Abstract
HIV-1 and other retroviruses occasionally undergo hypermutation, characterized by a high rate of G-to-A substitution. Recently, the human apolipoprotein B mRNA-editing, enzyme-catalytic, polypeptide-like 3G (APOBEC3G), first identified as CEM15, was shown to be packaged into retroviral virions and to deaminate deoxycytidine to deoxyuridine in newly synthesized viral minus-strand DNA, thereby inducing G-to-A hypermutation. This innate mechanism of resistance to retroviral infection is counteracted by the HIV-1 viral infectivity factor (Vif), which protects the virus by preventing the incorporation of APOBEC3G into virions by rapidly inducing its ubiquitination and proteasomal degradation. To gain insights into the mechanism by which Vif protects HIV-1 from APOBEC3G, we substituted several amino acids in human APOBEC3G with equivalent residues in simian APOBEC3Gs that are resistant to HIV-1 Vif and determined the effects of the mutations on HIV-1 replication in the presence and absence of Vif. We found that a single amino acid substitution mutant of human APOBEC3G (D128K) can interact with HIV-1 Vif but is not depleted from cells; thus, it inhibits HIV-1 replication in an HIV-1 Vif-resistant manner. Interestingly, rhesus macaque simian immunodeficiency virus 239 or HIV-2 Vif coexpression depleted the intracellular steady state levels of the D128K mutant and abrogated its antiviral activity, indicating that it can be a substrate for the proteasomal pathway. The HIV-1 Vif-resistant mutant APOBEC3G could provide a gene therapy approach to combat HIV-1 infection.
It has been observed for several years that genomes of HIV-1, other retroviruses, and hepatitis B viruses are occasionally subjected to a very high rate of G-to-A substitutions, a phenomenon named hypermutation (1–3). Early on, it was hypothesized that G-to-A hypermutation occurs through the action of an error-prone reverse transcriptase (2) or through the mutagenic effects of localized biased deoxynucleotide pool imbalances (4). However, recent studies clearly indicate that G-to-A hypermutation results from the action of a host cell cytidine deaminase (5–8).
Efforts to elucidate the role of HIV-1-encoded viral infectivity factor (Vif) (9, 10) led to the discovery of the human apolipoprotein B mRNA-editing, enzyme-catalytic, polypeptide-like 3G (APOBEC3G), first identified as CEM15 (11). HIV-1 genomes that do not express Vif (ΔVif) fail to replicate in primary T cells, macrophages, and some “nonpermissive” T cell lines (12, 13). Other “permissive” T cell lines, such as 293T cells, can support the replication of ΔVif HIV-1 genomes (14). ΔVif HIV-1 virions produced in nonpermissive cell lines are apparently normal with respect to their viral RNA and protein components (15, 16). Heterokaryons generated from cell-to-cell fusions between nonpermissive and permissive cells are nonpermissive, leading to the idea that a host cell factor inhibited replication of HIV-1 and that Vif was required to overcome its inhibitory effects. In a groundbreaking study, Sheehy et al. (11) used a complementation strategy to identify a cDNA (CEM15) that was responsible for the nonpermissive phenotype. CEM15 was shown to encode APOBEC3G, a member of a family of cytidine deaminases that convert cytidines to uridines in mRNA or DNA.
The cytidine deminase activity of APOBEC3G immediately suggested a mechanism for G-to-A hypermutation; deamination of cytidines to uridines in retroviral minus-strand DNA followed by copying of the uridines with the complementary adenines would result in G-to-A substitutions. Indeed, several recent reports have demonstrated that expression of APOBEC3G in virus producer cells but not target cells results in G-to-A hypermutation of the HIV-1 genomes in the absence of Vif (5–8).
The mechanism by which HIV-1 Vif overcomes the inhibitory effects of A POBEC3G is currently being elucidated. APOBEC3G has been shown to be packaged into retroviruses produced from nonpermissive cells and permissive cells that are modified to express APOBEC3G (7, 8, 11, 17). Several recent studies indicate that the intracellular steady-state levels of APOBEC3G are reduced in the presence of HIV-1 Vif (18–22). It was recently shown that HIV-1 Vif interacts with APOBEC3G and induces rapid degradation of APOBEC3G through the proteasomal pathway by recruiting cellular factors Cul5, elongins B and C, and Rbx1 to form an Skp1-cullin-F-box-like complex (19). However, in other studies, pulse–chase analyses did not indicate an increased rate of degradation of APOBEC3G in the presence of HIV-1 Vif (17, 23). It has also been reported that Vif reduces the intracellular levels of APOBEC3G by influencing its translation (20).
Mariani et al. (17) recently determined the properties of APOBEC3Gs derived from African green monkey (AGM), rhesus macaque (MAC), and mouse. The results of these studies indicated that these APOBEC3G proteins were able to inhibit HIV-1 replication and thus exhibited an HIV-1 Vif-resistant phenotype. To elucidate the mechanism by which the AGM and MAC APOBEC3Gs are resistant to HIV-1 Vif, we performed extensive mutational analysis of human APOBEC3G. Our results indicate that a single amino acid substitution in human APOBEC3G can confer the Vif-resistant phenotype.
Materials and Methods
Plasmids and Mutagenesis. HIV-1-based vector pHDV-EGFP (HIV-1-derived vector-virus enhanced GFP) was kindly provided by Derya Unutmaz (Vanderbilt University Medical Center, Nashville, TN) (24); pHCMV (human cytomegalovirus)-G expresses the vesiculostomatitis virus envelope glycoprotein G (25). pC-Help is an HIV-1 helper construct that lacks several cis-acting elements needed for viral replication, including the packaging signal and primer-binding site; it expresses all of the viral proteins except Nef and Env (14). pC-HelpΔVif is identical to pC-Help except that the Vif ORF has been disrupted with a deletion. Most of the amino acid substitution mutations were introduced into the pcDNA-APO3G either by using the QuikChange MultiSite-Directed Mutagenesis Kit (Stratagene) or by PCR-based mutagenesis. The presence of the desired mutations and the absence of undesired mutations were verified by DNA sequencing (Science Applications International, Frederick, MD). We constructed simian immunodeficiency virus (SIV)mac239 (26) and HIV-2 (27) and SIVagm (28) Vif expression constructs by PCR amplification of the Vif ORFs from the respective proviral constructs and by cloning the PCR products into pCR 3.1 (Invitrogen). The structure of the Vif expression constructs was verified by DNA sequencing (Science Applications International).
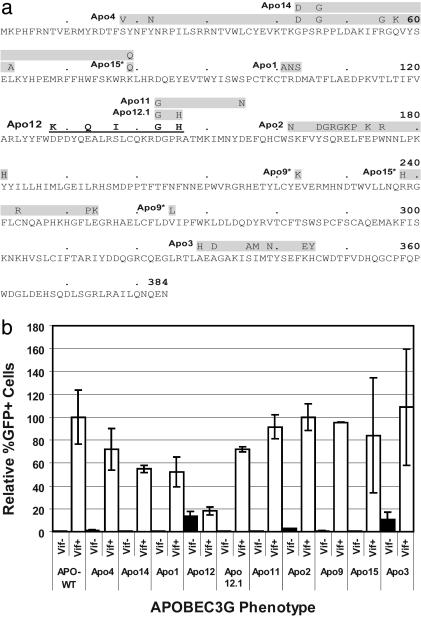
Mutations. Starting near the N-terminal end of APOBEC3G and proceeding toward the C-terminal end, the Apo4 mutant contained substitutions S18V, Y22N, G43D, R46G, R55G, Q57K, and L62A; the Apo14 mutant contained substitutions G43D, R46G, and K79Q; the Apo1 mutant contained substitutions T101A, R102N, and D103S; the Apo12 mutant contained substitutions D128K, E133Q, S137I, D143G, and R146H; the Apo12.1 mutant contained substitutions D143G and R146H; the Apo11 mutant contained substitutions D143G and D155N; the Apo2 mutant contained substitutions S162N, Y166D, S167G, Q168R, R169G, E170K, L171P, E173K, W175R, and Y181H; the Apo9 mutant contained substitutions E223K and V265L; the Apo15 mutant contained substitutions K79Q, R238H, C243R, L253P, and E254K; and the Apo3 mutant contained substitutions A329H, A331D, S336A I337M, T339N, K344E, and H345Y.
Transfections, Infections, and Flow Cytometry Analysis. We transfected 293T cells by using a CalPhos Mammalian Transfection Kit (BD Biosciences); 36–48 h later, we harvested the virus. The p24 capsid amounts in the culture supernatants were determined by ELISA (Perkin–Elmer). 293T cells were infected with virus that was equivalent to 100 ng of p24 capsid. The infected cells were analyzed by flow cytometry (FACScan, Becton Dickinson) for green fluorescence 36–48 h after infection, and the results were analyzed with cellquest software (Becton Dickinson). To determine the effects of wild-type or mutant APOBEC3Gs on HIV-1 replication in the absence of Vif (Vif–bar graphs in Fig. 1b; see also Fig. 3 a–c) the pHDV-EGFP, pC-HelpΔVif, pHCMV-G, and pcDNA-APO3G or mutants of pcDNA-APO3G were cotransfected by using 20:15:4:4 μg of DNA respectively. The molar ratios of pHDV-EGFP, pC-HelpΔVif, pHCMV-G, and the pcDNA-APO3G or APOBEC3G mutant plasmids were ≈1:1:0.4:0.4, respectively. To determine the effects of wild-type or mutant APOBEC3Gs on HIV-1 replication in the presence of Vif (Vif+ bar graphs in Fig. 1b; see also Fig. 3 a–c) the pHDV-EGFP, pC-Help, pHCMV-G, and pcDNA-APO3G or mutants of pcDNA-APO3G were cotransfected by using 20:15:4:4 μg of DNA, respectively. The molar ratios of pHDV-EGFP, pC-Help, and the pcDNA-APO3G wild-type or mutant plasmids were ≈1:1:0.4:0.4, respectively. To determine whether the Apo12 and D128K mutants are dominant over the wild-type APOBEC3G, pHDV-EGFP, pC-Help, and pHCMV-G plasmids were cotransfected with pcDNA-APO3G and/or mutant APOBEC3G plasmids (Apo12 or D128K). The total amount of the APOBEC3G plasmids was 4 μg in each experiment, and the molar ratio of pHDV-EGFP, pC-Help, pHCMV-G, and APOBEC3G plasmids was 1:1:0.4:0.4, respectively.
Fig. 1.
Effects of mutations in human APOBEC3G on inhibition of HIV-1 replication in the absence and presence of HIV-1 Vif. (a) Ten mutants containing clusters of amino acid substitutions, starting at the N-terminal end of APOBEC3G, are labeled Apo4, Apo14, Apo15, Apo1, Apo12, Apo12.1, Apo11, Apo2, Apo9, and Apo3 (the amino acid substitutions are listed in Materials and Methods). The clusters of substitution mutations are shown above the sequence in shaded boxes. The HIV-1 Vif-resistant Apo12 mutant is shown in bold and underlined letters. An asterisk next to Apo9 and Apo15 indicates discontinuous sets of mutations. (b) Flow cytometry analysis of the effects of mutations in APOBEC3G on inhibition of HIV-1 replication in the absence (Vif–) and presence (Vif+) of HIV-1 Vif. The proportion of GFP+ cells after infection with HDV-EGFP in the presence of wild-type APOBEC3G and HIV-1 Vif was set to 100%. Error bars represent the SEM of eight experiments for the Apo12 mutant and two experiments for the other mutants.
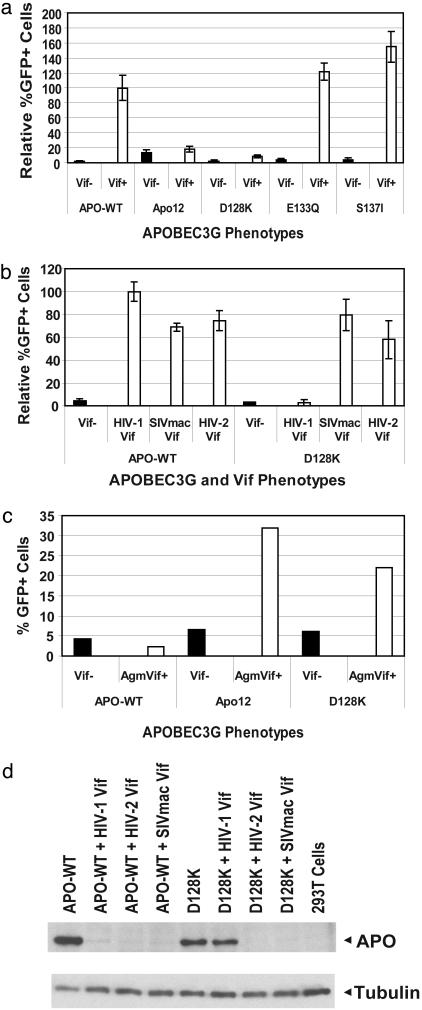
Fig. 3.
A single amino acid substitution renders human APOBEC3G resistant to HIV-1 Vif. (a) Flow cytometry analysis of the effects of amino acid substitutions in APOBEC3G on sensitivity to HIV-1 Vif. The relative proportion of GFP+ cells generated by infection with HDV-EGFP virion in the presence of wild-type or mutant APOBEC3G are shown. The proportion of GFP+ cells generated in the presence of wild-type APOBEC3G in the presence of HIV-1 Vif was set to 100%. The effects of the Apo12 mutant and the single amino acid substitution mutants (D128K, E133Q, and S137I) on viral replication were compared in the absence (Vif–) or presence (Vif+) of HIV-1 Vif. Error bars represent the SEM of two to seven experiments. (b) Sensitivity of wild-type and D128K mutant APOBEC3G to HIV-1, SIVmac239 (SIVmac), and HIV-2 Vif. The proportion of GFP+ cells generated in the presence of wild-type APOBEC3G in the presence of HIV-1 Vif was set to 100%. Error bars represent the SEM of two experiments. (c) Sensitivity of wild-type, Apo12, and D128K mutants of APOBEC3G to SIVagm Vif. The percent of GFP+ cells are shown in the absence of Vif (black bars) and in the presence of SIVagm Vif (white bars). (d) Western blot analysis of the steady-state levels of wild-type APOBEC3G and D128K mutant of APOBEC3G in the absence of any Vif or presence of HIV-1, SIVmac239 (SIVmac), and HIV-2 Vif proteins. The APOBEC3G proteins were detected by using an anti-myc tag antibody, and the tubulin proteins were detected with an anti-tubulin antibody to normalize the amounts of cell lysates analyzed.
Coimmunoprecipitation and Western Blotting Analysis. For coimmunoprecipitation, The anti-c-Myc antibody (Sigma–Aldrich) was coupled to paramagnetic beads according to manufacturer's instructions (Dynal Biotech, Oslo). We cotransfected 293T cells with APOBEC3G expressing plasmids and either pC-Help or pC-HelpΔVif. About 36 h after transfection, 2 × 106 cells were harvested, washed twice with ice-cold PBS, and lysed in 1 ml of cell extraction buffer (20 mM Tris·Cl, pH 8.0/137 mM NaCl/1 mM EDTA/1 mM NaVO3/10% glycerol/1% Triton X-100) and protease inhibitor mixture (Roche). Cell extracts were adjusted to equivalent protein concentration by using Bradford reagent (Bio-Rad), and equal aliquots were then used for coimmunoprecipitation and Western blotting analysis. The cell extracts were then centrifuged at 1,500 × g for 4 min, and the supernatants were incubated with anti-c-Myc antibody-conjugated (Sigma–Aldrich) paramagnetic beads (Dynal Biotech) for 3 h in slow rotation on RKDynal rotor (Dynal Biotech) at 4°C. After incubation, the paramagnetic beads were washed three times with 50 mM Tris·HCl, pH 7.5/500 mM LiCl/1 mM NaVO3/0.5% Triton X-100, three times with 50 mM Tris·HCl, pH 7.5/500 mM LiCl/1 mM NaVO3, and once with 1 mM NaVO3. The bound proteins were eluted from the beads by heating to 90°C for 5 min in SDS/PAGE loading buffer. For cell lysis, 2 × 106 cells were harvested, washed with ice-cold PBS 36 h after transfection, lysed in 1× SDS/PAGE loading buffer, and heated to 90°C for 5 min. For Western blot analysis, the myc epitope-tagged APOBEC3G proteins were detected by using the anti-c-Myc antibody (Sigma–Aldrich), the tubulin protein was detected by using the anti-tubulin antibody (Sigma–Aldrich), and the HIV-1 Vif protein was detected by using anti-HIV-1 HXB2 Vif antiserum (catalog no. 2221, Dana Gabuzda, Dana–Farber Cancer Institute) obtained through the AIDS Reagents and Reference Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Results
Mutational Analysis of Human APOBEC3G. Mariani et al. (17) recently reported that the HIV-1 Vif can overcome the inhibitory effects of human and chimpanzee APOBEC3Gs but not the homologous AGM or MAC APOBEC3Gs. One hypothesis suggested by this observation was that the AGM or MAC APOBEC3Gs do not interact with HIV-1 Vif and as a result are not degraded; alternatively, the HIV-1 Vif could interact with the AGM and MAC APOBEC3Gs but fail to induce its ubiquitination and/or proteasomal degradation. To determine the mechanism by which the AGM and MAC APOBEC3Gs are resistant to HIV-1 Vif, we generated 10 human APOBEC3G mutants in which clusters of amino acids were substituted with equivalent residues in AGM and MAC APOBEC3G (Fig. 1a). We targeted 40 amino acid residues that were the same in the human and the chimpanzee APOBEC3Gs but were substituted with identical residues in both AGM and MAC APOBEC3Gs.
Effects of Mutations in Human APOBEC3G on HIV-1 Replication. To determine the effects of mutations in human APOBEC3G on HIV-1 replication, we performed cotransfections with pHDV-EGFP (24), pC-Help-ΔVif, pHCMV-G (25), and wild-type or mutated APOBEC3G plasmids (Fig. 1b). pHDV-EGFP contains the cis-acting elements needed for packaging and replication and expresses only the Gag-Pol, Tat, and Rev viral proteins; pC-Help-ΔVif does not express Vif, Env, or packagable viral RNA but expresses all other viral proteins needed to complete one cycle of replication. pHCMV-G expresses the vesicular stomatitis virus envelope glycoprotein G, which can pseudotype HIV-1 vectors and produce infectious virion. Infection of 293T cells with HDV-EGFP results in a single cycle of replication and GFP expression, which can be detected by flow cytometry. The effects of wild-type and mutated APOBEC3G on replication of pHDV-EGFP are compared to the wild type APOBEC3G (Vif+ set to 100%) and summarized in Fig. 1b (bar graphs labeled Vif–). In the presence of wild-type APOBEC3G and HIV-1 Vif, an average of 30% of the pHDV-EGFP infected cells expressed GFP as determined by flow-cytometry (data not shown). In contrast, cotransfection with wild-type APOBEC3G resulted in severe reductions in GFP-positive cells to <1% in the absence of HIV-1 Vif expression (data not shown). The results indicated that all of the mutated APOBEC3G plasmids expressed an enzymatically active deoxycytidine deaminase that, in the absence of Vif, was incorporated into the HDV-EGFP virion, resulting in G-to-A hypermutation and diminished GFP expression.
To determine the effects of the mutations in APOBEC3G on HIV-1 replication in the presence of HIV-1 Vif, we performed cotransfections with pHDV-EGFP, pC-Help (14), pHCMV-G, and wild-type or mutated pc-DNA-APO3G. pC-Help is similar to pC-Help-ΔVif but also expresses HIV-1 Vif. The results obtained from these cotransfections are shown in Fig. 1b (bar graphs labeled Vif+). Unlike the cotransfections in which HIV-1 Vif was absent, expression of wild-type APOBEC3G did not diminish GFP expression in infected cells. The result indicated that the HIV-1 Vif expressed from pC-Help was able to protect HDV-EGFP replication from wild-type APOBEC3G. Similarly, pC-Help cotransfection and the resulting HIV-1 Vif expression protected HDV-EGFP replication from most of the mutated APOBEC3G proteins; the relative percent of GFP+ cells obtained for wild-type and mutant APOBEC3Gs were not significantly different (two sample t tests; P > 0.1). Thus, most of these mutant APOBEC3Gs did not influence the ability of HIV-1 Vif to counteract their inhibitory effects on HIV-1 replication. In sharp contrast, the Apo12 mutant inhibited the replication of HDV-EGFP, indicating that its antiviral activity was resistant to HIV-1 Vif (P < 0.03).
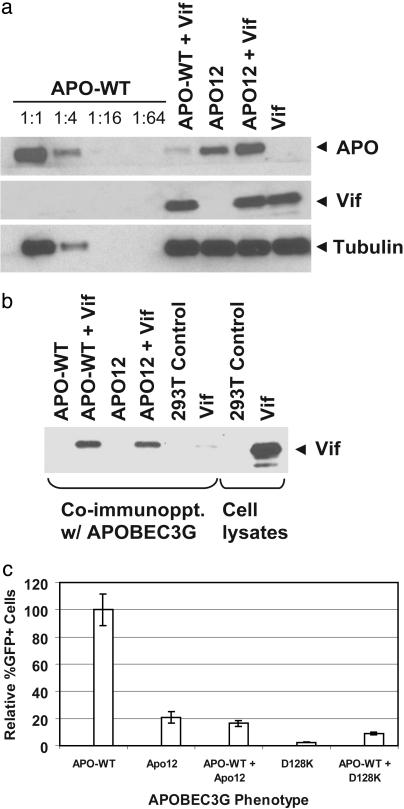
HIV-1 Vif Does Not Deplete Intracellular Steady-State Levels of HIV-1 Vif-Resistant Mutant APOBEC3G. Based on recent studies indicating that HIV-1 Vif abrogates the antiretroviral effects of APOBEC3G by inducing its degradation through the proteasomal pathway (18, 19, 21, 22), we hypothesized that the intracellular steady-state levels of the Apo12 Vif-resistant mutant will not be depleted in the presence of HIV-1 Vif. To test this hypothesis, we determined the intracellular steady-state levels of wild-type and Apo12 mutant APOBEC3G proteins by Western blot detection of a C-terminal myc tag in the presence and absence of HIV-1 Vif (Fig. 2a). The steady-state levels of wild-type APOBEC3G protein but not the Apo12 mutant APOBEC3G protein were significantly depleted in the presence of HIV-1 Vif; this result was consistent with the view that the HIV-1 Vif-resistant Apo12 mutant was not degraded in the presence of HIV-1 Vif. To quantify the extent to which steady-state levels of wild-type APOBEC3G were reduced in the presence of HIV-1 Vif, extracts of cells expressing wild-type APOBEC3G in the absence of HIV-1 Vif were serially diluted, and the intensities of the bands generated were compared to the band generated in the presence of HIV-1 Vif. The results indicated that the steady-state levels of wild-type APOBEC3G were reduced by 4- to 16-fold.
Fig. 2.
HIV-1 Vif reduces intracellular steady-state levels of wild-type APOBEC3G but not the Vif-resistant Apo12 mutant. (a) Western blotting quantitative analysis of wild-type APOBEC3G degradation in the presence of Vif. Serial dilution of cell lysates transfected with wild-type APOBEC3G in the absence of HIV-1 Vif (APO-WT) are compared to undiluted lysate from cells transfected with wild-type APOBEC3G and pC-Help (APO-WT + Vif). The APOBEC3G proteins were identified by using an anti-myc tag antibody, and the same lysates were analyzed by using an anti-tubulin antibody to ensure that equivalent aliquots were loaded onto gels. The cell lysates after transfection with wild type APOBEC3G and the Apo12 mutant were also analyzed for the presence of Vif by using an anti-Vif antibody. (b) Wild-type and Apo12 mutant APOBEC3G interact with HIV-1 Vif as determined by coimmunoprecipitation. An anti-myc antibody attached to magnetic beads was used to immunoprecipitate the wild-type APOBEC3G and the Apo12 mutant proteins from cell lysates. The cellular proteins that were coimmunoprecipitated were analyzed for the presence of Vif by using an anti-Vif antibody. (c) Flow cytometry analysis of the effect of wild-type APOBEC3G on inhibition of HIV-1 replication by the Apo12 and D128K mutants. Four micrograms of each APOBEC3G plasmid was cotransfected with pHDV-EGFP, C-Help, and HCMV-G for bar graphs labeled APO-WT, Apo12, and D128K, respectively. Two micrograms of each APOBEC3G plasmid was cotransfected with pHDV-EGFP, C-Help, and HCMV-G for bar graphs labeled APO-WT, APO-WT + Apo12, and APO-WT + D128K, respectively. The proportion of GFP+ cells after infection with HDV-EGFP in the presence of wild-type APOBEC3G and HIV-1 Vif was set to 100%.
We considered the possibility that the Apo12 mutant increased the turnover rate of HIV-1 Vif and, thus, was not depleted from cells. To test this hypothesis, we performed Western blotting analysis by using an anti-Vif antiserum (Fig. 2a). The results suggested that the intracellular steady-state levels of HIV-1 Vif were equivalent in the presence of wild-type APOBEC3G and Apo12 mutant proteins.
HIV-1 Vif-Resistant Human APOBEC3G Mutant Binds to HIV-1 Vif. We sought to determine whether the Apo12 mutant fails to bind to HIV-1 Vif and as a result is not depleted from cells. To address this issue, we performed coimmunoprecipitation analysis with the anti-myc antibody (Fig. 2b). All proteins that were coimmunoprecipitated with APOBEC3G were subjected to Western blotting analysis with the anti-Vif antiserum. Analysis of coimmunoprecipitated proteins from cells that were transfected with pC-Help alone indicated that a trace amount of HIV-1 Vif was nonspecifically coimmunoprecipitated in the absence of APOBEC3G; in contrast, significantly greater amounts of HIV-1 Vif coimmunoprecipitated with both the wild-type and the Apo12 mutant APOBEC3G proteins. Furthermore, the amounts of Vif that coimmunoprecipitated with the wild-type and the Apo12 mutant of APOBEC3G appeared to be equivalent. These results indicated that the Apo12 mutant APOBEC3G retained the ability to interact with HIV-1 Vif.
HIV-1 Vif-Resistant Mutant APOBEC3G Dominantly Inhibits HIV-1. Next, we wished to ascertain whether the Apo12 mutant APOBEC3G is dominant over the wild-type APOBEC3G protein and as a result is not depleted in the presence of wild-type APOBEC3G; such a mutant could be useful in developing a gene therapy approach to inhibit HIV-1 replication. Alternatively, potential interactions between the wild-type APOBEC3G and the Apo12 mutant could result in the depletion of the Apo12 mutant from cells in the presence of Vif when the wild-type APOBEC3G is coexpressed. To determine whether the Apo12 mutant dominantly inhibited HDV-EGFP replication in the presence of wild-type APOBEC3G, we cotransfected pHDV-EGFP, pC-Help, pHCMV-G, and pc-DNA-APO3G or Apo12 mutant plasmid (Fig. 2c). As expected, the presence of the Apo12 mutant APOBEC3G protein but not the wild-type APOBEC3G protein resulted in inhibition of HDV-EGFP replication when HIV-1 Vif was coexpressed. When both the wild-type and Apo12 mutant APOBEC3G proteins were coexpressed in the presence of HIV-1 Vif, the replication of HDV-EGFP was inhibited. This result indicated that the Apo12 mutant dominantly inhibited HIV-1 replication in a Vif-resistant manner in the presence of the wild-type APOBEC3G.
A Single Amino Acid Substitution Confers the HIV-1 Vif-Resistant Phenotype. The HIV-1 Vif-resistant Apo12 mutant contained five single amino acid substitutions (D128K, E133Q, S137I, D143G, and R146H). Because the Vif-sensitive Apo12.1 mutant contained the D143G and R146H substitutions, they were unlikely to be responsible for the Vif-resistant phenotype (Fig. 1). To determine which mutations in the Apo12 mutant resulted in the Vif-resistant phenotype, we generated mutants containing the single substitutions D128K, E133Q, and S137I and determined their ability to inhibit HDV-EGFP replication in the absence and presence of HIV-1 Vif (Fig. 3a). The results indicated that the D128K mutant inhibited HDV-EGFP replication at least as efficiently as the Apo12 mutant and therefore exhibited the HIV-1 Vif-resistant phenotype (P < 0.03). In contrast, the E133Q and S137I substitutions did not exhibit any ability to inhibit HDV-EGFP replication in the presence of HIV-1 Vif. Similar to the Apo12 mutant, the D128K mutant APOBEC3G also dominantly inhibited HDV-EGFP replication in the presence of wild-type APOBEC3G (Fig. 2c).
The D128K Mutant of Human APOBEC3G Is Depleted from Cells in the Presence of SIVmac239, HIV-2, and SIVagm Vif. One hypothesis suggested by our results was that the D128K mutant of human APOBEC3G is not depleted from cells in the presence of HIV-1 Vif because it is not a substrate for the ubiquitin/proteosomal degradation pathway. To determine whether the D128K mutation rendered APOBEC3G resistant to degradation by the proteasomal pathway, we determined the ability of SIVmac239 Vif and HIV-2 Vif to protect HDV-EGFP replication from the antiviral activity of the wild-type and D128K mutant APOBEC3G proteins (Fig. 3b). In the absence of Vif, both the wild-type and the D128K mutant APOBEC3G inhibited HDV-EGFP replication. However, in contrast to HIV-1 Vif, coexpression of the SIVmac239 Vif or HIV-2 Vif protected HDV-EGFP replication from both the wild-type and the D128K mutant APOBEC3G proteins. These observations suggested that the D128K mutant is a viable substrate for the proteasomal degradation pathway.
Human APOBEC3G is not depleted in the presence of SIVagm Vif (17). We sought to determine whether the substitutions in the Apo12 mutant and D128K substitution render the human APOBEC3G sensitive to depletion by SIVagm Vif (Fig. 3c). Preliminary results suggest that SIVagm Vif was unable to rescue HDV-EGFP replication from wild-type APOBEC3G. The results also suggest that SIVagm Vif is able to rescue the replication of HDV-EGFP from the inhibitory effects of the Apo12 mutant, implying that the substitutions in the Apo12 mutant enhance the binding of SIVagm Vif to human APOBEC3G.
We wished to verify that coexpression of SIVmac239 and HIV-2 Vif proteins resulted in reduction of the intracellular steady-state levels of the D128K mutant APOBEC3G from cells. We performed Western blotting analysis of cells that were cotransfected with either the wild-type or the D128K mutant APOBEC3G and HIV-1 Vif, HIV-2 Vif, or SIVmac239 Vif expression plasmids. As expected, the wild-type APOBEC3G was depleted from cells in the presence of all three Vif proteins. As also expected, the D128K mutant APOBEC3G was not depleted from cells in the presence of HIV-1 Vif. However, consistent with their ability to rescue HDV-EGFP replication, the SIVmac239 and HIV-2 Vif coexpression resulted in depletion of the intracellular steady-state levels of the D128K mutant APOBEC3G protein (Fig. 3d). These results indicated that the D128K mutant was resistant to the HIV-1 Vif but not the SIVmac239 or HIV-2 Vif, providing further evidence that it can be a substrate for the proteasomal pathway.
Discussion
In these studies, we found that a single D128K substitution can render the human APOBEC3G resistant to depletion induced by HIV-1 Vif. The results strongly support the model that HIV-1 Vif counteracts the inhibitory effects of APOBEC3G by enhancing its degradation. Contrary to our expectation that the D128K substitution would disrupt the Vif-APOBEC3G interaction, we observed that the Apo12 mutant containing the D128K substitution is capable of interacting with HIV-1 Vif. The D128K substitution is not within the recently reported N-terminal region involved in Vif binding, which is consistent with our observation that the Apo12 mutant containing the D128K substitution could interact with HIV-1 Vif (29). It is possible that the D128K mutation reduces the HIV-1 Vif binding affinity, and, as a result, the intracellular levels of the D128K mutant are not depleted. Although we did not observe a reduction in binding between the D128K mutant and HIV-1 Vif at a relatively high salt concentration (500 mM LiCl), it is possible that the Vif binding affinity of the D128K mutant is different from that of the wild-type APOBEC3G in the intracellular environment. Another possibility is that the D128K mutation alters the intracellular location of APOBEC3G and therefore it is no longer susceptible to depletion by the presence of Vif. Although this possibility cannot be ruled out, the D128K mutant was fully capable of inhibiting the replication of HDV-EGFP in the presence and absence of Vif, suggesting that its intracellular location is not drastically different from that of the wild-type APOBEC3G.
Interaction between the D128K mutant APOBEC3G and SIVmac239 or HIV-2 Vif, but not HIV-1 Vif, apparently triggers APOBEC3G degradation through ubiquitination and the proteasomal pathway (19, 30). Therefore, the D128K mutation did not disrupt a structural element that serves as a recognition signal for proteasomal degradation. The D128K residue is not a component of the previously described HECT (homologous to E6-AP C terminus) domain-containing or RING finger-containing ubiquitin-protein ligases (31). These results suggest that HIV-1 Vif interaction with the wild-type APOBEC3G is qualitatively different from its interaction with the D128K mutant and that the latter is not sufficient to initiate its degradation. We propose that HIV-1 Vif interaction triggers a conformational change in the wild-type APOBEC3G protein that is necessary to initiate the degradation, which is suppressed by the D128K mutation. Such a conformational change might be necessary to expose a target signal that is recognized by the ubiquitination and/or proteasomal degradation machinery. This hypothesis is also consistent with previous findings indicating that misfolded proteins are targeted for proteasomal degradation (32).
It was recently reported that degradation of HIV-1 Vif is also enhanced in the presence of human APOBEC3G (22). Our Western blotting analysis did not reveal a difference in the steady-state levels of HIV-1 Vif in the presence of APOBEC3G; a quantitative pulse–chase analysis of the rate of degradation of HIV-1 Vif will be required to confirm the effects of APOBEC3G–Vif interactions on the intracellular steady-state levels of HIV-1 Vif.
It will be of interest to determine whether the D128 residue of APOBEC3G (or other amino acids that might play a role in resistance to HIV-1 Vif) is polymorphic in the human population and whether it is associated with resistance to HIV-1 infection and/or pathogenesis. The D128K HIV-1 Vif-resistant mutant might be a useful tool in elucidating the pathway through which HI V-1 Vif induces the degradation of the wild-type APOBEC3G. Additionally, because the Vif-resistant mutant APOBEC3G is dominant and can inhibit HIV-1 replication in the presence of wild-type APOBEC3G, it could be useful in developing a gene therapy approach to the treatment of HIV-1 infection.
Acknowledgments
We thank John Coffin for his continued support and valuable discussions of results, Wei-Shau Hu for intellectual input throughout the project, and Eric Freed for critical reading of the manuscript.
Abbreviations: APOBEC3G, apolipoprotein B mRNA-editing, enzyme-catalytic, polypeptide-like 3G; EGFP, enhanced GFP; Vif, virion infectivity factor; AGM, African green monkey; MAC, rhesus macaque; SIV, simian immunodeficiency virus; HDV, HIV-1-derived vector.
References
- 1.Vartanian, J. P., Meyerhans, A., Asjo, B. & Wain-Hobson, S. (1991) J. Virol. 65, 1779–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pathak, V. K. & Temin, H. M. (1990) Proc. Natl. Acad. Sci. USA 87, 6019–6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gunther, S., Sommer, G., Plikat, U., Iwanska, A., Wain-Hobson, S., Will, H. & Meyerhans, A. (1997) Virology 235, 104–108. [DOI] [PubMed] [Google Scholar]
- 4.Martinez, M. A., Vartanian, J. P. & Wain-Hobson, S. (1994) Proc. Natl. Acad. Sci. USA 91, 11787–117891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lecossier, D., Bouchonnet, F., Clavel, F. & Hance, A. J. (2003) Science 300, 1112. [DOI] [PubMed] [Google Scholar]
- 6.Zhang, H., Yang, B., Pomerantz, R. J., Zhang, C., Arunachalam, S. C. & Gao, L. (2003) Nature 424, 94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mangeat, B., Turelli, P., Caron, G., Friedli, M., Perrin, L. & Trono, D. (2003) Nature 424, 99–103. [DOI] [PubMed] [Google Scholar]
- 8.Harris, R. S., Bishop, K. N., Sheehy, A. M., Craig, H. M., Petersen-Mahrt, S. K., Watt, I. N., Neuberger, M. S. & Malim, M. H. (2003) Cell 113, 803–809. [DOI] [PubMed] [Google Scholar]
- 9.Fisher, A. G., Ensoli, B., Ivanoff, L., Chamberlain, M., Petteway, S., Ratner, L., Gallo, R. C. & Wong-Staal, F. (1987) Science 237, 888–893. [DOI] [PubMed] [Google Scholar]
- 10.Strebel, K., Daugherty, D., Clouse, K., Cohen, D., Folks, T. & Martin, M. A. (1987) Nature 328, 728–730. [DOI] [PubMed] [Google Scholar]
- 11.Sheehy, A. M., Gaddis, N. C., Choi, J. D. & Malim, M. H. (2002) Nature 418, 646–650. [DOI] [PubMed] [Google Scholar]
- 12.Gabuzda, D. H., Lawrence, K., Langhoff, E., Terwilliger, E., Dorfman, T., Haseltine, W. A. & Sodroski, J. (1992) J. Virol. 66, 6489–6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sakai, H., Shibata, R., Sakuragi, J., Sakuragi, S., Kawamura, M. & Adachi, A. (1993) J. Virol. 67, 1663–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mochizuki, H., Schwartz, J. P., Tanaka, K., Brady, R. O. & Reiser, J. (1998) J. Virol. 72, 8873–8883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaddis, N. C., Chertova, E., Sheehy, A. M., Henderson, L. E. & Malim, M. H. (2003) J. Virol. 77, 5810–5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.von Schwedler, U., Song, J., Aiken, C. & Trono, D. (1993) J. Virol. 67, 4945–4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mariani, R., Chen, D., Schrofelbauer, B., Navarro, F., Konig, R., Bollman, B., Munk, C., Nymark-McMahon, H. & Landau, N. R. (2003) Cell 114, 21–31. [DOI] [PubMed] [Google Scholar]
- 18.Marin, M., Rose, K. M., Kozak, S. L. & Kabat, D. (2003) Nat. Med. 9, 1398–1403. [DOI] [PubMed] [Google Scholar]
- 19.Yu, X., Yu, Y., Liu, B., Luo, K., Kong, W., Mao, P. & Yu, X. F. (2003) Science 302, 1056–1060. [DOI] [PubMed] [Google Scholar]
- 20.Stopak, K., de Noronha, C., Yonemoto, W. & Greene, W. C. (2003) Mol. Cell 12, 591–601. [DOI] [PubMed] [Google Scholar]
- 21.Sheehy, A. M., Gaddis, N. C. & Malim, M. H. (2003) Nat. Med. 9, 1404–1407. [DOI] [PubMed] [Google Scholar]
- 22.Mehle, A., Strack, B., Ancuta, P., Zhang, C., McPike, M. & Gabuzda, D. (2004) J. Biol. Chem. 279, 7792–7798. [DOI] [PubMed] [Google Scholar]
- 23.Kao, S., Khan, M. A., Miyagi, E., Plishka, R., Buckler-White, A. & Strebel, K. (2003) J. Virol. 77, 11398–11407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Unutmaz, D., KewalRamani, V. N., Marmon, S. & Littman, D. R. (1999) J. Exp. Med. 189, 1735–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yee, J. K., Friedmann, T. & Burns, J. C. (1994) Methods Cell Biol. 43, 99–112. [DOI] [PubMed] [Google Scholar]
- 26.Regier, D. A. & Desrosiers, R. C. (1990) AIDS Res. Hum. Retroviruses 6, 1221–1231. [DOI] [PubMed] [Google Scholar]
- 27.Guyader, M., Emerman, M., Sonigo, P., Clavel, F., Montagnier, L. & Alizon, M. (1987) Nature 326, 662–669. [DOI] [PubMed] [Google Scholar]
- 28.Jin, M. J., Hui, H., Robertson, D. L., Muller, M. C., Barre-Sinoussi, F., Hirsch, V. M., Allan, J. S., Shaw, G. M., Sharp, P. M. & Hahn, B. H. (1994) EMBO J. 13, 2935–2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Conticello, S. G., Harris, R. S. & Neuberger, M. S. (2003) Curr. Biol. 13, 2009–2013. [DOI] [PubMed] [Google Scholar]
- 30.Deshaies, R. J. (1999) Annu. Rev. Cell. Dev. Biol. 15, 435–467. [DOI] [PubMed] [Google Scholar]
- 31.Pickart, C. M. (2001) Annu. Rev. Biochem. 70, 503–533. [DOI] [PubMed] [Google Scholar]
- 32.Kopito, R. R. (1997) Cell 88, 427–430. [DOI] [PubMed] [Google Scholar]