revealed that lf locus is slc2a15b and this encodes a solute career (Slc) protein required for coloration of xanthophores and leucophores, consistent with the previous description that lf mutants have deficiently-colored xanthophores and leucophores. These genetic data also suggest that leucophores are closely related to xanthophores. This left the major unanswered question of what mechanism allows selection of xanthophore or leucophore fate from these intermediate progenitors.
Medaka many leucophores-3 (ml-3) mutant embryos exhibit excessive formation of leucophores and absence of visible xanthophores [30]–[32]. To elucidate the molecular mechanisms regulating the fate choices of xanthophore and leucophore, we have investigated medaka ml-3 mutant by genetic and developmental approaches. Here we report that ml-3 locus encodes medaka sox5 and that Sox5 functions cell-autonomously to control specification of xanthophores from the shared xanthophore-leucophore progenitor.
Results
ml-3 mutants show combined ectopic and excessive formation of leucophores and absence of xanthophores
Medaka ml-3 is a spontaneous mutant with excessive formation of leucophores, and a lack of visible xanthophore pigmentation in the embryo/larva [30], [31], but with no apparent phenotypes of other neural crest derivatives (Figures S1A–S1J). We first re-examined all pigment cell types during embryonic development in ml-3 mutant. In wild-type (WT) embryos, the first differentiating leucophores appear ventral to the hindbrain/midbrain (ventral head leucophores) by 18–19 somite stage (50 hours post fertilization: hpf, data not shown) [31], [33]. The other population of leucophores, found on the dorsal surface of the head and trunk, begin to differentiate at 3.5–4 days post fertilization (dpf) when a few are visible (Figure S2A). By hatching stage (9 dpf), these leucophores have increased to approximately 30 cells scattered on the dorsal head and 40 cells in the dorsal midline of the trunk (Figures 1A, 1C, 1I). In ml-3 homozygous mutants, formation of ventral head leucophores was not markedly altered (data not shown); in contrast, there is already an excess compared with WT of leucophores in the dorsal head and trunk at 4 dpf, (Figures S2A, S2B). By 7 dpf, the trunk leucophores are scattered more laterally than in WT, forming three lines (Figures 1D, S2C, S2D). Subsequently, leucophores in the central line of the three become gradually reduced, the number in the more lateral lines become substantially increased after hatching. The number of leucophores in ml-3 homozygotes was significantly larger in both head and dorsal trunk than that in WT (Figures 1I, S2P).
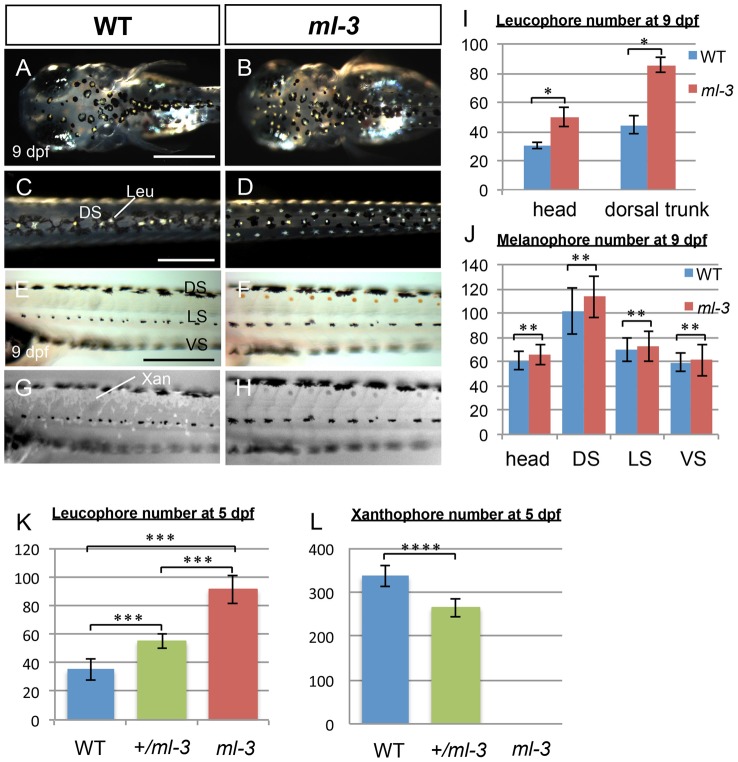
Figure 1. Larval pigment pattern phenotypes of medaka ml-3 mutants.
(A–H) 9 dpf (hatching stage). (A, C, E, G) WT. (B, D, F, H) ml-3 mutant. (E, F) Bright field. (G, H) UV light. (A–D) Dorsal views. (E–H) Lateral views. (A–D) Leucophores are formed in excess in mutants. Whereas in WT larvae leucophores lie over the head (A) and along dorsal midline of the trunk (C) in association with melanophores, in ml-3 larvae excess leucophores are localized ectopically to form bilateral lines along the anterior-posterior axis (B, D) in addition to those in the normal position in head (B) and trunk midline (D). (E–H) Xanthophores are absent from ml-3 mutants. In WT (E) as well as in ml-3 (F), xanthophores are not evident when observed in brightfield. Under UV illumination, however, auto-fluorescent xanthophores are readily visible in WT, covering over dorsal trunk (G). In ml-3 mutants, xanthophore auto-fluorescence is not detected (H). (A–H) Melanophore patterns are unaffected in ml-3 mutants. In both WT (E, G) and ml-3 mutants (F, H), melanophores are distributed in three longitudinal stripes (DS, LS and VS). (I, J) Counts of leucophores and melanophores in ml-3 mutants at 9 dpf. Mean counts (error bars give standard deviation) are shown for WT (blue) and ml-3 mutants (red). (I) The numbers of leucophores in head region and in dorsal trunk are larger in ml-3 than in WT (n = 12 for each group and region): two-way ANOVA, F(group) = 389.09, df = 1,42, p<0.0001; F(group×region) = 46.93, df = 1,42, p<0.0001 and Student's t-test for each region, *, p<0.0001. (J) There is no significant difference in melanophore number between WT and ml-3 (n = 12 for WT, n = 11 for ml-3): two-way ANOVA, F(group) = 3.72, df = 1,88, p>0.05; F(group×region) = 0.74, df = 1,88, p>0.05 and Student's t-test for each region, **, p>0.05. (K, L) Mean (±s.d.) counts of leucophores and xanthophores on dorsal trunk surface in WT, ml-3 heterozygotes (+/ml-3) and ml-3 homozygotes (ml-3) at 5 dpf (also see Figures S2E–S2J). (K) The leucophore numbers seem to be negatively correlated with the copy-number of ml-3 gene (n = 10 for each group). p<0.0001 by one-way ANOVA. ***, p<0.0001 by Student's t-test for WT versus ml-3 heterozygotes, WT versus ml-3 homozygotes, and ml-3 heterozygotes versus homozygotes. (L) The xanthophore numbers seem to be positively correlated with the copy-number of ml-3 gene (n = 10 for each group). p<0.0001 by one-way ANOVA. ****, p<0.0001 by Student's t-test. Leu, leucophore; Xan, xanthophore; DS, dorsal stripe; LS, lateral stripe; VS, ventral stripe. Scale bars: 250 µm.
Pigmented xanthophores are first observed at 6 dpf in WT, but differentiating xanthophores are difficult to detect at early embryonic stages. In order to detect them more readily, we examined them using their auto-fluorescence upon UV-light exposure (Figures 1E–1H) [34]. In WT hatchlings, numerous xanthophores cover the dorsal trunk and others are scattered around the melanophores in the lateral stripe (see below, Figure 1G). In ml-3 homozygotes, no xanthophores were detected on the body surface (Figure 1H).
To further elucidate the effect of ml-3 deficiency, we investigated the phenotypes of ml-3 heterozygotes. The ml-3 heterozygous leucophores were normally positioned in the dorsal midline of the trunk at 5 dpf, but significantly increased, compared with WT leucophores (Figures 1K, S2E–S2G). Xanthophores, scattered over the trunk surface, were significantly fewer at 5 dpf in ml-3 heterozygotes than in WT (Figures 1L, S2H–S2J). These results indicate that ml-3 is a semi-dominant mutation and the increase of leucophores and the decrease of xanthophores are dependent on ml-3 gene dosage. These data suggest that the ml-3 locus acts as a fate-switch for xanthophore versus leucophore lineages within a shared progenitor cell. Further experiments below were carried out using ml-3 homozygotes (described as ml-3 mutants) unless otherwise specified.
We found no evidence for a change in the number of the other two pigment cell-types, melanophores or iridophores. Melanophores first appeared in the trunk region at 12 somite stage (41 hpf) and formed three stripes in trunk: dorsal stripe (DS), lateral stripe (LS), and ventral stripe (VS) (Figure 1E). Whereas most melanophores in WT were associated with leucophores in the DS and head region (Figures 1A, 1C), there are no melanophores associated with the ectopic leucophores in ml-3 hatchlings (Figures 1B, 1D, 1F). Consistent with this, the number of melanophores was not significantly different between WT and ml-3 in either of the DS, LS, VS or head region (Figures 1J, S2O). Iridophores appeared on the eyes and the yolk sac at 5 dpf and could be readily observed at later stages in WT (data not shown and Figures S2K, S2M). Although we were unable to count numbers of iridophores due to ambiguity in their cell borders, there was no marked difference in the areas covered by iridophores in WT and ml-3 (Figures S2L, S2N). These findings indicate that the ml-3 mutation specifically affects the formation of xanthophores and leucophores, but not of melanophores and iridophores.
Xanthophore specification is defective in ml-3 mutants
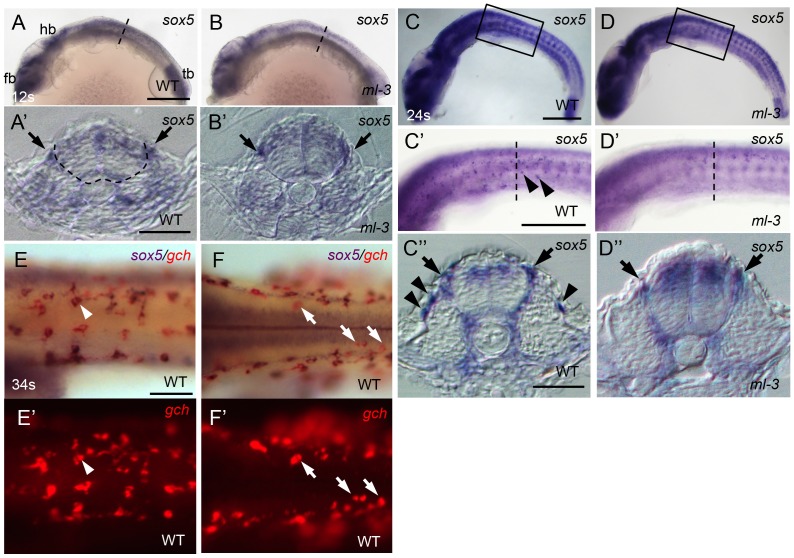
To address whether ml-3 mutation affects the specification of the xanthophore and leucophore lineages, we examined the expression of the earliest known markers for pigment cell precursors. GTP cyclohydrolase I (gch) converts GTP to dihydroneopterin triphosphate, which is a source material for pteridine pigments in xanthophore precursors [35], [36]. Dihydroneopterin triphosphate is also converted into H4biopterin, which functions as a cofactor for eumelanin synthesis in melanophores. Thus gch was reported to be expressed in both xanthophore and melanophore lineages in zebrafish [35]. Our in situ hybridization (ISH) analysis in WT embryos showed that gch (XM_004085058) was expressed in laterally migrating cells on the trunk surface and the ventral head leucophores at 24–27 somite stage (58–61 hpf) (Figures 2A, 2C, 2D). The gch-expressing cells were located on lateral (above somite) and dorsal (above neural tube) trunk surface at 34 somite stage (74 hpf, Figures 2H, 2H′). These cells included both unpigmented and differentiating pigment cells. Unlike in zebrafish, we saw no evidence for a transient overlap of gch expression with that the melanophore marker, dopachrome tautomerase (dct, XM_004081780) [37], indicating that medaka gch was not expressed in the melanophore lineage at the stages tested (Figures 2E, 2E′). Furthermore, at 7 dpf, gch mRNA was detected in all xanthophores and leucophores (Figures 2F, 2F′, 2G, 2G′). These data show that, in medaka, gch expression is a marker for xanthophore and leucophore linages from well before overt differentiation, but is not a marker for the melanophore lineage at these stages.
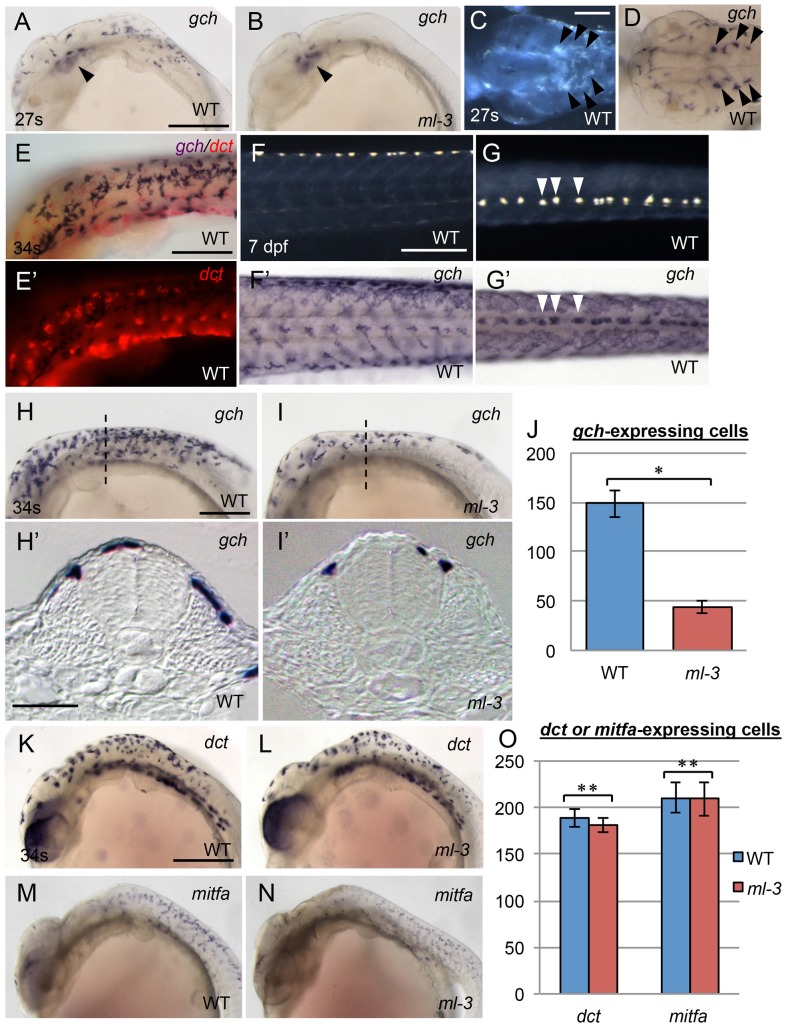
Figure 2. Embryonic pigment cell precursor formation in ml-3 mutants.
(A, B, D, F′, G′, H, I), gch. (K, L), dct.(M, N), mitfa. (E, E′) gch (blue) and dct (red). (A, C–H, K, M) WT. (B, I, L, N) ml-3 mutant. (A, B, E, F, H, I, K–N) Lateral views. (C, D, G) Dorsal views. (H′, I′) Transverse sections. (C, F, G) Pre-fixed samples in darkfield. (A–D) 27 somite stage (27 s, 61 hpf). (E, H, I, K–N) 34 somite stage (34 s, 74 hpf). (F, F′, G, G′) 7 dpf. (A–I) ISH analyses show gch expression in WT xanthophores and leucophores. Whereas gch-expressing cells are found over the lateral and anterior trunk surface and the head in WT at 27 s (A) as well as in ventral head leucophores (black arrowheads, C, D), ml-3 embryos have gch expression only in ventral head leucophores (black arrowheads, B). The embryo at 34 s is co-stained with gch riboprobe (purple) and dct riboprobe (red) (E, E′). gch signals show no overlap with dct signals. At 7 dpf, gch expression is detected on the surface of the whole length of trunk in WT (F′, G′). The dotted gch signals in the dorsal midline coincide with leucophore positions (compare F′ with F and G′ with G, some examples were represented by white arrowhead in G and G′). In WT at 34 s (H), gch-expressing cells are spread more posteriorly and scattered over the dorsal trunk surface (H′). In ml-3 mutants (I), gch expression is in fewer cells (J), and these are restricted to the dorsal trunk surface (I′) compared with WT. Transverse histological section from embryos at the level as indicated by dotted line in H and I. (J) Counts of gch-expressing cells at 34 s (n = 11 for WT, n = 10 for ml-3). The number is significantly fewer in ml-3 than in WT (*, p<0.0001 by Student's t-test). (K–N) Melanophore precursors, detected using dct (K, L) and mitfa (M, N) are not markedly altered in ml-3 (L, N) as compared with WT (K, M). (O) The number of dct- or mitfa-expressing cells is not significantly different between WT and ml-3 (dct, n = 11 for each group, **, p>0.05 by Student's t-test; mitfa, n = 12 for each group, **, p>0.05 by Student's t-test). (J, O) Mean (±s.d.) counts are shown as bars in blue (WT) and in red (ml-3). Scale bars: (A, F, H, K) 250 µm; (C) 100 µm; (E) 150 µm; (H′) 25 µm.
Similar results were obtained with xanthine dehydrogenase (xdh, XM_004066478), which is known as a xanthophore precursor marker in zebrafish (Figures S3A–S3D) [35]. Whereas xdh expression is specific to xanthophores in zebrafish, which do not have leucophores, xdh was clearly detectable in both xanthophores and leucophores in medaka. These observations are consistent with previous reports showing that medaka xanthophores and leucophores contain pteridines (sepiapterin in xanthophores and drosopterin in leucophores) and that the enzymes encoded by gch and xdh are required for pteridine synthesis [11]. Having defined gch (which was expressed more strongly than xdh) as an early marker of xanthophore and leucophore specification, we then used it to assess whether xanthophore specification, as opposed to visible differentiation, failed in ml-3 mutants.
At 24–27 somite stage (58–61 hpf), the ml-3 embryos lacked gch-expressing cells on the trunk although they had gch expression in the ventral head leucophores (Figure 2B). At 34 somite stage (74 hpf), many, but fewer than in WT embryos, gch-expressing cells were detected in ml-3 mutants (Figures 2H–2J). Transverse cross sections of these embryos revealed that, in ml-3 mutants, gch-expressing cells were detected on the dorsal trunk surface, but were fully absent from the lateral trunk surface (Figure 2I′). Differentiating leucophores and xanthophores are localized on the dorsal and lateral trunk surface, respectively, so that our data indicate that specification of the xanthophore lineage fails in ml-3 mutants. Expression of melanophore precursor markers, dct and mitfa (KC249980) was indistinguishable in ml-3 and WT siblings at 34 somite stage (74 hpf) (Figures 2K–2O), further confirming that ml-3 mutation specifically affects xanthophore/leucophore lineages.
ml-3 locus encodes sox5
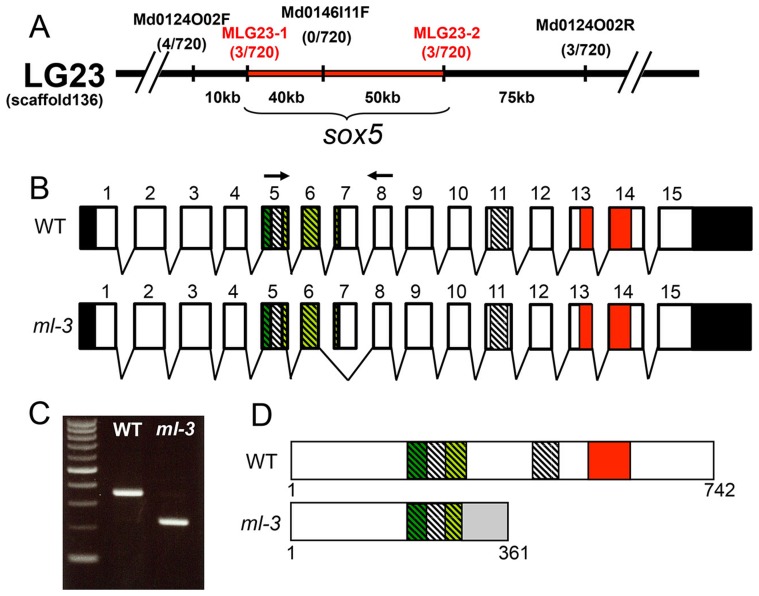
To identify the ml-3 locus, we carried out positional cloning. The ml-3 locus was mapped within a 90-kbp region on linkage group 23. sox5 (AB856413) was the only known gene in this genomic region (Figure 3A). cDNA cloning identified a medaka sox5 gene that consists of 15 exons with a 2229 bp open reading frame encoding a 742 amino acid protein (Figures 3B, 3D). Medaka Sox5 protein displayed strong similarities to human SOX5 (NP_008871.3) and zebrafish Sox5 (AY730586.1), (66% and 79% amino acid identities, respectively). Like Sox5 in other vertebrate species, medaka Sox5 contained several conserved domains, such as an HMG box DNA-binding domain, a leucine-zipper, a Q-box and two coiled-coil domains (Figure 3D). RT-PCR and sequencing of the ml-3 mutant cDNA identified a 124-bp deletion corresponding precisely to skipping of exon 7 (Figures 3B, 3C). The ml-3 mutation introduces a premature stop codon and results in generation of a truncated Sox5 protein lacking the HMG box domain; this is predicted to result in loss-of-function of Sox5 (Figure 3D).
Figure 3. Mapping of the ml-3 locus.
(A) ml-3 locus was mapped to a 90 kb region (red bar) between MLG23-1 and MLG23-2 on LG23, predicted to contain only one gene, sox5. Typing markers are indicated above the map, with the number of recombinants per total 720 haploid genomes examined at each position. (B) Medaka sox5 comprises 15 exons (upper). Sequencing of cDNAs showed that in ml-3, exon 7 is skipped (lower). Boxes represent exons. Angled lines represent introns. The 5′ and 3′ untranslated regions are colored in black. Diagonal stripes and colored regions correspond to regions encoding the protein domains described in (D). Black arrows show positions of the primer set used for RT-PCR in C. (C) RT-PCR detects the skipping of exon 7 in sox5 mRNA of ml-3 mutant. (D) WT sox5 gene encodes 742 amino acid protein consisting of two coiled-coil domains (diagonal stripe) and HMG box (red box). The first coiled-coil domain contains a leucine-zipper (green box) and a glutamine-rich domain (Q box, light green box). In ml-3, loss of exon 7 causes a premature stop codon leading to a truncated protein. Gray box represents an altered frame. The resultant truncated protein would lack the second coiled-coil domain and HMG box (lower).
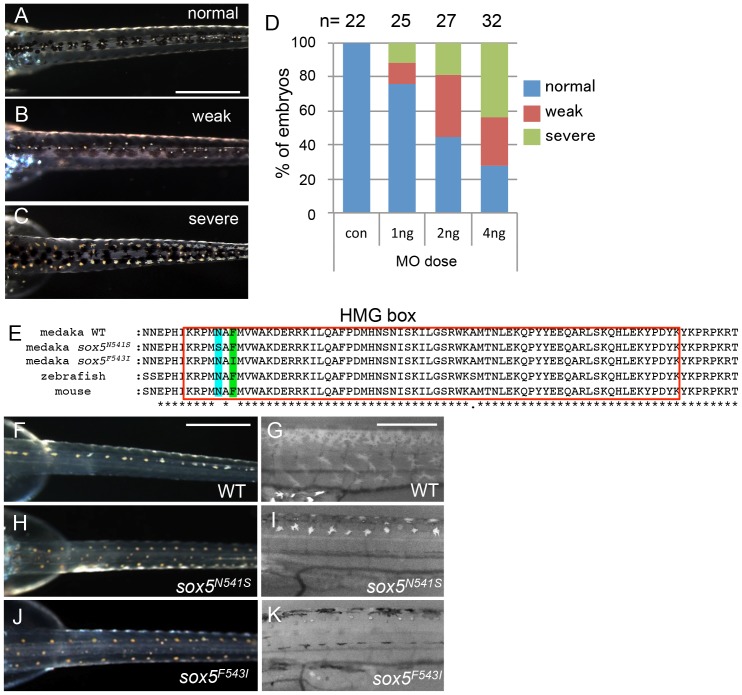
Despite sequencing introns 6 and 7 and exon 7 of the sox5 gene from ml-3 mutants, we were unable to identify any candidate mutations that might induce the aberrant splicing of exon 7. To confirm that sox5 is the causative gene for the ml-3 phenotype, we carried out knockdown of sox5 with an antisense morpholino (MO) targeted to the splice donor site of exon 7. The sox5 morphants exhibited the ectopic formation of leucophores at hatching stage while the control morphants injected with the standard control MO failed to mimic the ml-3 phenotypes (Figures 4A–4C). As expected, the severity of the morphants' pigment phenotype was dependent on the MO dosage, with higher doses resulting in a higher proportion of embryos closely resembling the ml-3 leucophore phenotype (Figure 4D). Sox5 morphants also mimicked the xanthophore phenotype of ml-3 (data not shown). To further confirm that ml-3 is sox5, we carried out TILLING screening for sox5 mutant alleles in medaka. We successfully isolated two mutant alleles that had a single nucleotide mutation in exon 13 causing an amino acid substitution in the HMG domain of Sox5 (sox5N541S and sox5F543I, Figure 4E). The homozygous mutants of both alleles were found to exhibit increased and ectopic formation of leucophores and complete absence of xanthophores, and thus closely phenocopied the ml-3 mutants (Figures 4F–4K). We conclude that the medaka sox5 gene is mutated in the ml-3 mutant.
Figure 4. sox5 morphant and allelic TILLING mutants show the ml-3 pigment phenotype.
(A–D) Injection of sox5 MO into WT embryo generates ml-3 mutant phenocopies with increased leucophores in the dorsal trunk compared with embryos injected with the control MO which all show a WT leucophore phenotype (A). The severity of the leucophore phenotype is classified into “weak” (B) and “severe” (C). In “weak” embryos, the majority of leucophores are located along the midline of the trunk, while fewer leucophores are ectopically positioned (B). “Severe” embryos show a phenotype indistinguishable from that of ml-3 (C). In most cases, general morphology is normal, although about half of sox5 morphants combined a “severe” pigment phenotype with many ectopic leucophores with gross morphological abnormalities (data not shown). (D) Using this phenotypic classification, we scored the severity of the ml-3 morphant phenotypes at three different doses (1 ng, 2 ng and 4 ng) and with control MO (con). The number of injected embryos is indicated above the bars (n). (E) Our TILLING screen for sox5 mutant alleles identified two distinct mutations causing amino acid substitutions at N541S (blue box) or F543I (green box) respectively in the HMG domain (red boxed). (F–K) Homozygotes for the TILLING mutations (sox5N541S and sox5F543I) are compared with WT siblings (F, G). These sox5 mutants exhibit ml-3 mutant phenocopies as manifested in ectopic and excessive formation of leucophores and complete absence of xanthophores. Leucophores are shown in the darkfield image (H, J). Xanthophores are not detectable under UV light (I, K). Scale bars: 500 µm.
sox5 is expressed in neural crest and xanthophore precursors
Medaka sox5 mRNA was detected throughout embryonic and early larval stages by RT-PCR (Figure S4). We used whole-mount ISH to determine the expression pattern of sox5. In WT embryos, sox5 mRNA was detected in head and tail bud regions before the onset of somite formation (data not shown). At the 12 somite stage (41 hpf), sox5 was expressed in the dorsal neural tube and premigratory NCCs (Figures 5A, 5A′). Slightly later sox5 expression was detected in migrating NCCs ventrally on a pathway between somite and neural tube (medial pathway). At 22–24 somite stage (54–58 hpf), some scattered sox5-expressing cells were observed on lateral trunk surface (Figures 5C–5C″). These signals became detectable before the onset of gch expression, but, at later stages, overlapped with that of gch in differentiating xanthophores on lateral trunk surface; in addition, a few gch-expressing sox5-negative cells were detectable on dorsal trunk surface (Figures 5E, 5E′, 5F, 5F′). We interpret these data as indicating that sox5-expressing cells on lateral trunk surface are differentiating xanthophores (xanthophore precursors), whereas leucophores (dorsal trunk cells) do not express sox5. The expression in the xanthophore lineages gradually faded after 4 dpf and became undetectable by 5 dpf. In ml-3 embryos, the sox5 expression was not significantly altered in the dorsal neural tube, premigratory NCCs, and the NCCs migrating on the medial pathway. However, the sox5-expressing cells on the lateral trunk surface were absent in ml-3 mutants (Figures 5B, 5B′, 5D–5D″). These results indicate that sox5 is expressed in the xanthophore precursors and sox5 is required exclusively for development of xanthophores.
Figure 5. Expression pattern of medaka sox5 in WT and ml-3 mutant embryos.
(A, C, E, F) WT. (B, D) ml-3 mutant. (A′, B′, C″, D″) Transverse sections. (E, F) sox5 (blue) and gch (red). (A–E, E′) Lateral views. (F, F′) Dorsal views. (A) In WT embryos at 12 somite stage (12 s, 41 hpf), sox5 is expressed in premigratory NCCs (see also section in A′, black arrow) as well as in dorsal neural tube and CNS ranging from forebrain (fb) to hindbrain (hb), and in tailbud (tb). The boundary between neural tube and somite is indicated by dotted line. (B) In ml-3, the sox5 expression pattern is not markedly altered. In particular, when observed in section (B′), premigratory NCCs are positive for sox5. (C, C′, C″) At 24 somite stage (24 s, 58 hpf), sox5-expressing cells are found in dorsal neural tube, premigratory NCCs (arrows in C″) and migrating NCCs between neural tube and somite and lateral trunk surface (black arrowheads in C′, C″) pathways in WT. sox5-expressing cells scattered on lateral trunk surface are prominent in WT (C′, C″). (D, D′, D″) In ml-3, sox5 expressing cells are absent from lateral trunk surface (D′, D″), whereas sox5 expression remains in dorsal neural tube, premigratory NCCs (black arrows) and migrating NCCs between neural tube and somite (D″). Boxed portion in C, D are magnified in C′, D′, respectively. (A′, B′, C″, D″) Transverse histological section from embryos at the level as indicated by dotted line in A, B, C′ and D′. (E, E′) sox5-expressing cells on lateral trunk surfaces at 34 somite stage (34 s, 74 hpf) also express gch. White arrowhead represents an example of gch-positive sox5-expressing cell. (F, F′) On dorsal trunk surface, some sox5-negative gch-positive cells were detected (white arrows). Scale bars: (A, C) 200 µm; (C′) 100 µm; (A′, C″) 20 µm; (E) 50 µm.
Sox5 functions cell-autonomously in xanthophore lineage
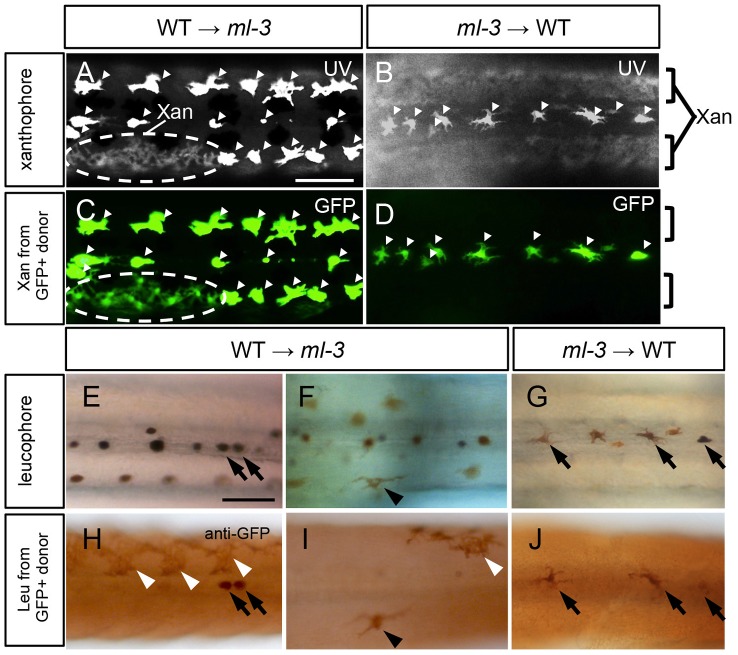
To test the hypothesis that Sox5 functions cell-autonomously in xanthophore specification, we performed cell transplantation between WT and ml-3 embryos. In this experiment, we used transgenic medaka Tg(slc2a15b:GFP), as a donor, which expresses GFP in both of differentiated xanthophores and leucophores (Figures S5A–S5C). First, WT donor cells from Tg(slc2a15b:GFP) embryos were transplanted into ml-3 host embryos. In some of these WT→ml-3 transplants, xanthophores were partially restored in patches of the body surface (15/31, Figure 6A, Table S1). All xanthophores in the transplants had GFP signals (15/15, Figure 6C), indicating that only WT donor cells were able to differentiate into xanthophores in ml-3 hosts. Next, we transplanted ml-3 donor cells with slc2a15b:GFP transgene into WT hosts. Of 25 ml-3→WT transplants tested, no embryo had GFP-positive xanthophores (0/25, Figures 6B, 6D, Table S1). We next examined whether ml-3 donor cells could develop into leucophores in WT hosts. Since GFP fluorescence could not be distinguished from auto-fluorescence in leucophores, we examined GFP expression by immunostaining with diaminobenzidine (DAB). Of 25 ml-3→WT transplants, 9 had a few leucophores expressing GFP in the dorsal midline, indicating that ml-3 donor cells can differentiate into leucophores at the correct position in WT hosts (9/25, Figures 6G, 6J, Table S1). We also tested if WT→ml-3 transplants had WT donor-derived leucophores. WT→ml-3 transplants had GFP-positive leucophores at normal and ectopic positions (2/15, Figures 6E, 6F, 6H, 6I, TableS1), suggesting that Sox5 does not control positioning of leucophores cell-autonomously, but rather that absence of xanthophores in ml-3 mutants may indirectly affect localization of leucophores. To further test this possibility, we performed cell transplantation using lf-2 mutants, which completely lack xanthophores and leucophores. Of 12 ml-3→lf-2 transplants, 3 had a few ectopic leucophores and no xanthophore (Figures S6A, S6B). This result together with that from ml-3→WT transplants supports the idea that Sox5 controls leucophore positioning indirectly by regulating specification of the xanthophore lineage. Considered together with the sox5 expression in xanthophore precursors, our data indicate that Sox5 controls specification of xanthophores cell-autonomously.
Figure 6. Cell transplantation.
(A, C, E, F, H, I) Transplants of WT donor cells into ml-3 hosts. (B, D, G, J) Transplants of ml-3 mutant donor cells into WT hosts. (A–D) Note that leucophores are fluorescent through both UV and GFP filters (white arrowheads) and xanthophores are fluorescent through UV filter but not through GFP filter. In both types of transplant experiments, xanthophores and leucophores express GFP when generated from donor cells having slc2a15b:GFP transgene, but in leucophores the GFP signal is masked by strong auto-fluorescence of these cells. However, GFP expression is readily detectable by immunostaining (see Figures S5A–S5C). (A, C) In WT→ml-3 transplants, ectopic leucophores are partially lost and instead a patch of xanthophores (marked with dotted circle) positive for UV (A) and GFP (C) fluorescence can be seen. (B, D) In ml-3→WT transplants, leucophores and xanthophores develop and are positioned normally although leucophores derived from donor cells cannot be identified because GFP fluorescence is masked by strong auto-fluorescence. No xanthophores show GFP fluorescence (compare UV image, B and GFP image, D). (E, H) In one of WT→ml-3 transplants, a few GFP positive leucophores are found along midline (black arrows). (F, I) In another WT→ml-3 transplants, leucophores are ectopically formed and positive for GFP (black arrowheads). White arrowheads represent donor-derived xanthophores detected by anti-GFP antibody. (G, J) In ml-3→WT transplants, GFP-positive leucophores develop and are positioned normally along midline (black arrows). All images are dorsal views at 7 dpf. Xan, xanthophore; Leu; leucophore. Scale bars: 100 µm.
Role of Sox5 and Pax7a in xanthophore specification
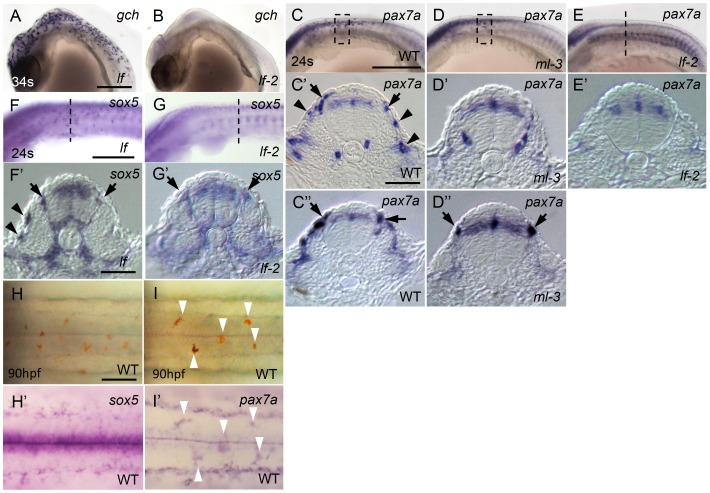
To begin to reveal the gene cascade underlying specification of xanthophore and leucophore lineages, we examined lf (slc2a15b) and lf-2 (pax7a) mutant embryos (Kimura et al, unpublished data), which each lacks discernible xanthophores and leucophores except for occasional escaper leucophores in lf-2 [30], [31]. gch expression was normal in lf but was absent in lf-2 at 34 somite stage (Figures 7A, 7B). This is consistent with our previous findings that slc2a15b is required for pigmentation of xanthophores and leucophores, whereas pax7a is required for specification of both xanthophore and leucophore lineages. We next examined the expression of pax7a (AB827303) in ml-3 and lf-2 mutants, and the expression of sox5 in lf and lf-2 mutants. Similar to sox5, pax7a was detected in the dorsal neural tube, premigratory NCCs, and migrating NCCs on the trunk surface in WT (Figures 7C–7C″). We found that pax7a was expressed in both differentiating xanthophores and leucophores, whereas sox5 expression was not detected in early leucophores (Figures 7C, 7C′, 7H, 7H′, 7I, 7I′). pax7a expression was absent in cells on the lateral trunk surface in ml-3 mutants (Figures 7D, 7D′), suggesting that the absence of pax7a on the trunk surface in ml-3 mutants reflects the absence of the xanthophore lineage. The pax7a-expressing premigratory NCCs were not altered in ml-3 mutants (Figure 7D″), whereas lf-2 mutants lack both of pax7a-expressing premigratory NCCs and differentiating xanthophores (Figures 7E, 7E′), suggesting that pax7a-positive premigratory NCCs are a shared precursor for both xanthophores and leucophores (shared xanthophore-leucophore progenitor).
Figure 7. Dual expression of sox5 and pax7a is required for xanthophore development.
(A, B) gch. (C–E, I′) pax7a. (F, G, H′) sox5. (A, F) lf mutant. (B, E, G) lf-2 mutant. (C, H, I) WT. (D) ml-3 mutant. (A–G) Lateral views. (H, I) Dorsal views. (C′, C″, D′, D″, E′, F′, G′) Transverse section. (A, B) gch expression in lf and lf-2 at 34 somite stage (34 s, 74 hpf). gch is normally expressed in lf (A), whereas in lf-2 gch-expressing cells are completely absent (B). (C–E) pax7a expression in WT, ml-3 and lf-2 at 24 somite stage (24 s, 58 hpf). In WT, pax7a is expressed in dorsal neural tube, premigratory NCCs (black arrows) in migratory NCCs between neural tube and somite and on lateral trunk surface (black arrowheads, C–C″). In ml-3, pax7a-expressing cells are lost from lateral trunk surface (D–D″). In lf-2, pax7a expression in premigratory and migratory NCCs was absent (E, E′). (F, G) sox5 expression in lf and lf-2 at 24 s. sox5 expression is normal in lf being detected in dorsal neural tube, premigratory NCCs (black arrows) and migrating NCCs between neural tube and somite and on lateral trunk surface (black arrowheads) as in WT (F, F′). In lf-2, premigratory NCC expression remains detectable, but sox5-expressing cells are lost from lateral trunk surface (G, G′). (E′–G′) The histological section from embryos at the level as indicated by dotted line in E, F and G. (C′, C″, D′, D″) Transverse sections from embryos were selected from the region boxed by dotted line in C and D. (H, I) Expression of sox5 and pax7a on dorsal trunk surface in WT at 90 hpf. (H, I) Pre-fixed WT embryos in bright field. The same embryos were processed for sox5 (H′) and pax7a (I′) ISH. Leucophores are positive for pax7a expression (I′, compare white arrowhead positions with I) but not for sox5 expression (H′). Scale bars: (A, C, E) 200 µm; (C′, E′) 20 µm; (G) 50 µm.
As expected, sox5 expression was normal in lf mutants but the sox5 expression in differentiating xanthophores on lateral trunk surface was not detected in lf-2 mutants (Figures 7F, 7F′, 7G, 7G′), suggesting sox5-expressing specified xanthophore precursors did not form in pax7a mutants. Although we expected that sox5-expressing premigratory NCCs would be absent in lf-2 (pax7a) mutants, sox5-expressing cells were observed at the premigratory position in lf-2. This is likely because sox5-positive premigratory NCCs contain other progenitor cells than the shared xanthophore-leucophore progenitors. In fact, sox5 was expressed prominently in cells on the medial pathway presumably giving rise to NCC derivatives other than xanthophores.
To test for genetic interaction between sox5 and pax7a, we generated compound mutants for sox5 and pax7a by crossing the sox5+/ml-3;pax7a+/lf-2 parents. The progeny could be categorized into three categories based on their leucophore phenotypes: WT (leucophore+, leucophores are positioned in the dorsal midline of the trunk, 128/233), ml-3 like (leucophore++, excess leucophores are ectopically located on the dorsal trunk surface, 36/233), lf-2 like (leucophore-, leucophores are completely absent, 69/233); we did not find any progeny showing phenotypes different from these three (Figures S7A–S7C). The ratio WT:ml-3 like:lf-2 like of the categorized larvae was approximately 9∶2.5∶4.8 (Figure S7D). According to Mendelian heredity, the expected ratio of WT:ml-3:lf-2:ml-3/lf-2 in the progeny is 9∶3∶3∶1. Our data suggest that sox5/pax7a double mutants showed the lf-2 (pax7a) type. Consistent with this, we confirmed that some of the lf-2 like larvae were homozygotes for the ml-3 mutation (Figure S7E). In sox5/pax7a double mutants, the shared xanthophore-leucophore progenitors did not form and therefore both xanthophores and leucophores did not develop. Our findings indicate that pax7a is required for development of the shared xanthophore-leucophore progenitors; sox5 is required for specification of xanthophore lineage from these progenitors.
Discussion
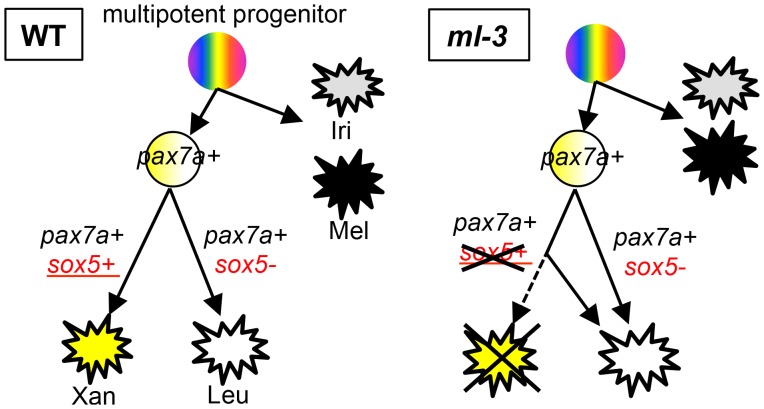
A diversity of pigment cell types in fish provides a unique opportunity to study the mechanisms controlling fate choices in multipotent NCCs. We take advantage of a novel trait in medaka, which have a fourth pigment cell type (leucophores), not seen in most fish species, to uncover the genetic basis for the greater complexity of fate choices. Our combination of studies using positional cloning, gene silencing and TILLING mutant screening have identified medaka sox5 as the gene responsible for the ml-3 mutant, which has no xanthophores but excess leucophores in the embryo/larva. Detailed observation of pigment cell phenotypes in sox5-deficient mutants revealed that the increase in leucophores and the decrease in xanthophores are sox5 dosage dependent. Our previous study revealed that pax7a is expressed in NCCs and required for development of both xanthophore and leucophore lineages (Kimura et al, unpublished data). In this study, we have demonstrated that sox5 is expressed in differentiating xanthophores and required cell-autonomously for development of the xanthophore lineage. Furthermore, the ISH analysis of pax7a in ml-3 and lf-2 mutants and the phenotype of sox5/pax7a double mutants suggest that pax7a is required for development of the shared xanthophore-leucophore progenitors from NCCs; sox5 then functions as a molecular switch for specification of xanthophores and leucophores from the progenitors, in which sox5-positive and sox5-negative cells become specified as and differentiate into xanthophores and leucophores, respectively (Figure 8).
Figure 8. Model for xanthophore and leucophore development from neural crest.
We propose that xanthophores and leucophores develop from shared progenitors. Sox5 functions to control fate specification of xanthophores in place of leucophores. In the progenitors, which are positive for pax7a, sox5-expressing cells are specified to xanthophore fate whereas pax7a-expressing sox5-negative cells give rise to leucophores. In ml-3 mutants, loss of functional Sox5 causes a failure of xanthophore specification, resulting in all progenitors becoming specified to leucophore fate. The phenotypes of lf-2 (pax7a) and ml-3 (sox5), which are independent of melanophore and iridophore lineages, suggest that xanthophore and leucophore share common progenitors. Mel, melanophore; Iri, iridophore; Xan, xanthophore; Leu, leucophore.
It might be expected that loss of Sox5 activity does not affect total cell number of xanthophores and leucophores if Sox5 determines a fate choice of leucophores versus xanthophores. Our data, however, showed that gch-expressing cells, which include both differentiating xanthophores and leucophores, significantly reduced in ml-3 mutants. Different proliferation rate of xanthophores and leucophores could explain the data. There are fewer leucophores than xanthophores in WT, suggesting that leucophores proliferate less than xanthophores (Figures 1K, 1L). Consistent with this, the pax7a-positive progenitors on the dorsal surface were not affected initially (at 18 somite stage) but later during development become significantly reduced in ml-3 compared to WT (Figures S8A–S8C).
In fish, the three or four pigment cell types are generated from multipotent NCCs, most likely through a progressive restriction mechanism. In zebrafish embryos, sox10 is required for specification of all non-ectomesenchymal NCC fates, so that loss of Sox10 function disturbs development not only of melanophores but also of xanthophores, iridophores and neural derivatives [38], [39]. Studies in zebrafish show that melanophore/iridophore progenitors are generated from sox10-positive NCCs. They initially all express mitfa, which is maintained in those that commit to melanophores fate [40]. In the case of iridophore specification, Foxd3 expression promotes iridophore cell fate by repressing mitfa expression. A previous study investigating zebrafish shady (ltk) mutants showed that the receptor tyrosine kinase leukocyte tyrosine kinase (Ltk) is necessary for iridophore specification [41]. Recently, Lopes et al. also provided evidence that sox10-positive/ltk-positive cells are likely multipotent progenitors for pigment cells, including melanophores and iridophores [41]. Some of sox10-positive/ltk-positive cells maintain ltk expression and eventually give rise to iridophores. Thus, in zebrafish, it appears that melanophore and iridophore lineages both diverge from a common progenitor cell. Consistent with these insights, ml-3 mutants have no melanophore nor iridophore phenotype, indicating that, in medaka, the xanthophore/leucophore lineage is specified independently from the melanophore/iridophore lineage.
The leucophore is poorly characterized partly because it has been described in only a few fish species including medaka, but not zebrafish [6]–[8]. Leucophores have been considered to be closely related to iridophores as the hue of both cell types is generated from light reflection due to intracellular purines, uric acid in leucophores and guanine in iridophores [42]. Furthermore, in medaka embryos, leucophores are positioned along the dorsal midline of the trunk and associated with melanophores in a similar manner to that of iridophores in zebrafish embryos [4], [33]. However, xanthophores and leucophores commonly contain pteridine-related substance and our in situ studies revealed that gch and xdh, which are associated with pteridine synthesis pathway, are expressed in both lineages. Medaka mutants lf (slc2a15b) and wl (slc2a11b) affect coloration of both xanthophore and leucophore lineages; Slc2a15b and Slc2a11b functions in the synthesis of pteridines in these pigment cells (Kimura et al, unpublished data). These biochemical traits of xanthophore and leucophore lineages imply a close relationship between xanthophore and leucophore lineages. Although purines are used for reflection of light in both leucophores and iridophores, we speculate that the purine-dependent system may have been co-opted from iridophores when leucophores diverged from the xanthophore lineage during evolution.
Sox5 belongs to the SoxD subgroup together with Sox6 and Sox13 [43]. SoxD proteins are unique in that they have no known transactivation or transrepression domain, despite their transcriptional regulatory actions. Thus, interaction with other transcription factor seems to be important for SoxD to exert its function. Several transcription factors have been suggested to interact with SoxD. In particular, Sox5 and Sox6 cooperate with Sox9 in transactivating chondrocyte-specific genes [44]. In mouse melanoblasts, Sox5 competes against Sox10 for DNA binding sequences and thus antagonistically modulates Sox10 activity to regulate transcription of mitf and dct [45]. Assuming that Sox10 is generally involved in development of all pigment cell lineages in fish, antagonistic activity of Sox5 against Sox10 may contribute to promote xanthophore lineage in medaka. Previous reports suggest that the interaction between Sox and Pax is important for pigment cell development in mammals. In melanocyte development, Pax3 cooperates with Sox10 in induction of mitf expression at early stage [22], [46] but later Pax3 competes with Sox10 to induce the expression of dct for terminal differentiation of melanocytes [47]. Pax3 and Pax7 are closely related Pax-family transcription factors with a high degree of sequence homology and a similar genomic organization. These proteins have a similar DNA-binding ability in vitro [48]. Given Pax7a functions similar to Pax3 in medaka, Pax7a may function cooperatively with Sox10 during xanthophore/leucophore lineage development in medaka. In this context, Sox5 may act antagonistically against Sox10 interacting with Pax7a within xanthophore progenitor cells in medaka. To address this issue, studies of medaka sox10 mutants and compound mutants of sox5, sox10, and pax7a are necessary. Further identification of target genes of these transcription factors and master genes for specification and differentiation of xanthophores should help us to understand the molecular mechanism by which Sox5 controls xanthophore specification.
The ectopic localization of leucophores in ml-3 implies that Sox5 controls NCC migration. However, in our cell transplantation analysis, no ml-3 mutant donor-derived leucophores were ectopically positioned in WT host embryos. In contrast, we found WT donor-derived leucophores at an ectopic position in ml-3 host embryo. We noticed that there were no xanthophores around the ectopic leucophores, whereas WT or ml-3 donor-derived leucophores were observed at the normal positions and laid medially adjacent to xanthophores. Consistent with this, in ml-3 heterozygotes, excess leucophores are formed at the normal position in the presence of reduced but numerous xanthophores. It has been suggested that the normal pigment pattern formation requires the interactions between distinct pigment cell types [4], [49], [50]. For stripe formation in adult zebrafish, the repulsive behavior between melanophores and xanthophores is suggested to be involved in segregation of the two cell types in the skin. Our findings suggest that the presence of xanthophores prevents leucophores from being positioned laterally close to xanthophores and the absence of xanthophores in ml-3 mutants allows leucophores to scatter more laterally. Supporting this idea, cell transplantation analysis by using lf-2 mutants as host embryos has shown that ml-3 donor-derived leucophores are often formed at ectopic positions in the complete absence of xanthophores. These results imply that repulsive interaction between xanthophores and leucophores likely determines the positions of the two pigment cell types in the medaka embryo.
We show that medaka Sox5 functions as a molecular switch in specification of xanthophores versus leucophores. However, it is not clear whether sox5 is used for specification of xanthophores in other fish species that have only three types of pigment cells, such as zebrafish. If so, repression of the sox5 expression in the intermediate progenitor might be linked to appearance of leucophore lineage. Future studies on the role of sox5 gene in pigment cell development of various vertebrate species should shed light on the mechanism of diversification of pigment cell development.
Materials and Methods
Ethics statement
This study was performed with the approval of Nagoya University ethics committee and in accordance with local and Japanese ethical guidelines.
Medaka strains and husbandry
The Nagoya strain of medaka fish, Oryzias latipes, having all of the normal four pigment cell types, was used as WT. ml-3, identified in Orange-red stocks (devoid of visible melanophores) by Hideo Tomita [30], was crossed with Nagoya several times to equip the mutant with melanophore prior to experimental use. HNI-I inbred strain (HNI-I/NAGOYA inbred substrain, which was obtained after brother-sister mating of HNI-I inbred strain in more than 15 generations at Nagoya University) was used for crossing in genetic mapping. lf and lf-2 have been described [51]. Fish stocks were maintained in 16-L tanks with a water circulating system (Meitosuien) at 26°C under a 14 h light/10 h dark cycle. In some experiments, embryos were treated with 0.003% phenylthiourea to block melanin synthesis.
Observation of pigment cells
Embryos and larvae were imaged on a Leica MZ APO or an AxioPlan 2 (Zeiss) microscope using an AxioCam camera (Zeiss). Xanthophores were illuminated with UV light to observe their auto-fluorescence through DAPI filter. Prior to counting melanophore number, embryos were treated with 2 mg/ml epinephrine to aggregate melanin. Significant differences were calculated using Student's t-test, one-way ANOVA or two-way ANOVA with GraphPad Prism (version 5.00 for Windows, GraphPad Software).
Whole-mount in situ hybridization (ISH)
ISH was performed as previously described [52] with modification. A digoxygenin (DIG)- or fluorescein (FITC)-labeled riboprobe was made from a cloned cDNA in pDrive (Qiagen), pCRII-TOPO (Invitrogen) or pGEM-T easy (Promega) by using SP6 or T7 polymerase after restriction enzyme digestion. The brief procedure is as follows. Fixed embryos were treated with Proteinase K (10 µg/ml) in PBS for 10 min at 30°C. Hybridization was performed at 65°C overnight. Signals were detected with alkaline-phosphate conjugated anti-DIG or anti-FITC Fab fragments (Roche) using NBT/BCIP (Roche) or FastRed (Roche) as a chromogenic substrate. For histology, the specimens were embedded in a Technovit 8100 (Heraeus Kulzer) and sectioned at 8-µm thickness.
Positional cloning
By crossing F1 generation of ml-3 and WT HNI-I hybrids, 360 F2 offspring displaying the ml-3 phenotype were collected and subjected to bulked segregant analysis with the M-marker system [53]. For further recombination analysis, polymorphic markers were isolated in reference to medaka genome database (http://medakagb.lab.nig.ac.jp/Oryzias_latipes/index.html). Detailed information on the markers used is shown in Table S2.
Antisense oligonucleotide-mediated knockdown
Antisense morpholino (MO) oligonucleotide (Gene Tools) for sox5 was designed on the splice donor site of exon 7. The MO sequence is: 5′-GATGCTACTCACTGCATCCGGGCTT-3′. The non-specific standard control MO was used as a negative control. The sequence is 5′-CCTCTTACCTCAGTTACAATTTATA-3′. MO was microinjected in one blastomere of 1-cell stage embryos.
TILLING mutant screening
TILLING screening was performed as described [54], [55]. Genomic fragment of exon 13 in sox5 was amplified with primers; 5′-AATCTTCAGTCGTTTGAGTG-3′ (forward) and 5′-TAACGCTTGCTGAGGAAC-3′ (reverse).
Generation of transgenic fish
The fosmid clone containing lf/slc2a15b region (GOLWFno17_n04) was modified (Kimura et al., unpublished data). Exon 1 of slc2a15b was replaced by GFP cDNA (slc2a15b:GFP). The fosmid construct slc2a15b:GFP (50 pg) was injected into 1-cell stage medaka embryos to establish a transgenic line Tg(slc2a15b:GFP). Detailed characterization of Tg(slc2a15b:GFP) will be published elsewhere.
Cell transplantation
Transplanting cells from donor to host was done at late blastula stage. Donors, either of WT or of ml-3, were given with the transgene slc2a15b:GFP by crossing. Donor and host blastulae were dechorionated with hatching enzyme and placed in groove on a 1.5% agarose plate in 3.5 cm plastic dish filled with balanced salt solution containing 20 U/ml penicillin and 100 µg/ml streptomycin. Approximately a hundred donor cells were transplanted with glass microcapillary. Transplants were incubated at 26°C until 7 dpf.
Immunohistochemistry
Embryos were fixed with 4% paraformaldehyde in PBS overnight at room temperature. The primary antibody (rat anti-GFP IgG, Nacalai tesque or mouse anti-HuC IgG, Santa Cruz Biotechnology) and the secondary antibody (biotin-conjugated anti-rat IgG or biotin-conjugated anti-mouse IgG, VECTOR) were used at a 1∶1000 dilution. Staining was done by using VECTASTAIN ABC kit (VECTOR) with DAB (MUTO PURE CHEMICALS) or Alexa Fluor 488 tyramide (Invitrogen). In the experiment that detects HuC-positive enteric neurons, immnostaining was performed on cryostat sections made from frozen samples mounted in O.C.T. (Sakura Finetek Japan).
Alcian blue staining
Hatching larvae were fixed with 4% paraformaldehyde overnight. The samples were washed in acid-alcohol (1% HCl, 70% ethanol) and incubated for 2 hours in 0.1% alcian blue in acid-alcohol, thereafter washed in acid-alcohol and stored in 90% glycerol.
Supporting Information
Formation of peripheral neurons and craniofacial cartilage in medaka ml-3 mutants. (A, C, E, G, I) WT. (B, D, F, H, J) ml-3 mutant. (A, B) Dorsal root ganglions (DRGs), 5 dpf. (C, D) Enteric neurons, 10 dpf. (E–J) Craniofacial cartilages, 9 dpf. (E, F) Lateral views. (G, H) Dorsal views. (I, J) Ventral views. (A, B) Immunohistochemistry with anti-HuC antibody reveals a normal segmental pattern of DRGs (white arrowheads) through the trunk in ml-3 (B) as compared with WT (A) at 5 dpf. The number of DRGs is indistinguishable between WT and ml-3 (mean±s.d., 59.2±1.9, n = 10 in WT; 59.3±2.4; n = 6 in ml-3; Student's t-test, p>0.05). (C, D) Transverse sections of the trunk are stained with anti-HuC antibody. ml-3 larva (D) shows enteric neurons around the gut whose numbers per section are comparable to those in WT (C) (mean±s.d., 8.58±0.33, n = 4 for WT; 8.91±0.27, n = 4 for ml-3; Student's t-test, p>0.05). Means of HuC-positive enteric neurons per section were calculated by counting on sequential 15 sections for each sample. (E–J) The structures of craniofacial skeleton at 9 dpf appear to be normal in ml-3 as is seen by alcian blue staining. Scale bars: (A) 100 µm; (C) 25 µm; (E) 150 µm.
(TIF)
Embryonic pigment patterns. (A, B) 4 dpf. (C, D, M, N) 7 dpf. (E–L) 5 dpf. (A, C, E, H, K, M) WT. (B, D, G, J, L, N) ml-3 homozygotes (ml-3). (F, I) ml-3 heterozygotes (+/ml-3). (A–G, K–N) Dorsal views. (H–J) Lateral views. (H–J) UV light. (A, C) In WT, leucophores first appear along the dorsal midline at 3.5–4 dpf (A, white arrowheads). By 7 dpf, leucophore become aligned on the midline associated with melanophores (C). (B, D) In ml-3 mutants, excess leucophores are already observed at 4 dpf (B, white arrowheads) and then form three lines by 7 dpf (D). (E, F, G) In ml-3 heterozygotes (F), like in WT (E), leucophores are located in the dorsal midline whereas in ml-3 homozygotes leucophores are scattered over the dorsal trunk (G). The leucophore number in ml-3 heterozygotes is significantly larger than that in WT and significantly smaller than that in ml-3 homozygotes (Figure 1K). (H, I, J) In ml-3 heterozygotes (I), like in WT (H), most xanthophores are positioned laterally to leucophores and melanophores. The xanthophore number in ml-3 heterozygotes is significantly reduced as compared with WT (Figure 1L). No xanthophores are observed on the trunk surface in ml-3 homozygotes (J). (K–N) Iridophores appear on the yolk sac (white arrows) at 5 dpf in WT (K) and ml-3 (L), and increase in number at 7 dpf (M, N). (O, P) Mean (±s.d.) counts of melanophores in DS (O) and leucophores in dorsal trunk (P) in WT (blue) and in ml-3 (red) are plotted against age. (O) Melanophore number in dorsal stripe is indistinguishable between WT and ml-3 during embryogenesis (n = 10 for each group and time point): two-way ANOVA, F(group) = 0.42, df = 1,144, p>0.05; F(group×time point) = 1.18, df = 7,144, p>0.05 and Student's t-test for each time point, **, p>0.05). (P) Leucophore number in dorsal trunk is significantly larger in ml-3 than in WT throughout embryonic stages examined (n = 10 for each group and time point): two-way ANOVA, F(group) = 3777.85, df = 1,108, p<0.0001; F(group×time point) = 47.06, df = 5,108, p<0.0001 and Student's t-test for each time point, *,p<0.0001. Xan, xanthophore; Leu; leucophore. Scale bars: (A) 250 µm; (C) 500 µm; (E) 300 µm; (H) 150 µm; (K, M) 200 µm.
(JPG)
Medaka xdh expression is restricted to leucophore and xanthophore lineages. (A, D) Dorsal views. (B, C) Lateral views. At 27 somite stage (27 s, 58 hpf), xdh is expressed in ventral head leucophores (white arrowheads) (A) and on lateral trunk surface (B). At 34 somite stage (34 s, 74 hpf), xdh expression consists of broadly scattered cells on lateral trunk surface (C). At 5 dpf, xdh mRNA is detected in differentiated leucophores (black arrows) and xanthophores (black arrowheads) (D). Scale bars: 100 µm.
(TIF)
RT-PCR of sox5. The fragment amplified contains the region from start codon to exon 5. sox5 mRNA was expressed at all stages examined. Medaka ef1α is shown as a positive control.
(TIF)
Tg(slc2a15b:GFP). (A) Live image. (B) Fluorescent image. (C) Immunostained image. (A–C) All images are at 7 dpf and dorsal views. In Tg(slc2a15b:GFP) embryos, GFP signals are observed in xanthophores on lateral trunk surfaces (black arrows) (B). Leucophores in the midline (black arrowheads, A) show strong auto-fluorescence (B). In these leucophores, GFP signals are also detected by immunostaining (C). Scale bar: 100 µm.
(TIF)
ml-3→lf-2 transplants. (A) Live image in darkfield. (B) UV image. (A,B) At 7 dpf and dorsal views. In ml-3→lf-2 transplants, some leucophores were formed at normal positions in the dorsal midline (arrowheads) and at ectopic positions (arrows, A, B), whereas no xanthophores developed (B). Dorsal midline was shown by dotted line. Scale bar: 250 µm.
(TIF)
Epistasis analysis between sox5 and pax7a. (A–C) Hatching stage (9 dpf) and dorsal views. The offspring from sox5+/ml-3 and pax7a+/lf-2 heterozygote intercross could be classified into three categories based on their leucophore phenotype; WT, leucophores positioned in the dorsal midline (A); ml-3 like, ectopic leucophores bilaterally scattered (B); and lf-2 like, leucophores completely lost (C). Numbers of each category are shown in pie graph (D). The ratio WT:ml-3 like:lf-2 like of the categorized larvae was 9∶2.5∶4.8 (approximately 9∶3∶4). RT-PCR analysis by using the primer set described in Figure 3B revealed that ml-3 homozygotes (showing just the small sized fragment resulting from loss of exon 7), were included in some of the lf-2 like embryos (red boxed, E). Scale bar: 500 µm.
(TIF)
Comparison of the numbers of pax7a-positive NCCs between WT and ml-3. (A, B) At 18 somite stage (18 s, 50 hpf), pax7a expression in NCCs is restricted to the premigratory positions in both WT (A) and ml-3 (B). (C) The pax7a-positive NCCs are counted and shown as means in WT (blue) and ml-3 (red). At 18 s, the counts have no significant difference between WT and ml-3. At 22 somite stage (22 s, 54 hpf) and 24 somite stage (24 s, 58 hpf, also see Figures 7C and 7D), the pax7a-positive cells are fewer in ml-3 than in WT. AT 18 s: WT, 8.3±2.2 (mean±s.d., n = 6); ml-3, 7.4±2.3 (n = 5) (Student's t-test, p>0.05). At 22 s: WT, 21.0±0.9 (n = 4); ml-3, 11.0±2.1 (n = 4) (Student's t-test, p<0.0001). At 24 s: WT, 27.4±4.1 (n = 10); ml-3, 13.5±1.7 (n = 8) (Student's t-test, p<0.0001). Scale bar: 200 µm.
(TIF)
The ratio of transplants' phenotypes.
(DOCX)
The list of typing markers used in positional cloning.
(DOCX)
Acknowledgments
We thank NBRP medaka for supplying lf and lf-2 lines and fosmid clone.
Funding Statement
YN was supported by grant number 5881 from the Japan Society for the Promotion of Science (JSPS) Research Fellowship (http://www.jsps.go.jp/english/index.html). HH was supported in part by grant (No. 23570077) from JSPS KAKENHI (https://www.jsps.go.jp/english/e-grants/). MH and HH were supported in part from strategic young researcher overseas visits program for vitalizing brain circulation (http://www.jsps.go.jp/english/e-zunoujunkan2/index.html). This work was supported by the National Institute for Basic Biology Cooperative Research Project (12-354 and 13-337) to HH and the National Institute for Basic Biology Priority Collaborative Research Project (12-105 and 13-103) to YT (http://www.nibb.ac.jp/en/collabo/collabo.html). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Donoghue PC, Graham A, Kelsh RN (2008) The origin and evolution of the neural crest. Bioessays 30: 530–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Le Douarin NM, Creuzet S, Couly G, Dupin E (2004) Neural crest cell plasticity and its limits. Development 131: 4637–4650. [DOI] [PubMed] [Google Scholar]
- 3. Fujii R (1993) Cytophysiology of fish chromatophores. Int Rev Cytol 143: 191–255. [Google Scholar]
- 4. Kelsh RN (2004) Genetics and evolution of pigment patterns in fish. Pigment Cell Res 17: 326–336. [DOI] [PubMed] [Google Scholar]
- 5. Kelsh RN, Brand M, Jiang YJ, Heisenberg CP, Lin S, et al. (1996) Zebrafish pigmentation mutations and the processes of neural crest development. Development 123: 369–389. [DOI] [PubMed] [Google Scholar]
- 6. Odiorne JM (1933) The Occurrence of Guanophores in Fundulus. Proc Natl Acad Sci U S A 19: 750–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fries EF (1942) White Pigmentary Effectors (Leucophores) in Killifishes. Proc Natl Acad Sci U S A 28: 396–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Takeuchi IK (1976) Electron microscopy of two types of reflecting chromatophores (iridophores and leucophores) in the guppy, Lebistes reticulatus Peters. Cell Tissue Res 173: 17–27. [DOI] [PubMed] [Google Scholar]
- 9.Hama T (1975) In: Yamamoto T, editor. Medaka (KILLFISH): Biology and Strains. Tokyo: Keigaku Publishing Company. pp. 138–153. [Google Scholar]
- 10. Oliphant LW, Hudon J (1993) Pteridines as reflecting pigments and components of reflecting organelles in vertebrates. Pigment Cell Res 6: 205–208. [DOI] [PubMed] [Google Scholar]
- 11. Braasch I, Schartl M, Volff JN (2007) Evolution of pigment synthesis pathways by gene and genome duplication in fish. BMC Evol Biol 7: 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Le Douarin NM, Dupin E (2003) Multipotentiality of the neural crest. Curr Opin Genet Dev 13: 529–536. [DOI] [PubMed] [Google Scholar]
- 13. Calloni GW, Glavieux-Pardanaud C, Le Douarin NM, Dupin E (2007) Sonic Hedgehog promotes the development of multipotent neural crest progenitors endowed with both mesenchymal and neural potentials. Proc Natl Acad Sci U S A 104: 19879–19884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Calloni GW, Le Douarin NM, Dupin E (2009) High frequency of cephalic neural crest cells shows coexistence of neurogenic, melanogenic, and osteogenic differentiation capacities. Proc Natl Acad Sci U S A 106: 8947–8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bhatt S, Diaz R, Trainor PA (2013) Signals and switches in Mammalian neural crest cell differentiation. Cold Spring Harb Perspect Biol 5 (2) pii: a008326. doi: 10.1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tassabehji M, Newton VE, Read AP (1994) Waardenburg syndrome type 2 caused by mutations in the human microphthalmia (MITF) gene. Nat Genet 8: 251–255. [DOI] [PubMed] [Google Scholar]
- 17. Mochii M, Mazaki Y, Mizuno N, Hayashi H, Eguchi G (1998) Role of Mitf in differentiation and transdifferentiation of chicken pigmented epithelial cell. Dev Biol 193: 47–62. [DOI] [PubMed] [Google Scholar]
- 18. Lister JA, Robertson CP, Lepage T, Johnson SL, Raible DW (1999) nacre encodes a zebrafish microphthalmia-related protein that regulates neural-crest-derived pigment cell fate. Development 126: 3757–3767. [DOI] [PubMed] [Google Scholar]
- 19. Hodgkinson CA, Moore KJ, Nakayama A, Steingrimsson E, Copeland NG, et al. (1993) Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell 74: 395–404. [DOI] [PubMed] [Google Scholar]
- 20. Lee M, Goodall J, Verastegui C, Ballotti R, Goding CR (2000) Direct regulation of the Microphthalmia promoter by Sox10 links Waardenburg-Shah syndrome (WS4)-associated hypopigmentation and deafness to WS2. J Biol Chem 275: 37978–37983. [DOI] [PubMed] [Google Scholar]
- 21. Elworthy S, Lister JA, Carney TJ, Raible DW, Kelsh RN (2003) Transcriptional regulation of mitfa accounts for the sox10 requirement in zebrafish melanophore development. Development 130: 2809–2818. [DOI] [PubMed] [Google Scholar]
- 22. Bondurand N, Pingault V, Goerich DE, Lemort N, Sock E, et al. (2000) Interaction among SOX10, PAX3 and MITF, three genes altered in Waardenburg syndrome. Hum Mol Genet 9: 1907–1917. [DOI] [PubMed] [Google Scholar]
- 23. Lacosta AM, Muniesa P, Ruberte J, Sarasa M, Dominguez L (2005) Novel expression patterns of Pax3/Pax7 in early trunk neural crest and its melanocyte and non-melanocyte lineages in amniote embryos. Pigment Cell Res 18: 243–251. [DOI] [PubMed] [Google Scholar]
- 24. Watanabe A, Takeda K, Ploplis B, Tachibana M (1998) Epistatic relationship between Waardenburg syndrome genes MITF and PAX3. Nat Genet 18: 283–286. [DOI] [PubMed] [Google Scholar]
- 25. Potterf SB, Furumura M, Dunn KJ, Arnheiter H, Pavan WJ (2000) Transcription factor hierarchy in Waardenburg syndrome: regulation of MITF expression by SOX10 and PAX3. Hum Genet 107: 1–6. [DOI] [PubMed] [Google Scholar]
- 26. Curran K, Raible DW, Lister JA (2009) Foxd3 controls melanophore specification in the zebrafish neural crest by regulation of Mitf. Dev Biol 332: 408–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ignatius MS, Moose HE, El-Hodiri HM, Henion PD (2008) colgate/hdac1 Repression of foxd3 expression is required to permit mitfa-dependent melanogenesis. Dev Biol 313: 568–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Thomas AJ, Erickson CA (2009) FOXD3 regulates the lineage switch between neural crest-derived glial cells and pigment cells by repressing MITF through a non-canonical mechanism. Development 136: 1849–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nitzan E, Pfaltzgraff ER, Labosky PA, Kalcheim C (2013) Neural crest and Schwann cell progenitor-derived melanocytes are two spatially segregated populations similarly regulated by Foxd3. Proc Natl Acad Sci U S A 110: 12709–12714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tomita H (1992) The lists of the mutants and strains of the medaka, common gambusia, silver crucian carp, goldfish, and golden venus fish maintained in the Laboratory of Freshwater Fish Stocks, Nagoya University. Fish Biol J MEDAKA 4: 45–47. [Google Scholar]
- 31. Kelsh RN, Inoue C, Momoi A, Kondoh H, Furutani-Seiki M, et al. (2004) The Tomita collection of medaka pigmentation mutants as a resource for understanding neural crest cell development. Mech Dev 121: 841–859. [DOI] [PubMed] [Google Scholar]
- 32.Tomita H (1975) In: Yamamoto T, editor. Medaka (KILLFISH): Biology and Strains. Tokyo: Keigaku Publishing Company. pp. 251–272. [Google Scholar]
- 33. Lynn Lamoreux M, Kelsh RN, Wakamatsu Y, Ozato K (2005) Pigment pattern formation in the medaka embryo. Pigment Cell Res 18: 64–73. [DOI] [PubMed] [Google Scholar]
- 34. Odenthal J, Rossnagel K, Haffter P, Kelsh RN, Vogelsang E, et al. (1996) Mutations affecting xanthophore pigmentation in the zebrafish, Danio rerio. Development 123: 391–398. [DOI] [PubMed] [Google Scholar]
- 35. Parichy DM, Ransom DG, Paw B, Zon LI, Johnson SL (2000) An orthologue of the kit-related gene fms is required for development of neural crest-derived xanthophores and a subpopulation of adult melanocytes in the zebrafish, Danio rerio. Development 127: 3031–3044. [DOI] [PubMed] [Google Scholar]
- 36. Pelletier I, Bally-Cuif L, Ziegler I (2001) Cloning and developmental expression of zebrafish GTP cyclohydrolase I. Mech Dev 109: 99–103. [DOI] [PubMed] [Google Scholar]
- 37. Kelsh RN, Schmid B, Eisen JS (2000) Genetic analysis of melanophore development in zebrafish embryos. Dev Biol 225: 277–293. [DOI] [PubMed] [Google Scholar]
- 38. Dutton KA, Pauliny A, Lopes SS, Elworthy S, Carney TJ, et al. (2001) Zebrafish colourless encodes sox10 and specifies non-ectomesenchymal neural crest fates. Development 128: 4113–4125. [DOI] [PubMed] [Google Scholar]
- 39. Kelsh RN, Eisen JS (2000) The zebrafish colourless gene regulates development of non-ectomesenchymal neural crest derivatives. Development 127: 515–525. [DOI] [PubMed] [Google Scholar]
- 40. Curran K, Lister JA, Kunkel GR, Prendergast A, Parichy DM, et al. (2010) Interplay between Foxd3 and Mitf regulates cell fate plasticity in the zebrafish neural crest. Dev Biol 344: 107–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lopes SS, Yang X, Muller J, Carney TJ, McAdow AR, et al. (2008) Leukocyte tyrosine kinase functions in pigment cell development. PLoS Genet 4: e1000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Obika M (1996) Morphology of chromatophores of the medaka. Fish Biol J MEDAKA 8: 21–27. [Google Scholar]
- 43. Lefebvre V (2010) The SoxD transcription factors–Sox5, Sox6, and Sox13–are key cell fate modulators. Int J Biochem Cell Biol 42: 429–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lefebvre V, Li P, de Crombrugghe B (1998) A new long form of Sox5 (L-Sox5), Sox6 and Sox9 are coexpressed in chondrogenesis and cooperatively activate the type II collagen gene. EMBO J 17: 5718–5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Stolt CC, Lommes P, Hillgartner S, Wegner M (2008) The transcription factor Sox5 modulates Sox10 function during melanocyte development. Nucleic Acids Res 36: 5427–5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Watanabe K, Takeda K, Yasumoto K, Udono T, Saito H, et al. (2002) Identification of a distal enhancer for the melanocyte-specific promoter of the MITF gene. Pigment Cell Res 15: 201–211. [DOI] [PubMed] [Google Scholar]
- 47. Lang D, Lu MM, Huang L, Engleka KA, Zhang M, et al. (2005) Pax3 functions at a nodal point in melanocyte stem cell differentiation. Nature 433: 884–887. [DOI] [PubMed] [Google Scholar]
- 48. Schafer BW, Czerny T, Bernasconi M, Genini M, Busslinger M (1994) Molecular cloning and characterization of a human PAX-7 cDNA expressed in normal and neoplastic myocytes. Nucleic Acids Res 22: 4574–4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Inaba M, Yamanaka H, Kondo S (2012) Pigment pattern formation by contact-dependent depolarization. Science 335: 677. [DOI] [PubMed] [Google Scholar]
- 50. Parichy DM (2007) Homology and the evolution of novelty during Danio adult pigment pattern development. J Exp Zool B Mol Dev Evol 308: 578–590. [DOI] [PubMed] [Google Scholar]
- 51. Wada H, Shimada A, Fukamachi S, Naruse K, Shima A (1998) Sex-Linked Inheritance of the lf Locus in the Medaka Fish (Oryzias latipes). Zoolog Sci 15: 123–126. [DOI] [PubMed] [Google Scholar]
- 52. Hashimoto H, Rebagliati M, Ahmad N, Muraoka O, Kurokawa T, et al. (2004) The Cerberus/Dan-family protein Charon is a negative regulator of Nodal signaling during left-right patterning in zebrafish. Development 131: 1741–1753. [DOI] [PubMed] [Google Scholar]
- 53. Kimura T, Jindo T, Narita T, Naruse K, Kobayashi D, et al. (2004) Large-scale isolation of ESTs from medaka embryos and its application to medaka developmental genetics. Mech Dev 121: 915–932. [DOI] [PubMed] [Google Scholar]
- 54. Ishikawa T, Kamei Y, Otozai S, Kim J, Sato A, et al. (2010) High-resolution melting curve analysis for rapid detection of mutations in a Medaka TILLING library. BMC Mol Biol 11: 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Taniguchi Y, Takeda S, Furutani-Seiki M, Kamei Y, Todo T, et al. (2006) Generation of medaka gene knockout models by target-selected mutagenesis. Genome Biol 7: R116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Formation of peripheral neurons and craniofacial cartilage in medaka ml-3 mutants. (A, C, E, G, I) WT. (B, D, F, H, J) ml-3 mutant. (A, B) Dorsal root ganglions (DRGs), 5 dpf. (C, D) Enteric neurons, 10 dpf. (E–J) Craniofacial cartilages, 9 dpf. (E, F) Lateral views. (G, H) Dorsal views. (I, J) Ventral views. (A, B) Immunohistochemistry with anti-HuC antibody reveals a normal segmental pattern of DRGs (white arrowheads) through the trunk in ml-3 (B) as compared with WT (A) at 5 dpf. The number of DRGs is indistinguishable between WT and ml-3 (mean±s.d., 59.2±1.9, n = 10 in WT; 59.3±2.4; n = 6 in ml-3; Student's t-test, p>0.05). (C, D) Transverse sections of the trunk are stained with anti-HuC antibody. ml-3 larva (D) shows enteric neurons around the gut whose numbers per section are comparable to those in WT (C) (mean±s.d., 8.58±0.33, n = 4 for WT; 8.91±0.27, n = 4 for ml-3; Student's t-test, p>0.05). Means of HuC-positive enteric neurons per section were calculated by counting on sequential 15 sections for each sample. (E–J) The structures of craniofacial skeleton at 9 dpf appear to be normal in ml-3 as is seen by alcian blue staining. Scale bars: (A) 100 µm; (C) 25 µm; (E) 150 µm.
(TIF)
Embryonic pigment patterns. (A, B) 4 dpf. (C, D, M, N) 7 dpf. (E–L) 5 dpf. (A, C, E, H, K, M) WT. (B, D, G, J, L, N) ml-3 homozygotes (ml-3). (F, I) ml-3 heterozygotes (+/ml-3). (A–G, K–N) Dorsal views. (H–J) Lateral views. (H–J) UV light. (A, C) In WT, leucophores first appear along the dorsal midline at 3.5–4 dpf (A, white arrowheads). By 7 dpf, leucophore become aligned on the midline associated with melanophores (C). (B, D) In ml-3 mutants, excess leucophores are already observed at 4 dpf (B, white arrowheads) and then form three lines by 7 dpf (D). (E, F, G) In ml-3 heterozygotes (F), like in WT (E), leucophores are located in the dorsal midline whereas in ml-3 homozygotes leucophores are scattered over the dorsal trunk (G). The leucophore number in ml-3 heterozygotes is significantly larger than that in WT and significantly smaller than that in ml-3 homozygotes (Figure 1K). (H, I, J) In ml-3 heterozygotes (I), like in WT (H), most xanthophores are positioned laterally to leucophores and melanophores. The xanthophore number in ml-3 heterozygotes is significantly reduced as compared with WT (Figure 1L). No xanthophores are observed on the trunk surface in ml-3 homozygotes (J). (K–N) Iridophores appear on the yolk sac (white arrows) at 5 dpf in WT (K) and ml-3 (L), and increase in number at 7 dpf (M, N). (O, P) Mean (±s.d.) counts of melanophores in DS (O) and leucophores in dorsal trunk (P) in WT (blue) and in ml-3 (red) are plotted against age. (O) Melanophore number in dorsal stripe is indistinguishable between WT and ml-3 during embryogenesis (n = 10 for each group and time point): two-way ANOVA, F(group) = 0.42, df = 1,144, p>0.05; F(group×time point) = 1.18, df = 7,144, p>0.05 and Student's t-test for each time point, **, p>0.05). (P) Leucophore number in dorsal trunk is significantly larger in ml-3 than in WT throughout embryonic stages examined (n = 10 for each group and time point): two-way ANOVA, F(group) = 3777.85, df = 1,108, p<0.0001; F(group×time point) = 47.06, df = 5,108, p<0.0001 and Student's t-test for each time point, *,p<0.0001. Xan, xanthophore; Leu; leucophore. Scale bars: (A) 250 µm; (C) 500 µm; (E) 300 µm; (H) 150 µm; (K, M) 200 µm.
(JPG)
Medaka xdh expression is restricted to leucophore and xanthophore lineages. (A, D) Dorsal views. (B, C) Lateral views. At 27 somite stage (27 s, 58 hpf), xdh is expressed in ventral head leucophores (white arrowheads) (A) and on lateral trunk surface (B). At 34 somite stage (34 s, 74 hpf), xdh expression consists of broadly scattered cells on lateral trunk surface (C). At 5 dpf, xdh mRNA is detected in differentiated leucophores (black arrows) and xanthophores (black arrowheads) (D). Scale bars: 100 µm.
(TIF)
RT-PCR of sox5. The fragment amplified contains the region from start codon to exon 5. sox5 mRNA was expressed at all stages examined. Medaka ef1α is shown as a positive control.
(TIF)
Tg(slc2a15b:GFP). (A) Live image. (B) Fluorescent image. (C) Immunostained image. (A–C) All images are at 7 dpf and dorsal views. In Tg(slc2a15b:GFP) embryos, GFP signals are observed in xanthophores on lateral trunk surfaces (black arrows) (B). Leucophores in the midline (black arrowheads, A) show strong auto-fluorescence (B). In these leucophores, GFP signals are also detected by immunostaining (C). Scale bar: 100 µm.
(TIF)
ml-3→lf-2 transplants. (A) Live image in darkfield. (B) UV image. (A,B) At 7 dpf and dorsal views. In ml-3→lf-2 transplants, some leucophores were formed at normal positions in the dorsal midline (arrowheads) and at ectopic positions (arrows, A, B), whereas no xanthophores developed (B). Dorsal midline was shown by dotted line. Scale bar: 250 µm.
(TIF)
Epistasis analysis between sox5 and pax7a. (A–C) Hatching stage (9 dpf) and dorsal views. The offspring from sox5+/ml-3 and pax7a+/lf-2 heterozygote intercross could be classified into three categories based on their leucophore phenotype; WT, leucophores positioned in the dorsal midline (A); ml-3 like, ectopic leucophores bilaterally scattered (B); and lf-2 like, leucophores completely lost (C). Numbers of each category are shown in pie graph (D). The ratio WT:ml-3 like:lf-2 like of the categorized larvae was 9∶2.5∶4.8 (approximately 9∶3∶4). RT-PCR analysis by using the primer set described in Figure 3B revealed that ml-3 homozygotes (showing just the small sized fragment resulting from loss of exon 7), were included in some of the lf-2 like embryos (red boxed, E). Scale bar: 500 µm.
(TIF)
Comparison of the numbers of pax7a-positive NCCs between WT and ml-3. (A, B) At 18 somite stage (18 s, 50 hpf), pax7a expression in NCCs is restricted to the premigratory positions in both WT (A) and ml-3 (B). (C) The pax7a-positive NCCs are counted and shown as means in WT (blue) and ml-3 (red). At 18 s, the counts have no significant difference between WT and ml-3. At 22 somite stage (22 s, 54 hpf) and 24 somite stage (24 s, 58 hpf, also see Figures 7C and 7D), the pax7a-positive cells are fewer in ml-3 than in WT. AT 18 s: WT, 8.3±2.2 (mean±s.d., n = 6); ml-3, 7.4±2.3 (n = 5) (Student's t-test, p>0.05). At 22 s: WT, 21.0±0.9 (n = 4); ml-3, 11.0±2.1 (n = 4) (Student's t-test, p<0.0001). At 24 s: WT, 27.4±4.1 (n = 10); ml-3, 13.5±1.7 (n = 8) (Student's t-test, p<0.0001). Scale bar: 200 µm.
(TIF)
The ratio of transplants' phenotypes.
(DOCX)
The list of typing markers used in positional cloning.
(DOCX)