Abstract
Ceramide, the central molecule of sphingolipid metabolism, is an important bioactive molecule participating in cellular regulatory events and having implications for disease. A challenge in deciphering ceramide signaling emanates from the myriad of ceramide species that exist and the possibility that many of them may have distinct functions. Here, we applied systems biology and molecular approaches to perturb ceramide metabolism in the yeast (Saccharomyces cerevisiae) and inferred causal relationships between ceramide species and their potential targets by combining lipidomic, genomic, and transcriptomic analyses. We find that during heat stress distinct metabolic mechanisms control the abundance of different groups of ceramide species. Additionally, distinct groups of ceramide species regulated different sets of functionally related genes, indicating that specific sub-groups of lipids participated in different regulatory pathways. These results indicate a previously unrecognized complexity and versatility of lipid-mediated cell regulation.
INTRODUCTION
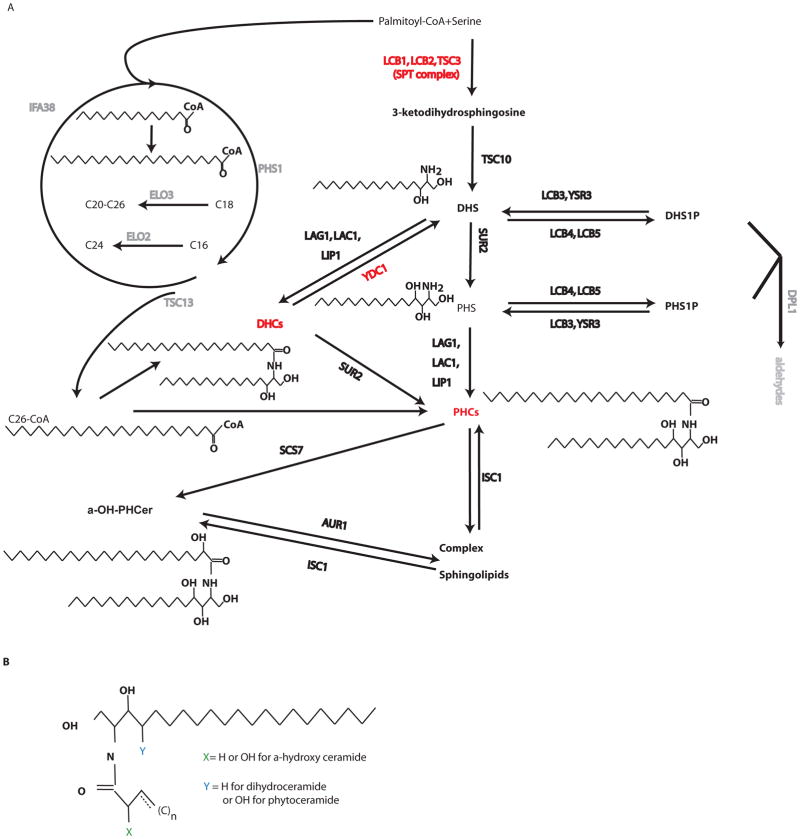
Ceramides constitute a family of structurally related molecules that form the core structure of the broader family of bioactive lipids found in all eukaryotes, the sphingolipids (1). These structural variants of ceramide arise from the condensation of one or more sphingoid bases and several fatty acids. These, in turn, can be modified by the addition of distinct hydroxyl groups on either the sphingoid backbone or the fatty acid. Thus, the biosynthesis of ceramides is the product of the combinatorial action of multiple enzymes that control the structural variations of the ceramide products. In yeast (Saccharomyces cerevisiae), ceramide biosynthesis (Fig. 1) generates more than 30 distinct species that can be identified by contemporary mass spectroscopy-based lipidomic approaches (2); in mammals, the total number of ceramide species may exceed 200 (3).
Fig. 1. S. Cerevisiae sphingolipid metabolism.
(A) Complete sphingolipid metabolic pathway with explicit examples of ceramide structures of each ceramide subspecies investigated. Enzymes manipulated or mentioned in the text are highlighted with red color. (B) Generic ceramide structure with a C18 sphingoid base indicating placement of hydroxyl groups of alpha-hydroxy and phytoceramide species. A double bond is indicated at the third carbon of the fatty acid, but ceramide species with monounsaturated fatty acids may vary in placement of the double bond.
In humans, ceramides are collectively involved in physiological processes, such as growth regulation and apoptosis, and in pathological conditions, such as diabetes and cancer (2). However, a fundamental question of ceramide-mediated signaling is whether the structural diversity of ceramides underlies functional diversity. In other words, do the distinct ceramides encode specific signals? Although manipulation of individual enzymes of ceramide metabolism has enabled assignment of specific functions to these enzymes (1, 4, 5), these approaches do not clearly delineate the specific lipid species involved in the process, because sphingolipid metabolism constitutes a highly connected network such that perturbing the function of an enzyme can lead to broad changes in sphingolipid species beyond the substrates and products of the enzyme (metabolic ripple effects) (3, 6). Pinpointing the functions of the lipid or lipids implicated by manipulating a sphingolipid metabolic enzyme is critical in deciphering the specific downstream pathways and the mechanisms that mediate the changes in cellular behavior, because it is the lipid product and not the enzyme per se that propagates the downstream signal. Therefore, new tools and approaches capable of delineating connections between specific ceramide structures and diverse downstream signaling pathways are needed.
S. cerevisiae has emerged as a powerful model to dissect metabolic and functional pathways of sphingolipids. Activation of de novo sphingolipid synthesis is essential for yeast to survive heat stress (7, 8), and sphingolipids mediate specific downstream processes in response to heat stress, such as cell cycle arrest (9–11), mRNA sequestration (12), and inhibition of nutrient uptake (13). Microarray analysis revealed that de novo synthesis of sphingolipids mediates the regulation of several hundred genes in response to heat stress (14). This simultaneous sphingolipid-dependent regulation of diverse processes provides an opportunity to identify functions of diverse ceramide species, but also requires the development and application of novel methodology.
RESULTS
Systematic perturbation of sphingolipid metabolism decouples the biosynthesis of some groups of lipids
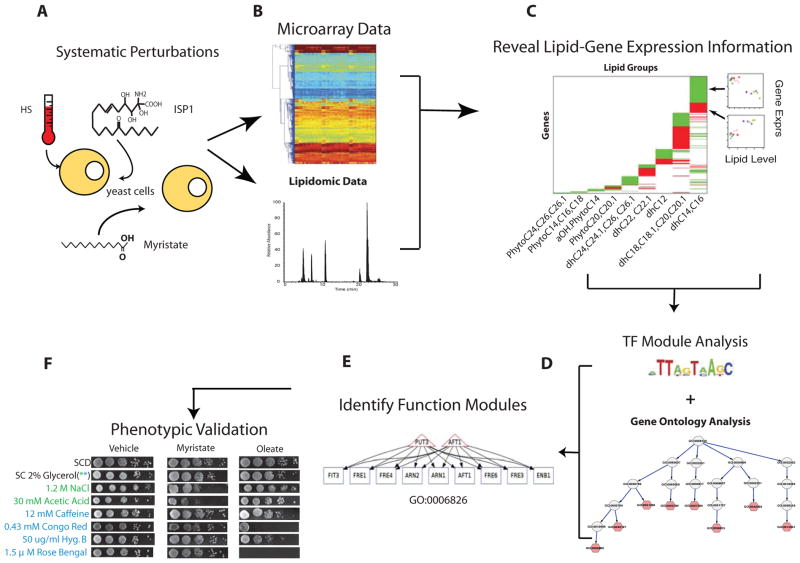
Our overall framework of dissecting the functions of specific ceramide species in yeast proceeded as follows: 1) systematically perturb ceramide metabolism using physiological and pharmacological treatments, 2) monitor lipidomic and transcriptomic responses to the treatments, and 3) apply systems biology analysis to deconvolute the signaling roles of ceramide species in these responses. Figure 2 shows the flow of our approach: Yeast cells were subjected to different combinations (see supplementary methods for detail) of heat stress, ISP1 treatment and myristate treatment (Fig. 2A), with each perturbation affecting different part(s) of the lipid metabolic network and leading to diverse lipid profiles. We measured the relative abundance of the ceramide species by mass spectrometry and the changes in gene expression in response to these perturbations using microarrays (Fig. 2B). We then performed a systems biology analysis to identify correlated changes in ceramide species and gene expression and identified lipid groups that showed similar profiles under all perturbations (Fig. 2C). We then applied ontology-based function analysis and transcription factor analysis (Fig. 2D, E) to identify functional modules among the genes that were potential targets regulated by a specific ceramide species (or a lipid group). Selected predicted functional associations were validated using phenotypic and transcriptomic experiments (Fig. 2F).
Fig. 2. Overall strategy of the study.
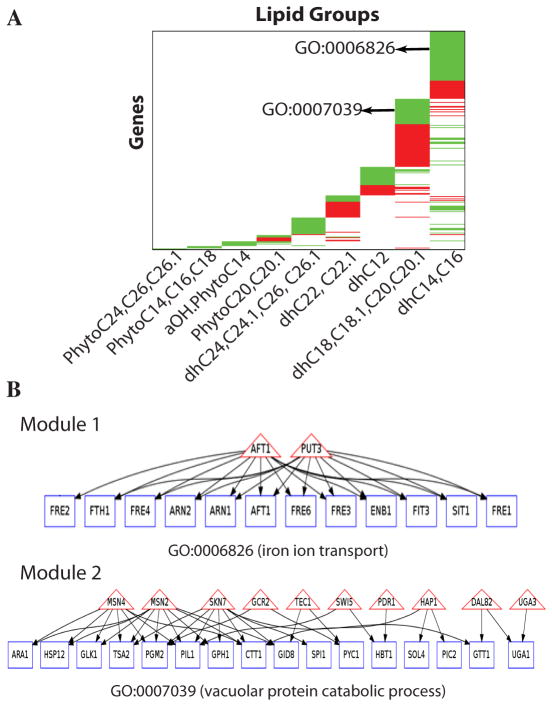
A. Perturbing sphingolipid metabolism in different experimental conditions: heat stress, ISP1 and myristate treatments. B. Collecting lipidomic and gene expression data. C. Modeling the relationship between lipids and genes. The pseudo-colored matrix shows that different lipid groups (columns) are significantly correlated with different genes (rows). The scatter plots illustrate that genes in green region of the matrix are negatively correlated to a lipid, and those in a red region are positively correlated to a lipid. D. Performing ontology based function analysis and transcription factor analysis. E. Identifying functional modules associated with lipid groups. Triangles represent genes encoding transcription factors, rectangles depict genes, and an edge indicates a gene is regulated by a transcription factor. F. Validating prediction using phenotype assays.
Figure guidelines:
Prepare 5.0 in wide..
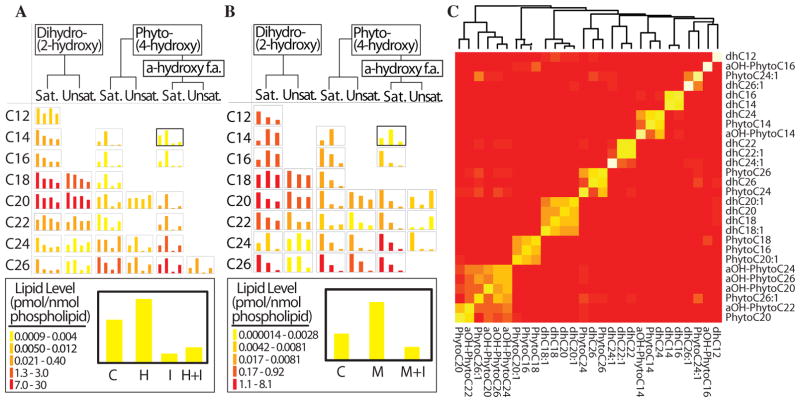
We first studied ceramide profiles when cells were subjected to heat stress and investigated the impact of blocking de novo synthesis using ISP1 (myriocin), which inhibits the serine-palmitoyl transferase (SPT) complex (Fig. 1), the first committed reaction in the de novo pathway of sphingolipid biosynthesis. Many ceramide species, especially the phytoceramide family (PHC), responded to heat stress through increased de novo synthesis (Fig. 3A; table S1). These included C14, C16, and C18 PHC and α-hydroxy-PHCs (as an example, see inset in Fig. 3A for C14-α-hydroxy-PHC). In contrast, several members of the dihydroceramide family (DHC) such as saturated C24 and C26 DHC, decreased during heat stress in the presence or absence of ISP1 (Fig. 3A). The decrease of DHCs during heat stress is a novel finding, and the mechanism of how heat stress affects these species has therefore not been defined.
Fig. 3. Lipidomic Analysis.
A: Lipidomic response to heat stress/ISP1 treatment. Control (C), heat (H), ISP1 (I), heat plus ISP1 (H+I). B. Lipidomic response to myristate treatment. Control (C), myristate (M), myristate plus ISP1 (M+I). In A and B, rows represent N-acyl chain length; columns represent single combination of hydroxyl groups for each ceramide. Saturated (Sat.), monounsaturated (Unsat.) N-acyl chain. Bar height is averaged triplicate ceramide level; the range of each chart is color-coded. Legend inset: C14-alpha-hydroxy phytoceramide. C. Consensus clustering of lipidomic data. The heatmap of the consensus matrix reflects how frequently a pair of lipids is assigned to a common cluster during repeated sampling and clustering. A red cell in the matrix indicates that a pair of lipids tends to be assigned to mutually exclusive clusters and a yellow cell indicates that a pair tends to be assigned to a common cluster. Lipid name abbreviations: ‘dh’ dihydro, ‘aOH’ alpha hydroxy, and ‘C’ followed by a number indicates fatty acid chain length.
To test the hypothesis that different ceramides regulate distinct cellular signals to mediate cell stress responses, we sought to infer the signaling roles of different ceramide species using gene expression data as readouts of cellular signals. Because of the high connectivity of the sphingolipid metabolic network (6), many species, for example DHCs differing only in N-acyl chain length, showed correlated changes during heat stress (Fig. 2A), which obscured potential contributions of individual ceramides or subsets of ceramides. To further dissect and segregate specific ceramide responses, we treated cells with the fatty acid myristate, coupled with treatment with ISP1, to define more specific ceramide responses.
Fatty acid treatment changes the concentration of lipid species with a particular fatty acid side chain (15–17). Matmati et al showed that adding different fatty acids with different chain lengths to the media enriches the PHC pool with those PHC species that correspond to the same chain length (18). Using this methodology, we treated yeast cells with the long-chain (C14) fatty acid myristate to trigger an acute increase of ceramides with the corresponding C14 acyl chains. Additionally, we also treated the cells with ISP1 to block the incorporation of myristate or palmitate (derived from myristate elongation) into the sphingoid backbone, which would lead to an indiscriminate increase in sphingolipids. Upon myristate treatment, C14, C16 and C24 DHC and C14 PHC species increased (Fig. 3B). Moreover, several other ceramide species (Fig. 3B) and the sphingoid bases (C0) (Fig. S1, table S1) decreased in response to myristate, suggesting selective channeling of sphingoid bases to C14 DHC and C14 PHC at the expense of other ceramides. Thus, the C14 and C16 ceramides were effectively decoupled from other ceramides, creating a contrast that would help to resolve the signaling role of these species from other ceramides.
To reveal biologically meaningful patterns from the complex lipidomics datasets collected from the systematic perturbations, we applied consensus-clustering analysis (19, 20) to the pooled lipidomic data sets to identify distinct lipid groups. The consensus clustering method repeatedly performs clustering among randomly drawn subsets of the samples in order to identify intrinsic subgroups of samples, in the current case, the lipids that were inseparable during the repeated clustering. The results showed that ceramides could be further segregated into distinct subgroups (the yellow blocks in Fig. 3C), identifying lipid subgroups, such as one containing C16, C18 and C20:1 PHCs and one containing C18 and C20 DHCs (Fig. 2C). Generally, the lipid species that co-segregated into individual ceramide clusters share similar structures and are mostly products of a common set of specific enzymatic reactions in the sphingolipid pathway. For example, the cluster consisting C16, C18, C20.1-PHCs is separated from the cluster comprised of C18, C18.1, C20, C20.1-DHCs, and synthesis of these ceramide species are metabolically separated by the function of the hydroxylase, Sur2. Clear separation of these clusters indicated that the perturbations induced distinct profiles and decoupled lipids that would exhibit a similar profile if the yeast had only been exposed to a single perturbation, for example heat stress.
On the basis of the results of clustering analysis and knowledge of ceramide metabolism, we divided the ceramides into 9 major groups (table S1) within which group members were statistically inseparable in the clustering analysis and metabolically inseparable on the basis of biosynthetic pathways. Identification of these clusters lends credence to the theory that enzymes in the sphingolipid metabolism network respond to cellular changes, thus producing distinct profiles for different species. Therefore, we hypothesized that each group functions as a single metabolic, signaling, and functional unit and attempted to identify their corresponding downstream targets using gene expression data and statistical analyses.
Transcriptomic responses are specific to perturbations in sphingolipid metabolism
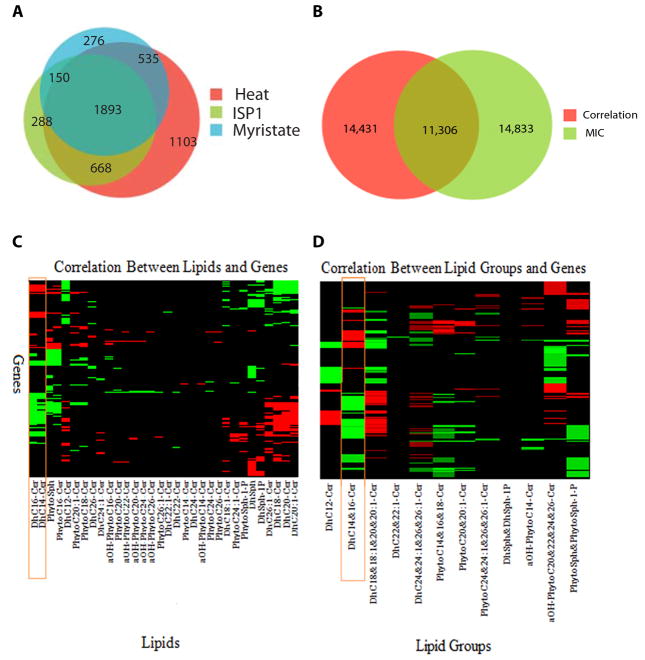
From the microarray data collected in parallel to the lipidomic data, we identified differentially expressed genes responding to different perturbations (Fig. 4A, table S2). We identified 1,893 lipid-mediated stress-responding genes that represented the intersections of the heat-sensitive genes with the ISP1-sensitive and with the myristate-sensitive genes. The members of the union gene set were ISP1-sensitive and thus dependent on de novo synthesis of sphingolipids, corroborating previous findings that sphingolipids play an important role in the yeast stress responses (21–24).
Fig. 4. Assessing the correlation between lipid abundance and gene expression.
A. Venn diagram illustrating number of genes sensitive to different treatments. B. Venn Diagram illustrating number of lipid-gene pairs with significant association assessed using MIC and Pearson correlation analyses. C and D. Heatmaps representation of Pearson correlation coefficient between lipids (C) or lipid groups (D) and gene expression. In the figures, rows correspond to genes that have significant correlation with at least one lipid species; the columns correspond to lipid species. In both figures, a black cell indicates the corresponding lipid-gene pair is not significantly correlated; a red cell represents that the pair is positively correlated; a green cell indicates the positively the pair is negatively correlated. Left panel shows the correlation of genes with respect to all lipid species; the right panel shows that correlation of genes with respect to lipid groups.
To test the hypothesis that distinct ceramides encode disparate signals, which can be detected through the regulation of distinct target gene sets, we studied the relationship between lipidomic and transcriptomic data using three distinct methodologies: (i) the maximum information coefficient (MIC) (25), (ii) the Pearson correlation analysis, and (iii) a Bayesian regression model. The MIC quantifies the information between a pair of variables, such as a lipid species profile and a gene expression profile. MIC can capture both linear and nonlinear relationships between variables in a form similar to the familiar correlation coefficient, although the measured association (positively or negatively associated) lack directionality. We assessed the significance of MIC and the Pearson’s correlation of all lipid-versus-gene pairs. A total of 26,139 lipid-gene pairs had significant MIC values (p < 0.01, Fig 4.); 25,737 lipid-gene pairs had significant Pearson’s correlation coefficients (p < 0.01) with a false discovery threshold q-value (26) set at q < 0.05. There were non-overlapping portions of the MIC and Pearson sets (Fig. 4B), which likely reflect the difference in assessing statistical significance between the two methods. We also performed a series of permutation tests in which lipidomic data were randomly permuted (see supplementary methods) to assess false discovery rate (27). None of the Pearson correlation coefficients derived from the permutation experiment passed the threshold of p < 0.01 and q < 0.05, indicating that the observed relationships between lipids and gene expression were not false discoveries that could result from multiple statistical testing.
For the third method, we employed a regularized regression model (28), which represents the expression value (log2-based) of a gene as a linear function of lipids. It progressively shrinks the weighting coefficient of each lipid predictor towards zero if that predictor is not statistically associated with the gene expression, until leaving only a single predictor with a non-zero coefficient. With this model, we achieved the following goals: (i) identifying the most informative ceramide with respect to a gene, (ii) representing the direction of a lipid influence (stimulate or inhibit), and (iii) providing a mathematical means to predict gene expression as a function of lipid concentration. We pooled the genes potentially regulated by each lipid cluster and further grouped them according to the direction of regulation (Fig. 5A). Each ceramide group had statistically significant parameters with respect to a set of genes, and the gene sets associated with different ceramides were largely non-overlapping, thus supporting the hypothesis that each species plays roles in distinct pathways regulating different gene sets. We also noticed that distinct gene sets were associated with ceramides with the same head group but different acyl chain lengths, for example, those associated with long-chain (C14, C16) were different from those associated with very-long-chain (C18, C18.1, C20, C20.1) DHCs (referred to as LC-DHCs and VLC-DHCs, respectively).
Fig. 5. Modeling relationship between lipidomic and gene expression data.
A. Organizing genes demonstrating significant correlation with specific ceramides. Genes (rows) are organized according to their association with the different lipid subgroups. A green block represents a set of genes negatively correlated to a lipid, and a red block represents a set of genes positively correlated to a lipid. Examples of major enriched GO terms within gene blocks are shown. B. Defining pathways of specific biologic modules that respond to specific ceramides, perform related functions, and share transcription factors. Two example modules are shown (all modules can be found at supplementary website). Rectangles represent lipid-correlated genes, triangles indicate the transcription factors shared by the genes; an edge from a transcription factor to a gene indicates that the gene has the binding sites for the transcription factor in its promoter. The function performed by the genes in a module is represented with a GO term.
To better define ceramide-dependent biological processes and to provide mechanistic understanding of ceramide-specific pathways, we performed ontology-based, semantic-driven function analysis and transcription-factor analysis of potential target genes. We divided the genes significantly associated with a lipid group into modules (certain genes can be in more than one module) by mining their Gene Ontology (GO) annotations (29), such that each module contains genes that participate in coherently related biological processes, which can be encompassed by a GO term that retains as much as possible the semantic meaning of their original annotations. We then applied a graph-based algorithm to search for a set of transcription factors that regulate the members of a module in a cooperative fashion, thus producing a transcription factor module. Analysis of the transcription factors that associated with gene modules that were negatively correlated with LC-DHCs or VLC-DHCs illustrates the results of this approach (Fig. 5B) and supplementary website). The genes in the LC-DHC-associated module are classified as involved in iron ion transportation (GO:0006826); whereas the genes in the VLC-DHC-associated module are classified as involved in vacuolar protein catabolic process (GO:0007039). The analyses revealed that the genes in these modules not only performed related functions but also shared transcription factors, which provided mechanistic evidence that the genes in a module were regulated by a common signal. Our functional analyses project the molecular findings from a gene level onto a conceptual level. For example, the results in Figure 5B can be translated into the following prediction: “LC-DHCs regulate the genes involved in iron ion transportation.” Thus, these gene modules produce testable hypotheses regarding functions of specific groups of ceramide species. All gene modules identified by our analyses—a function map of the ceramide-dependent genes—are available at the website http://www.dbmi.pitt.edu/publications/YeastCeramideSignaling.
Heat stress affects DHC metabolism through activation of Ydc1
Heat stress resulted in a decrease in several DHCs through a mechanism that was not inhibited by ISP1 and thus did not require de novo synthesis of sphingolipids (Fig. 3A). In turn, these changes in DHCs affected the expression of a large number of genes (Fig. 5A), reflecting their important role in mediating the cellular response to heat stress. Therefore, we investigated the molecular mechanism through which heat stress affected DHC metabolism, more specifically to identify the enzyme(s) that mediates the effect of heat stress.
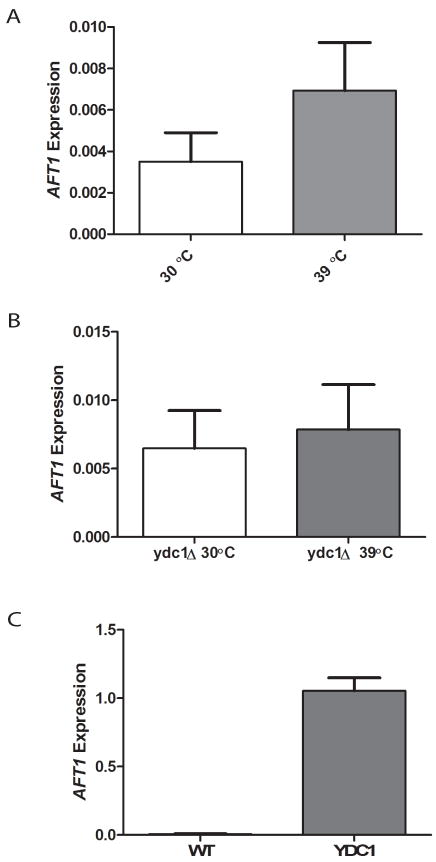
The alkaline dihydroceramidase (encoded by the YDC1 gene) is a good candidate enzyme to mediate the impact of heat stress on long chain DHCs. Ydc1 hydrolyzes dihydroceramides preferentially over phytoceramides (30) to a free fatty acid and dihydrosphingosine, thus reducing the concentration of all DHCs. Aft1 is one of the transcription factors associated with the LC-DHC negatively correlated genes, and the AFT1 gene is in the gene module, thus forming a positive feedback loop. Therefore, we analyzed the expression of AFT1 as an indicator of the transcriptional activity of Aft1 in the YDC1 deletion and overexpression yeast strains. We measured AFT1 expression to assess whether Ydc1 was required to mediate changes in gene expression in response to heat stress (Fig. 6A and 6B). Heat stress induced AFT1 expression (Fig. 6A), and deletion of YDC1 attenuated the response (Fig 6B). Overexpression of YDC1 should decrease DHCs and, thus, mimic the reduction in DHCs caused by the heat stress. Strikingly, overexpression of YDC1 induced AFT1 more than a hundred fold compared with that in wild-type yeast (Fig. 6C). Thus, our results confirm that DHCs regulated the expression of AFT1 in Module 1 and Aft1 is likely involved in this response. The results also indicated that activation of Ydc1 is sufficient to induce gene expression changes similar to those induced by heat stress, thus it is likely one of the enzymes that mediate the impact of heat stress on DHC metabolism.
Fig. 6. The role of Ydc1 in mediating the impact of heat stress on gene expression.
A. Effect of heat stress on AFT1 expression in wild-type (WT) yeast cells (n = 6 and 4, for 30 °C and 39 °C respectively, and p-value = 0.26). B. Effect of heat stress on AFT1 expression in the ydc1Δ strain (n = 4, for 30 °C and 39 °C, p-value = 0.76). C. Effect of over expression of YDC1 on AFT1 expression at 30°C (n = 6 and 2, for wt and +YDC1 respectively, p-value = 0.058)
Phenotypic experiments validate distinct signaling roles of different DHCs
The integrative analyses of lipidomic and transcriptomic data led to the following hypothesis: DHC species with different side chains participate in different signaling pathways. To investigate whether specific transcriptional regulations by distinct DHCs had functional impacts on cells, we examined the effects of perturbing DHCs on cell phenotypes. We focused on the two gene modules shown in Figure 3C, which are suggested to be regulated distinctly by LC-DHCs or VLC-DHCs. Because these modules were negatively correlated with the specific DHC groups, we predicted that increasing the respective lipids would repress genes in the corresponding modules and would produce phenotypes mimicking those resulting from deletion of module genes. We identified 17 phenotypes (XXX) associated with deletion of the genes in the two modules (Table S3), and we then evaluated yeast cell growth after treatment with myristate or oleate to increase production of the LC-DHCs and VLC-DHCs, respectively.
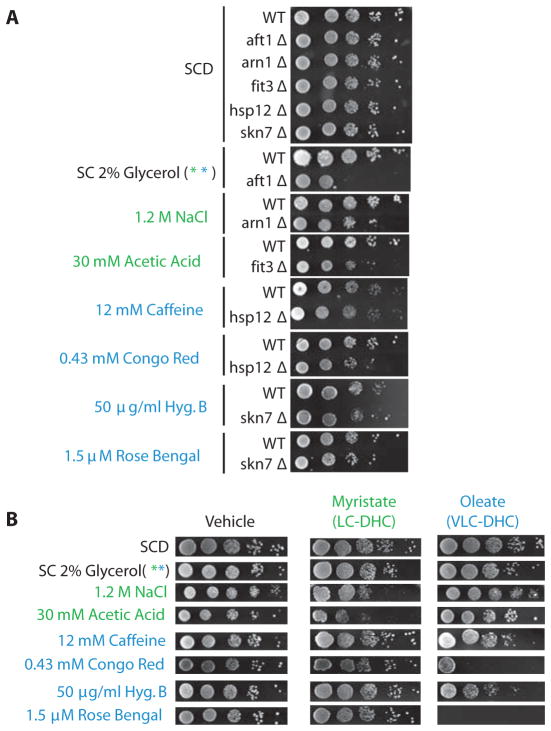
We analyzed in detail 7 phenotypes based on deletion mutant phenotypes (31–37) for the genes within the LC-DHC-sensitive gene module or the VLC-DHC-sensitive gene module (Fig. 7 and Fig S2). For example, the genes ARN1, ARN2, and FRE3 were among iron-transport genes that should be negatively regulated by LC-DHCs as predicted by our analysis (Fig 5B), and their corresponding deletion mutant strains arn1Δ, arn2Δ, and fre3Δ are all sensitive to high sodium. Increased production of LC-DHCs by myristate treatment, but not increased VLC-DHCs induced by oleate treatment, reproduced this growth defect in the wild-type strain (Fig. 5). Conversely, increased production of VLC-DHCs by oleate treatment, and not by myristate treatment, reproduced the Congo red and Rose Bengal sensitivity phenotypes associated with HSP12 and SKN7 deletions ((35, 38), respectively. We expected oleate and the corresponding increase in VLC-DHCs would mimic the phenotypes of hsp12Δ because this gene was identified from the microarray data as negatively correlated with VLC-DHCs. Skn7 is a transcription factor required to induce the genes involved in oxidative responses, and a profound sensitivity of skn7Δ to the singlet oxygen-producing chemical rose bengal was reported (38). Our transcription factor analysis indicated that Skn7 likely stimulates the transcription of seven genes in Module 2, thus leading to the hypothesis that VLC-DHCs regulate these genes through suppression of the transcriptional activity of Skn7. Oleate treatment led to marked increase in sensitivity to rose bengal in wild-type cells in a lipid-specific manner, which is consistent with the hypothesis that VLC-DHCs inhibited the transcriptional activity of Skn7. The results from these phenotypic experiments demonstrate the identification of specific functional responses to specific groups of ceramides.
Fig. 7. Experimental validation of lipid-dependent phenotypes.
(A) Confirmation of published genetic phenotypes as positive controls. Published phenotypes for deletion mutants from the LC-DHC-sensitive gene module (green font) or the VLC-DHC-sensitive gene module (blue font) were used to predict ceramide and fatty-acid-specific growth defects. One deletion mutant phenotype was confirmed for each treatment employed. Conditions were selected from the literature based on phenotypes of genes within each module (31–37).
(B) Validation of LC-DHC- or VLC-DHC-sensitive phenotypes. Rows: specific phenotypes predicted to manifest in response to C14 or C18:1 dihydroceramides, given the indicated treatment condition. Cells were spotted onto agar containing specified treatment plus vehicle (0.1 % ethanol), or saturating (1 mM) myristate or oleate. SCD is no treatment. Spots represent 1:10 serial dilutions of a single mid-log culture. Green font: phenotypes of the LC-DHC-sensitive module predicted to be induced by myristate treatment. Blue font: phenotypes of the VLC-DHC-sensitive module predicted to be induced by oleate treatment, and 2% glycerol is associated with both modules. Images are representative of triplicate experiments (Fig. S5).
DISCUSSION
Here, we addressed the challenging task of determining specific signaling roles of distinct ceramides in yeast. In general, a well-established approach to infer causal relationship between two objects (or events) is to manipulate the potential causal object (or event) in a random trial, while investigating whether the target object (or event) consistently responds to such manipulations (39). Adopting this principle to lipid-mediated signaling, we applied a series of perturbations to manipulate sphingolipid metabolism, with each leading to unique changes in both ceramide metabolism and gene expression through distinct mechanisms. The results showed significant correlations (linear or nonlinear) between specific ceramide species or ceramide groups and gene expression despite the diversity of lipid and gene response to these perturbations, thus supporting the hypotheses that causal relationships exist between the ceramides and genes studied in this report. Although it is possible that each perturbation may exert effects on gene expression (or phenotypes) through additional confounding mechanisms other than through ceramides, systematic perturbation experiments reduced the likelihood of such effects. For example, the combination of multiple approaches to manipulate LC-DHC—reducing these lipids by heat stress, myrocin treatment, or overexpression of YDC1, and inducing these lipids by myristate treatment—effectively minimize the impacts of potential confounding factors associated with each individual manipulation. Thus, we confidently concluded that LC-DHCs regulated the genes in Module 1.
In conclusion, ceramides mediated a multitude of distinct cellular signals in the yeast stress responses. Additionally, this study revealed that the abundance of DHCs was decreased during the yeast response to heat stress, likely through activation of the dihydroceramidase (Ydc1). Functionally, the various DHCs regulated distinct subsets of target genes predicted to participate in distinct biologic processes. Overall, we provided evidence that distinct ceramide species with different N-acyl chains, functional groups, and hydroxylation participate in regulatory processes. The structural complexity of ceramides underscores the potential diversity of the functions that they can play in cellular systems, because even closely related ceramides (such as LC-DHCs vs. VLC-DHCs) regulated distinct sets of functionally related genes. These findings suggest new research directions in the study of ceramide-mediated signaling, including their roles in human physiology and disease.
MATERIALS AND METHODS
Yeast Strains and Culture Conditions
Yeast strains used in this study including genotypes are listed in table S5. YPD medium was used for the heat stress experiment, and for fatty acid treatment, synthetic complete (SC) media containing: 0.17 % yeast nitrogen base (US Biological), 0.5 % ammonium sulfate, 2 mM sodium hydroxide, and 0.07 % synthetic complete supplement was used, SCD is SC containing 2 % dextrose. SCD dropout medium lacking uracil was used in cells transformed with pYES2 plasmid, SC with galactose lacking uracil was used to induce YDC1 open reading frame in pYES2 plasmid for the overexpression studies. For all experiments, cells were treated during mid-log growth at 30 °C. Heat stress was performed by shifting cells to a 39 °C water bath after 45 min pretreatment with myriocin (Sigma) or vehicle. Cultures were harvested by centrifugation at 3000 × g for 3 min and stored at −80 °C. For spot tests, compounds required for specified treatments including glycerol, sodium chloride, acetic acid, caffeine, congo red, hygromycin B, and rose bengal were purchased from Sigma. All compounds, including fatty acid (myristate or oleate) or vehicle (0.1 % ethanol), were dissolved in media by warming to 50 °C for 10 min. After media was re-equilibrated to room temperature, it was mixed with 2x agar at 50 °C to make SC with 2 % agar; 25 ml of media was poured into 100 mm Petri dishes. Solidified plates were dried for 20 min at 37 °C before use. Mid-log cultures in SCD were diluted (OD600 0.3), then 5 μl of 4 serial 1:10 dilutions were spotted and incubated at 30 °C for 3–5 days.
Heat stress and ISP1 treatment
Cells grown to mid-log (OD600 0.6) from overnight cultures were pretreated with 5 μM ISP1 or vehicle (0.1 % methanol) for 45 min, and then heat stress samples were shifted from 30 to 39 °C for 15 min. 100 ml samples were divided into 10 and 90 ml aliquots for microarray and lipidomic analysis, respectively, then harvested at room temperature by centrifugation at 3000 × g for 3 min, and then flash frozen in a dry ice methanol bath.
Myristate treatment
Cells grown to mid-log (OD600 0.6) from overnight cultures were pretreated with 5 μM myriocin or vehicle (0.1 % methanol) for 45 min, and then treated with 1 mM myristate (Sigma) or fatty acid vehicle (0.05 % ethanol) for 15 min. 100 ml samples were divided into 10 and 90 ml aliquots for microarray and lipidomic analysis, respectively, and then harvested at room temperature for 3 min and flash frozen in a dry ice methanol bath.
Systematic Perturbations and Collection of Lipidomic and Microarray Data
Yeast cells (JK9-3dα) were subjected to the following combinations of perturbations: 1) control condition; 2) ISP1 treatment at 30 °C; 3) heat stress; 4) heat stress plus ISP1 treatment; 5) control condition for fatty acid supplement experiment, 30 °C in SC medium; 6) myristate treatment at 30 °C; 7) myristate plus ISP1 treatment at 30 °C. Experiments were repeated three times under each of the above condition.
RNA was extracted from 108 cells using the hot acid phenol method (40). Synthesis of cDNA, in vitro transcription labeling and hybridization onto the Yeast2.0 chip were conducted using the Affymetrix GeneChip kit.
Cells were grown, treated, extracted, and total protein was measured, all according to (41), and relative lipid concentrations were quantified according to the method of (42), and normalized to total protein.
YDC1 experiments
Wildtype (BY4741) or ydc1Δ were used to perform experiments. Growth conditions and heat stress were done as described above. To achieve YDC1 overexpression, a BY4741 strain was transformed with pYES2 plasmid containing an open reading frame of YDC1 under galactose promoter. For galactose induction, cells were harvested from a dextrose-containing medium by centrifugation at 3000 × g for 3 min., pellets were washed with sterile water, then inoculated into a galactose-containing medium and grown for 6 hours before treatment with heat stress. After heat stress, cells were harvested by centrifugation, washed with sterile water and centrifuged again. The pellets were snap frozen in liquid nitrogen until ready for RNA extraction.
Quantitative Real Time reverse transcriptase-polymerase chain reaction
Total RNA was harvested using hot acid phenol method, described in Short Protocols in Molecular Biology, unit 13.10 (43). First strand cDNA was produced as described previously (44). Real time analysis was done using 7500 Real Time PCR system (Life Technology), SYBR Green Supermix protocol (Bio-Rad) was used to perform the analysis. Primers used in the rt-PCR are AFT1 forward primer: TCAAAAGCACACATTCCCTCA, AFT1 Reverse primer: AACTTTAAATGCGTCCGACC. The expression of target genes was normalized to the expression of RDN18, ALG9, TAF10. The primers are as follows: RDN18 forward primer: CCA TGG TTT CAA CGG GTA ACG, RDN18 reverse primer: GCC TTC CTT GGA TGT GGT AGC C; ALG9 forward primer: CACGGATAGTGGCTTTGGTGAACAATTAC, ALG9 reverse primer: TATGATTATTCTGGCAGCAGGAAAGAACTTGGG; TAF10 forward primer: ATATTCCAGGATCAGGTCTTCCGTAGC, TAF10 reverse primer: GTAGTCTTCTCATTCTGTTGATGTTGTTGTTG.
Ontology-based gene function analysis
Given a set of genes that are significantly correlated to a specific lipid species and their annotations in the form of the Gene Ontology (GO) (45) (www.geneontology.org) terms, we aimed to group genes into non-disjoint subsets, such that each module contained genes with closely related GO annotations and the overall function of the module was represented by a GO term that captured most of the semantic information of the original GO annotations of the genes. We represented genes and their annotations using a data structure referred to as GOGene graph (29, 46). In such a graph, a node represents a GO term and a directed edge between a pair of nodes reflects an “is a” (ISA) relationship between the GO terms, that is parent term subsumes that of the child term. In addition, each node kept track of the genes it annotated, therefore the graph contained information of both GO terms and genes. We constructed a canonical graph using all GO terms in the Biological Process namespace, according to the ontology definition from the GO consortium (www.geneontology.org). When given a set of genes and their annotations, we associated the genes to GO terms based on their annotations, and then we trimmed leaf nodes that had no genes associated. This produced a subgraph in which leaf nodes were a subset of the original GO annotations associated with the genes of interest. Under such a setting, the task of finding functionally coherent gene modules can be achieved by grouping genes according to their annotations through collapsing GOGene graph in a manner that leads to minimal information loss, and we stopped merging when the p-value of assessing the functional coherence of a gene module was equal or greater than 0.05 (29).
Microarray and data analysis
Affymetrix CEL files of the microarray experiments were processed using the “affy” package (v 1.24.2) and differential expression was assessed using the “limma” package (v3.2.3) of the Bioconductor Suite (http://www.bioconductor.org/). The threshold for detecting differential expression was set at p-value < 0.01 and q-value < 0.05.
Consensus clustering of lipidomic data
The R implementation of the ClusterCons (20) was downloaded from the CRAN (http://cran.r-project.org/web/packages/clusterCons/). Lipidomic data (32 species in 21 experimental conditions) were used as input for the program in multiple runs. For each lipid species, the concentration is normalized to a standard normal distribution (zero mean and unit standard deviation). The partition around medoids (PAM) and K-means algorithms were used as base clustering algorithms to run the ConsensusPlus algorithm. The cluster size (K) is set through a range (6–13) to explore optimal number of clusters to group the lipids.
Correlation analysis of ceramides concentrations and gene expression
The software for calculating maximal information coefficient was downloaded from the website: http://www.exploredata.net/ (accessed Dec, 2012), which was maintained by the authors of the report by Reshef et al (25). The statistical significance of the MIC values was determined using the significance table provided by the authors at the threshold of p < 0.01. Due to the fact that the authors only provide the p-values for MIC values that are sufficiently large, it is not possible to perform false discovery correction of these p-values using the q-values (26) package in such setting.
The Pearson correlation analysis was performed the standard R language package. The returned p-values for all lipid-vs-gene pairs were further subjected to false discovery correction using the Q-value package. The significance threshold is set at p-value ≤ 0.01 and q-value ≤ 0.05.
Supplementary Material
Figure S1: Lipidomics Data with Myristate Treatment Including Sphingoids
Figure S2. Validation of Lipid-Specific Growth Phenotypes in Triplicate
Table S1: Lipidomics Data Heat Stress (xlxs)
Table S2: Microarray Data Heat Stress (xlxs)
Table S3. All Treatments Phenotypes Tested for Lipid-Specific Growth Defects
Table S4: Lipid Groups
Table S5: Yeast Strains Used in this Study
Table S6: List of lipid-gene pairs with significant association assessed with Pearson correlation analysis
Table S7: List of lipid-gene pairs with significant association assessed with MIC methods.
Table S8: List of lipid lipid-gene pairs with significant association assessed with Bayesian regression.
Acknowledgments
We would like to acknowledge the MUSC lipidomics facility as well as the MUSC Proteogenomics facility for their services. We would also like to thank L. A. Cowart for commenting on the manuscript, and Cungui Mao for providing the plasmid overexpressing YDC1 and for his suggestions and comments.
Funding: This work is partially supported by NIH grants R01LM 010144, R01LM011155 (LX), R01LM010020 (GFC), and R01GM063265 (YAH).
Footnotes
Author contributions: YAH and LX conceived and directed the study; DJM, NM and BN performed yeast experiments, lipidomics, microarray data collection and yeast validation experiments; LC, SL, GFC and LX performed data analysis and modeling; DJM, LC, NM, LX and YAH drafted and edited the manuscript.
Competing interests: The authors declare that there is no conflicting interests.
Data and materials availability: Data collected in this study is made available as supplementary tables and the supplementary website.
References and Notes
- 1.Hannun YA, Obeid LM. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat Rev Mol Cell Biol. 2008;9:139–150. doi: 10.1038/nrm2329. [DOI] [PubMed] [Google Scholar]
- 2.Kolter T. A view on sphingolipids and disease. Chemistry and Physics of Lipids. 2011;164:590–606. doi: 10.1016/j.chemphyslip.2011.04.013. [DOI] [PubMed] [Google Scholar]
- 3.Hannun YA, Obeid LM. Many Ceramides. Journal of Biological Chemistry. 2011;286:27855–27862. doi: 10.1074/jbc.R111.254359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mullen TD, Spassieva S, Jenkins RW, Kitatani K, Bielawski J, Hannun YA, Obeid LM. Selective knockdown of ceramide synthases reveals complex interregulation of sphingolipid metabolism. J Lipid Res. 2011;52:68–77. doi: 10.1194/jlr.M009142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spassieva SD, Rahmaniyan M, Bielawski J, Clarke CJ, Kraveka JM, Obeid LM. Cell density-dependent reduction of dihydroceramide desaturase activity in neuroblastoma cells. J Lipid Res. 2012;53:918–928. doi: 10.1194/jlr.M019075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cowart LA, Shotwell M, Worley ML, Richards AJ, Montefusco DJ, Hannun YA, Lu X. Mol Syst Biol. EMBO and Macmillan Publishers Limited; 2010. Revealing a signaling role of phytosphingosine-1-phosphate in yeast. 16 February 2010 ed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wells GB, Dickson RC, Lester RL. Heat-induced Elevation of Ceramide in Saccharomyces cerevisiae via de Novo Synthesis. J Biol Chem. 1998;273:7235–7243. doi: 10.1074/jbc.273.13.7235. [DOI] [PubMed] [Google Scholar]
- 8.Cowart LA, Hannun YA. Selective Substrate Supply in the Regulation of Yeast de Novo Sphingolipid Synthesis. J Biol Chem. 2007;282:12330–12340. doi: 10.1074/jbc.M700685200. [DOI] [PubMed] [Google Scholar]
- 9.Cowart LA, Okamoto Y, Pinto FR, Gandy JL, Almeida JS, Hannun YA. Roles for Sphingolipid Biosynthesis in Mediation of Specific Programs of the Heat Stress Response Determined through Gene Expression Profiling. Journal of Biological Chemistry. 2003;278:30328–30338. doi: 10.1074/jbc.M300656200. [DOI] [PubMed] [Google Scholar]
- 10.Jenkins GM, Hannun YA. Role for de Novo Sphingoid Base Biosynthesis in the Heat-induced Transient Cell Cycle Arrest of Saccharomyces cerevisiae. J Biol Chem. 2001;276:8574–8581. doi: 10.1074/jbc.M007425200. [DOI] [PubMed] [Google Scholar]
- 11.Matmati N, Kitagaki H, Montefusco D, Mohanty BK, Hannun YA. Hydroxyurea Sensitivity Reveals a Role for ISC1 in the Regulation of G2/M. Journal of Biological Chemistry. 2009;284:8241–8246. doi: 10.1074/jbc.M900004200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cowart LA, Gandy JL, Tholanikunnel B, Hannun YA. Sphingolipids mediate formation of mRNA processing bodies during the heat-stress response of Saccharomyces cerevisiae. Biochem J. 2010;431:31–38. doi: 10.1042/BJ20100307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guenther GG, Peralta ER, Rosales KR, Wong SY, Siskind LJ, Edinger AL. Ceramide starves cells to death by downregulating nutrient transporter proteins. Proc Natl Acad Sci U S A. 2008;105:17402–17407. doi: 10.1073/pnas.0802781105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cowart LA, Okamoto Y, Lu X, Hannun YA. Distinct roles for de novo versus hydrolytic pathways of sphingolipid biosynthesis in Saccharomyces cerevisiae. The Biochemical journal. 2006;393:733–740. doi: 10.1042/BJ20050643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petschnigg J, Wolinski H, Kolb D, Zellnig Gn, Kurat CF, Natter K, Kohlwein SD. Good Fat, Essential Cellular Requirements for Triacylglycerol Synthesis to Maintain Membrane Homeostasis in Yeast. Journal of Biological Chemistry. 2009;284:30981–30993. doi: 10.1074/jbc.M109.024752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Toke DA, Martin CE. Isolation and Characterization of a Gene Affecting Fatty Acid Elongation in Saccharomyces cerevisiae. J Biol Chem. 1996;271:18413–18422. doi: 10.1074/jbc.271.31.18413. [DOI] [PubMed] [Google Scholar]
- 17.Al-Feel W, DeMar JC, Wakil SJ. A Saccharomyces cerevisiae mutant strain defective in acetyl-CoA carboxylase arrests at the G2/M phase of the cell cycle. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:3095–3100. doi: 10.1073/pnas.0538069100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matmati N, Metelli A, Tripathi K, Yan S, Mohanty BK, Hannun YA. Identification of c18:1-phytoceramide as the candidate lipid mediator for hydroxyurea resistance in yeast. The Journal of biological chemistry. 2013;288:17272–17284. doi: 10.1074/jbc.M112.444802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Monti S, Tamayo P, Mesirov J, Golub T. Consensus Clustering: A Resampling-Based Method for Class Discovery and Visualization of Gene Expression Microarray Data. Machine Learning. 2003;52:91–118. [Google Scholar]
- 20.Simpson TI, Armstrong JD, Jarman AP. Merged consensus clustering to assess and improve class discovery with microarray data. BMC Bioinformatics. 2010;11:590. doi: 10.1186/1471-2105-11-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mousley CJ, Tyeryar K, Ile KE, Schaaf G, Brost RL, Boone C, Guan X, Wenk MR, Bankaitis VA. Trans-Golgi Network and Endosome Dynamics Connect Ceramide Homeostasis with Regulation of the Unfolded Protein Response and TOR Signaling in Yeast. Molecular Biology of the Cell. 2008;19:4785–4803. doi: 10.1091/mbc.E08-04-0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dickson RC, Sumanasekera C, Lester RL. Functions and metabolism of sphingolipids in Saccharomyces cerevisiae. Progress in Lipid Research. 2006;45:447–465. doi: 10.1016/j.plipres.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 23.Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, Ballabio A, Bamber BA, Bassham DC, Bergamini E, Bi X, Biard-Piechaczyk M, Blum JS, Bredesen DE, Brodsky JL, Brumell JH, Brunk UT, Bursch W, Camougrand N, Cebollero E, Cecconi F, Chen Y, Chin LS, Choi A, Chu CT, Chung J, Clarke PG, Clark RS, Clarke SG, Clave C, Cleveland JL, Codogno P, Colombo MI, Coto-Montes A, Cregg JM, Cuervo AM, Debnath J, Demarchi F, Dennis PB, Dennis PA, Deretic V, Devenish RJ, Di Sano F, Dice JF, Difiglia M, Dinesh-Kumar S, Distelhorst CW, Djavaheri-Mergny M, Dorsey FC, Droge W, Dron M, Dunn WA, Jr, Duszenko M, Eissa NT, Elazar Z, Esclatine A, Eskelinen EL, Fesus L, Finley KD, Fuentes JM, Fueyo J, Fujisaki K, Galliot B, Gao FB, Gewirtz DA, Gibson SB, Gohla A, Goldberg AL, Gonzalez R, Gonzalez-Estevez C, Gorski S, Gottlieb RA, Haussinger D, He YW, Heidenreich K, Hill JA, Hoyer-Hansen M, Hu X, Huang WP, Iwasaki A, Jaattela M, Jackson WT, Jiang X, Jin S, Johansen T, Jung JU, Kadowaki M, Kang C, Kelekar A, Kessel DH, Kiel JA, Kim HP, Kimchi A, Kinsella TJ, Kiselyov K, Kitamoto K, Knecht E, Komatsu M, Kominami E, Kondo S, Kovacs AL, Kroemer G, Kuan CY, Kumar R, Kundu M, Landry J, Laporte M, Le W, Lei HY, Lenardo MJ, Levine B, Lieberman A, Lim KL, Lin FC, Liou W, Liu LF, Lopez-Berestein G, Lopez-Otin C, Lu B, Macleod KF, Malorni W, Martinet W, Matsuoka K, Mautner J, Meijer AJ, Melendez A, Michels P, Miotto G, Mistiaen WP, Mizushima N, Mograbi B, Monastyrska I, Moore MN, Moreira PI, Moriyasu Y, Motyl T, Munz C, Murphy LO, Naqvi NI, Neufeld TP, Nishino I, Nixon RA, Noda T, Nurnberg B, Ogawa M, Oleinick NL, Olsen LJ, Ozpolat B, Paglin S, Palmer GE, Papassideri I, Parkes M, Perlmutter DH, Perry G, Piacentini M, Pinkas-Kramarski R, Prescott M, Proikas-Cezanne T, Raben N, Rami A, Reggiori F, Rohrer B, Rubinsztein DC, Ryan KM, Sadoshima J, Sakagami H, Sakai Y, Sandri M, Sasakawa C, Sass M, Schneider C, Seglen PO, Seleverstov O, Settleman J, Shacka JJ, Shapiro IM, Sibirny A, Silva-Zacarin EC, Simon HU, Simone C, Simonsen A, Smith MA, Spanel-Borowski K, Srinivas V, Steeves M, Stenmark H, Stromhaug PE, Subauste CS, Sugimoto S, Sulzer D, Suzuki T, Swanson MS, Tabas I, Takeshita F, Talbot NJ, Talloczy Z, Tanaka K, Tanaka K, Tanida I, Taylor GS, Taylor JP, Terman A, Tettamanti G, Thompson CB, Thumm M, Tolkovsky AM, Tooze SA, Truant R, Tumanovska LV, Uchiyama Y, Ueno T, Uzcategui NL, van der Klei I, Vaquero EC, Vellai T, Vogel MW, Wang HG, Webster P, Wiley JW, Xi Z, Xiao G, Yahalom J, Yang JM, Yap G, Yin XM, Yoshimori T, Yu L, Yue Z, Yuzaki M, Zabirnyk O, Zheng X, Zhu X, Deter RL. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu M, Huang C, Polu SR, Schneiter R, Chang A. Regulation of sphingolipid synthesis via Orm1 and Orm2 in yeast. Journal of Cell Science. 2012 doi: 10.1242/jcs.100578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reshef DN, Reshef YA, Finucane HK, Grossman SR, McVean G, Turnbaugh PJ, Lander ES, Mitzenmacher M, Sabeti PC. Detecting Novel Associations in Large Data Sets. Science. 2011;334:1518–1524. doi: 10.1126/science.1205438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Storey J. The positive false discovery rate: A Bayesian interpretation and the q-value. Annals of Statistics. 2003;31:2013–2035. [Google Scholar]
- 27.Good P. Permutation Tests: A Practical Guide to Resampling Methods for Testing Hypotheses. Springer; 1994. [Google Scholar]
- 28.Friedman J, Hastie T, Tibshirani R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J Stat Softw. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- 29.Chen V, Lu X. Conceptualization of molecular findings by mining gene annotations. BMC Proceedings. 2013 doi: 10.1186/1753-6561-7-S7-S2. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mao C, Xu R, Bielawska A, Obeid LM. Cloning of an alkaline ceramidase from Saccharomyces cerevisiae. An enzyme with reverse (CoA-independent) ceramide synthase activity. The Journal of biological chemistry. 2000;275:6876–6884. doi: 10.1074/jbc.275.10.6876. [DOI] [PubMed] [Google Scholar]
- 31.Sambade M, Alba M, Smardon AM, West RW, Kane PM. A Genomic Screen for Yeast Vacuolar Membrane ATPase Mutants. Genetics. 2005;170:1539–1551. doi: 10.1534/genetics.105.042812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, Arkin AP, Astromoff A, El-Bakkoury M, Bangham R, Benito R, Brachat S, Campanaro S, Curtiss M, Davis K, Deutschbauer A, Entian KD, Flaherty P, Foury F, Garfinkel DJ, Gerstein M, Gotte D, Guldener U, Hegemann JH, Hempel S, Herman Z, Jaramillo DF, Kelly DE, Kelly SL, Kotter P, LaBonte D, Lamb DC, Lan N, Liang H, Liao H, Liu L, Luo C, Lussier M, Mao R, Menard P, Ooi SL, Revuelta JL, Roberts CJ, Rose M, Ross-Macdonald P, Scherens B, Schimmack G, Shafer B, Shoemaker DD, Sookhai-Mahadeo S, Storms RK, Strathern JN, Valle G, Voet M, Volckaert G, Wang CY, Ward TR, Wilhelmy J, Winzeler EA, Yang Y, Yen G, Youngman E, Yu K, Bussey H, Boeke JD, Snyder M, Philippsen P, Davis RW, Johnston M. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 33.Mira N, Teixeira M, Sa-Correia I. Adaptation and tolerance to weak acid stress in Saccharomyces cerevisiae: a genome-wide view. OMICS: A Journal of Integrative Biology. 2010;14:525–540. doi: 10.1089/omi.2010.0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Motshwene P, Karreman R, Kgari G, Brandt W, Lindsey G. LEA (late embryonic abundant)-like protein Hsp 12 (heat-shock protein 12) is present in the cell wall and enhances the barotolerance of the yeast Saccharomyces cerevisiae. Biochem J. 2004;377:769–774. doi: 10.1042/BJ20031301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karreman RJ, Lindsey GG. Modulation of Congo-red-induced aberrations in the yeast Saccharomyces cerevisiae by the general stress response protein Hsp12p. Can J Microbiol. 2007;53:1203–1210. doi: 10.1139/W07-090. [DOI] [PubMed] [Google Scholar]
- 36.Li S, Dean S, Li Z, Horecka J, Deschenes RJ, Fassler JS. The Eukaryotic Two-Component Histidine Kinase Sln1p Regulates OCH1 via the Transcription Factor, Skn7p. Molecular Biology of the Cell. 2002;13:412–424. doi: 10.1091/mbc.01-09-0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brombacher K, Fischer BB, Rüfenacht K, Eggen RIL. The role of Yap1p and Skn7p-mediated oxidative stress response in the defence of Saccharomyces cerevisiae against singlet oxygen. Yeast. 2006;23:741–750. doi: 10.1002/yea.1392. [DOI] [PubMed] [Google Scholar]
- 38.Brombacher K, Fischer BB, Rufenacht K, Eggen RI. The role of Yap1p and Skn7p-mediated oxidative stress response in the defence of Saccharomyces cerevisiae against singlet oxygen. Yeast. 2006;23:741–750. doi: 10.1002/yea.1392. [DOI] [PubMed] [Google Scholar]
- 39.Glymour C, Cooper G. Computation, Causation, and Discovery. MIT Press; Cambridge, MA: 1999. [Google Scholar]
- 40.Collart MA, Oliviero S. Current Protocols in Molecular Biology. John Wiley & Sons, Inc; 2001. Preparation of Yeast RNA. [Google Scholar]
- 41.Montefusco DJ, Newcomb B, Gandy JL, Brice SE, Matmati N, Cowart LA, Hannun YA. Sphingoid Bases and the Serine Catabolic Enzyme CHA1 Define a Novel Feedforward/Feedback Mechanism in the Response to Serine Availability. Journal of Biological Chemistry. 2012;287:9280–9289. doi: 10.1074/jbc.M111.313445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bielawski J, Szulc ZM, Hannun YA, Bielawska A. Simultaneous quantitative analysis of bioactive sphingolipids by high-performance liquid chromatography-tandem mass spectrometry. Methods. 2006;39:82–91. doi: 10.1016/j.ymeth.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 43.Ausubel FM. Short protocols in molecular biology : a compendium of methods from Current protocols in molecular biology. Wiley; New York: 2002. [Google Scholar]
- 44.Kitagaki H, Cowart LA, Matmati N, Montefusco D, Gandy J, de Avalos SV, Novgorodov SA, Zheng J, Obeid LM, Hannun YA. ISC1-dependent metabolic adaptation reveals an indispensable role for mitochondria in induction of nuclear genes during the diauxic shift in Saccharomyces cerevisiae. The Journal of biological chemistry. 2009;284:10818–10830. doi: 10.1074/jbc.M805029200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene Ontology: tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muller B, Richards AJ, Jin B, Lu X. GOGrapher: A Python library for GO graph representation and analysis. BMC Res Notes. 2009;2:122. doi: 10.1186/1756-0500-2-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Lipidomics Data with Myristate Treatment Including Sphingoids
Figure S2. Validation of Lipid-Specific Growth Phenotypes in Triplicate
Table S1: Lipidomics Data Heat Stress (xlxs)
Table S2: Microarray Data Heat Stress (xlxs)
Table S3. All Treatments Phenotypes Tested for Lipid-Specific Growth Defects
Table S4: Lipid Groups
Table S5: Yeast Strains Used in this Study
Table S6: List of lipid-gene pairs with significant association assessed with Pearson correlation analysis
Table S7: List of lipid-gene pairs with significant association assessed with MIC methods.
Table S8: List of lipid lipid-gene pairs with significant association assessed with Bayesian regression.