Abstract
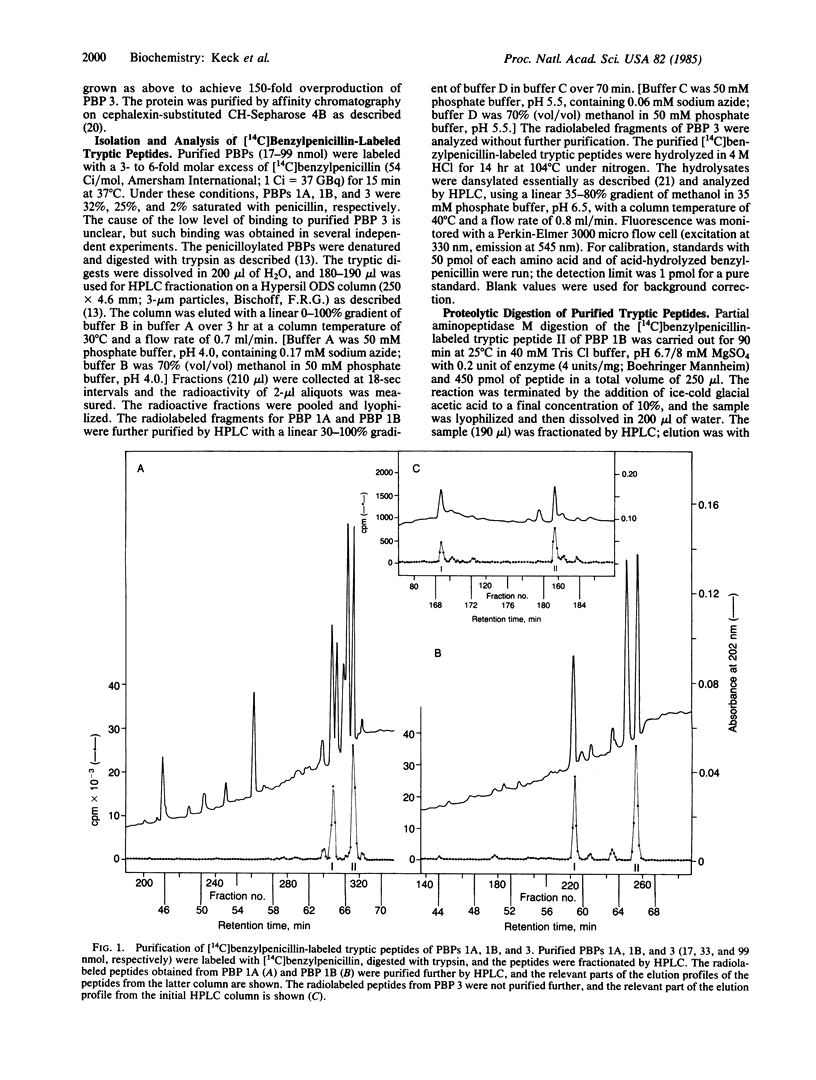
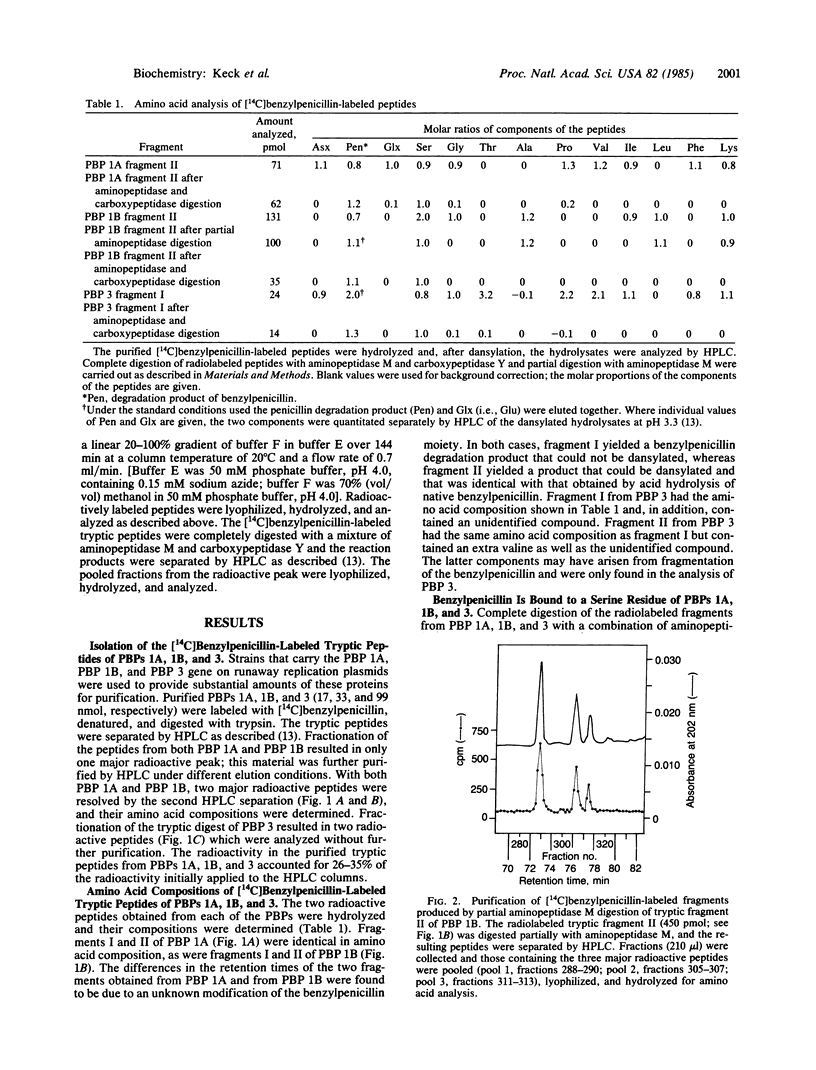
The amino acid compositions of the radioactive peptides obtained from trypsin digestion of [14C]benzylpenicillin-labeled penicillin-binding proteins (PBPs) 1A, 1B, and 3 of Escherichia coli have been obtained. Complete digestion of these peptides with a combination of aminopeptidase M and carboxypeptidase Y showed that benzylpenicillin was bound to a serine residue in each of these proteins. Comparison of the compositions of the penicillin-labeled peptides with the complete amino acid sequences of PBPs 1A, 1B, and 3 showed that the acylated serine occurs near the middle of each of the proteins, within the conserved sequence Gly-Ser-Xaa-Xaa-Lys-Pro. The sequence around the acylated serine of these high Mr PBPs shows little similarity to that around the acylated serine of the low-Mr PBPs (D-alanine carboxypeptidases) or of the class A or class C beta-lactamases, except that in all of these enzymes which interact with penicillin the acylated serine residue occurs within the sequence Ser-Xaa-Xaa-Lys.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ambler R. P. The structure of beta-lactamases. Philos Trans R Soc Lond B Biol Sci. 1980 May 16;289(1036):321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- Bizzozero S. A., Baumann W. K., Dutler H. Kinetic investigation of the alpha-chymotrypsin-catalyzed hydrolysis of peptide substrates. The relationship between the peptide structure C-terminal to the cleaved bond and reactivity. Eur J Biochem. 1982 Feb;122(2):251–258. doi: 10.1111/j.1432-1033.1982.tb05874.x. [DOI] [PubMed] [Google Scholar]
- Duez C., Joris B., Frère J. M., Ghuysen J. M., Van Beeumen J. The penicillin-binding site in the exocellular DD-carboxypeptidase-transpeptidase of Actinomadura R39. Biochem J. 1981 Jan 1;193(1):83–86. doi: 10.1042/bj1930083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher J., Belasco J. G., Khosla S., Knowles J. R. beta-Lactamase proceeds via an acyl-enzyme intermediate. Interaction of the Escherichia coli RTEM enzyme with cefoxitin. Biochemistry. 1980 Jun 24;19(13):2895–2901. doi: 10.1021/bi00554a012. [DOI] [PubMed] [Google Scholar]
- Frère J. M., Duez C., Ghuysen J. M., Vandekerkhove J. Occurrence of a serine residue in the penicillin-binding site of the exocellular DD-carboxy-peptidase-transpeptidase from Streptomyces R61. FEBS Lett. 1976 Nov;70(1):257–260. doi: 10.1016/0014-5793(76)80770-3. [DOI] [PubMed] [Google Scholar]
- Ghuysen J. M., Frère J. M., Leyh-Bouille M., Coyette J., Dusart J., Nguyen-Distèche M. Use of model enzymes in the determination of the mode of action of penicillins and delta 3-cephalosporins. Annu Rev Biochem. 1979;48:73–101. doi: 10.1146/annurev.bi.48.070179.000445. [DOI] [PubMed] [Google Scholar]
- Hedge P. J., Spratt B. G. A gene fusion that localises the penicillin-binding domain of penicillin-binding protein 3 of Escherichia coli. FEBS Lett. 1984 Oct 15;176(1):179–184. doi: 10.1016/0014-5793(84)80936-9. [DOI] [PubMed] [Google Scholar]
- Jackson G. E., Strominger J. L. Synthesis of peptidoglycan by high molecular weight penicillin-binding proteins of Bacillus subtilis and Bacillus stearothermophilus. J Biol Chem. 1984 Feb 10;259(3):1483–1490. [PubMed] [Google Scholar]
- Joris B., Van Beeumen J., Casagrande F., Gerday C., Frère J. M., Ghuysen J. M. The complete amino acid sequence of the Zn2+-containing D-alanyl-D-alanine-cleaving carboxypeptidase of streptomyces albus G. Eur J Biochem. 1983 Jan 17;130(1):53–69. doi: 10.1111/j.1432-1033.1983.tb07116.x. [DOI] [PubMed] [Google Scholar]
- Knott-Hunziker V., Petursson S., Jayatilake G. S., Waley S. G., Jaurin B., Grundström T. Active sites of beta-lactamases. The chromosomal beta-lactamases of Pseudomonas aeruginosa and Escherichia coli. Biochem J. 1982 Mar 1;201(3):621–627. doi: 10.1042/bj2010621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott-Hunziker V., Petursson S., Waley S. G., Jaurin B., Grundström T. The acyl-enzyme mechanism of beta-lactamase action. The evidence for class C Beta-lactamases. Biochem J. 1982 Nov 1;207(2):315–322. doi: 10.1042/bj2070315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott-Hunziker V., Waley S. G., Orlek B. S., Sammes P. G. Penicillinase active sites: labelling of serine-44 in beta-lactamase I by 6beta-bromopenicillanic acid. FEBS Lett. 1979 Mar 1;99(1):59–61. doi: 10.1016/0014-5793(79)80248-3. [DOI] [PubMed] [Google Scholar]
- Larsen J. E., Gerdes K., Light J., Molin S. Low-copy-number plasmid-cloning vectors amplifiable by derepression of an inserted foreign promoter. Gene. 1984 Apr;28(1):45–54. doi: 10.1016/0378-1119(84)90086-6. [DOI] [PubMed] [Google Scholar]
- Nakamura M., Maruyama I. N., Soma M., Kato J., Suzuki H., Horota Y. On the process of cellular division in Escherichia coli: nucleotide sequence of the gene for penicillin-binding protein 3. Mol Gen Genet. 1983;191(1):1–9. doi: 10.1007/BF00330881. [DOI] [PubMed] [Google Scholar]
- Spratt B. G. Distinct penicillin binding proteins involved in the division, elongation, and shape of Escherichia coli K12. Proc Natl Acad Sci U S A. 1975 Aug;72(8):2999–3003. doi: 10.1073/pnas.72.8.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spratt B. G. Penicillin-binding proteins and the future of beta-lactam antibiotics. The Seventh Fleming Lecture. J Gen Microbiol. 1983 May;129(5):1247–1260. doi: 10.1099/00221287-129-5-1247. [DOI] [PubMed] [Google Scholar]
- Tamura T., Suzuki H., Nishimura Y., Mizoguchi J., Hirota Y. On the process of cellular division in Escherichia coli: isolation and characterization of penicillin-binding proteins 1a, 1b, and 3. Proc Natl Acad Sci U S A. 1980 Aug;77(8):4499–4503. doi: 10.1073/pnas.77.8.4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapuhi Y., Schmidt D. E., Lindner W., Karger B. L. Dansylation of amino acids for high-performance liquid chromatography analysis. Anal Biochem. 1981 Jul 15;115(1):123–129. doi: 10.1016/0003-2697(81)90534-0. [DOI] [PubMed] [Google Scholar]
- Tipper D. J., Strominger J. L. Mechanism of action of penicillins: a proposal based on their structural similarity to acyl-D-alanyl-D-alanine. Proc Natl Acad Sci U S A. 1965 Oct;54(4):1133–1141. doi: 10.1073/pnas.54.4.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlin B. E., Molin S., Gustafsson P., Nordström K. Plasmids with temperature-dependent copy number for amplification of cloned genes and their products. Gene. 1979 Jun;6(2):91–106. doi: 10.1016/0378-1119(79)90065-9. [DOI] [PubMed] [Google Scholar]
- Waxman D. J., Strominger J. L. Penicillin-binding proteins and the mechanism of action of beta-lactam antibiotics. Annu Rev Biochem. 1983;52:825–869. doi: 10.1146/annurev.bi.52.070183.004141. [DOI] [PubMed] [Google Scholar]
- Waxman D. J., Strominger J. L. Sequence of active site peptides from the penicillin-sensitive D-alanine carboxypeptidase of Bacillus subtilis. Mechanism of penicillin action and sequence homology to beta-lactamases. J Biol Chem. 1980 May 10;255(9):3964–3976. [PubMed] [Google Scholar]
- Yocum R. R., Amanuma H., O'Brien T. A., Waxman D. J., Strominger J. L. Penicillin is an active-site inhibitor for four genera of bacteria. J Bacteriol. 1982 Mar;149(3):1150–1153. doi: 10.1128/jb.149.3.1150-1153.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yocum R. R., Rasmussen J. R., Strominger J. L. The mechanism of action of penicillin. Penicillin acylates the active site of Bacillus stearothermophilus D-alanine carboxypeptidase. J Biol Chem. 1980 May 10;255(9):3977–3986. [PubMed] [Google Scholar]