Abstract
Elongation is becoming increasingly recognized as a critically controlled step in transcriptional regulation. While traditional genetic and biochemical studies have identified major players of transcriptional elongation, our understanding of the importance and roles of these factors is evolving rapidly through the recent advances in genome-wide and single-molecule technologies. Here we focus on how elongation can modulate the transcriptional outcome through the rate-liming step of RNA polymerase II pausing near promoters, and how the participating factors were identified. Among the factors we describe are NELF and DSIF, the pausing factors, and P-TEFb, the key player in pause release. We also describe non-exclusive models for how pausing is achieved by making use of high resolution genome-wide mapping of paused Pol II relative to promoter elements and the first nucleosome. We also discuss Pol II elongation through the bodies of genes and the roles of FACT and Spt6, the factors that allow Pol II to move through nucleosomes.
Keywords: RNA polymerase II, gene regulation, pausing, promoter, nucleosome, P-TEFb
1. INTRODUCTION
Advances in genome-wide, high-throughput technology have taken our analysis of transcriptional elongation into a new era. The distribution of RNA polymerases and their regulatory or auxiliary factors are being examined on all genes under steady-state cellular conditions or in response to regulatory signals and systematic perturbations. Computational analyses of these massive data sets along with more traditional genetic and biochemical analyses are transforming our understanding of transcription and its control. With all these advances, transcriptional elongation is becoming more widely recognized as a significant step in gene regulation. In this review, we describe and connect the ideas from traditional studies with results from newer methodologies, and we provide high-resolution models of how transcription is controlled at elongation.
We begin by illustrating one classic example of how traditional genetics have contributed to the study of transcriptional elongation. In early 80’s, Winston and colleagues selected genes that are critical for the transcription of a retrotransposon element called Ty in yeast (140, 141). These Suppressor of Ty (Spt) genes turned out to be essential elements in Pol II transcription in general. In particular, a group of Spt genes characterized by genetic complementation patterns later turned out to be genuine elongation factors acting directly on elongating Pol II using biochemical analyses (141). Through this convergence between genetics and biochemistry, some of the key players of elongation have been identified, and are currently being extensively studied using the newest methods such as genome-wide or single-molecule technologies.
To further discuss the control mechanisms of transcriptional elongation in depth, we divided the stages of elongation into two parts, ‘early elongation’ and ‘productive elongation’. After initiation, Pol II starts the iterative incorporation of nucleotides at its 3′ end to extend the nascent RNA. This elongation by Pol II is not uniform throughout the gene. During the early elongation stage, Pol II first transcribes the initial 20~60 nucleotides (nt) of RNA and pauses (106). Progressing beyond this point is rate-limiting for approximately half of all active Drosophila and mammalian genes, and the escape beyond the pause into productive elongation is often highly regulated.
Once Pol II makes the transition from the paused state into productive elongation, it then progresses through the body of the gene. In the gene body, Pol II still faces other barriers and requires additional elongation factors to overcome them (116). Finally, Pol II transcribes through the 3′ end of the gene, where nascent RNA cleavage and poly-adenylation defines the end of the mRNA transcript. The actual termination of a transcribing Pol II molecule takes place on average 8 kb downstream of the 3′ end of the gene in human cells (27). This 3′ end processing of the nascent RNA and Pol II termination can add an extra dimension to the co-transcriptional and post-transcriptional control of gene expression (97).
In this review, we will focus on the distinct stages of early elongation and the productive elongation and discuss the factors and mechanisms controlling these processes (Table 1). Other recent reviews deal with other complementary aspects transcription elongation control including the functions of promoter-proximal pausing, conflicts between transcription and DNA replication, and co-transcriptional RNA processing (see Related Resources).
Table 1.
A growing list of the factors participating in transcriptional elongation
| Factor | Function | Related factors/notes | |
|---|---|---|---|
| GAGA-factor | Generate nucleosome free region (132) and promoter structure for pausing (121) | NURF(133) | |
| GTFs | TFIID | Generate promoter structure for pausing (121) | |
| TFIIF | Increase elongation rate (10, 130) | Near promoters (101) | |
| TFIIS | Rescue backtracked Pol II (10, 57) | Pol III (38) | |
| Pausing factors | NELF | Stabilize Pol II pausing (86, 144) | |
| DSIF | Stabilize Pol II pausing (135)/facilitate elongation (46, 143) | ||
| P-TEFb | Phosphorylates NELF (36), DSIF (58), Pol II CTD (105) for pause release | ||
| Processivity factors | Elongin | Increase elongation rate (5) | |
| ELL | Increase elongation rate (120) | AFF4 (71) | |
| SEC | Contains P-TEFb and ELL (71) | Mediator (129), PAF (47) | |
| Activator | c-Myc | Can directly recruit P-TEFb (32) | |
| NF-κB | Can directly recruit P-TEFb (7) | ||
| Coactivator | Brd4 | Recruits P-TEFb (53, 147) | |
| Mediator | Recruits P-TEFb via SEC (129) | ||
| Capping machinery | CE | Facilitates P-TEFb recruitment (70), counters NELF/DSIF (77) | |
| RNMT a | Methylates RNA 5′ end to complete capping | Myc (29) | |
| Premature termination factors | Dcp2 | De-caps nascent RNA for Xrn2 digestion (14) | Dcp1a/Edc3 (14) |
| Microprocessor | Cleaves hairpin structure for Xrn2 digestion (136) | Tat, Senx (136) | |
| Xrn2 | ‘Torpedo’s Pol II with RNA 5′-3′ exonucleation (14, 136) | ||
| TTF2 | Release Pol II from DNA (14, 136) | ||
| Gdown1 | Anti-termination and stabilize paused Pol II (22, 50, 54) | TFIIF (54), Mediator (50) | |
| Histone chaperone | FACT | H2A/H2B eviction and chaperon (9, 94) | Tracks with Pol II (117) |
| Nap1 a | H2A/H2B chaperon (51) | RSC (73), CHD (137) | |
| Spt6 | H3/H4 chaperon (13) | Tracks with Pol II (117) | |
| Asf1 a | H3/H4 chaperon (118) | H3K56ac (113) | |
| Chromatin remodeler | RSC a | SWI/SNF remodeling in gene body(20) | H3K14ac (56) |
| Chd1 a | Maintain gene body nucleosome organization (42) | FACT, DSIF (137) | |
| NURF | ISWI remodeling at promoter (133) | GAGA factor (132) | |
| PARP | Transcription independent nucleosome loss (98) | Tip60 (99) | |
| PAF complex | Loading dock for elongation factors (119) | SEC (47), FACT (96) | |
| Histone tail modifiers | MOF | H4K16ac, recruit Brd4 (153) | H3S10ph, 14-3-3 (153) |
| Tip60 | H2AK5ac, activate PARP (99) | ||
| Elongator a | Acetylates H3, facilitate nucleosomal elongation (59) | Also in cytoplasm (59) | |
| Rpd3C (Eaf3) a | Deacetylates and inhibit spurious initiation in gene body (21) | H3K36me3 (21) | |
| Set1 a | H3K4me3 (60, 88) | MLL/COMPASS | |
| Set2 a | H3K36me3 (63), regulate acetylation-deacetylation cycle (21) | Rpd3C (21) | |
| PIM1 | H3S10ph, recruit 14-3-3 and MOF (152) | ||
| RNF20/40 | H2BK123ub1, facilitate nucleosomal DNA unwrapping (96) | UbcH6, PAF | |
Not directly covered in our main text.
2. IDENTIFICATION OF THE REGULATORY STEP DURING EARLY ELONGATION
Early elongation can be defined as the transition of Pol II between promoter escape and fully productive elongation, and is often accompanied by an intermediate step of promoter proximal pausing (116). This transition of initiated Pol II to paused occurs as nascent RNA with a length from ~10 nucleotides (nt) is extended to 20–60 nt, as measured most precisely in Drosophila (65, 87, 106), and the bulk of this paused Pol II in mammalian genes occurs in a similar position (82). When the RNA chain is less than 10 nt long, Pol II is still considered to be within the initiation stage; it is associated with TFIIB and can terminate prematurely (abortive initiation), which may provide checkpoints for promoter control (72). Once the RNA grows longer than 12 nt and TFIIB is displaced, the Pol II elongation complex is stably engaged (115) and is now in the early elongation stage. Early elongation is not a simple smooth transition of Pol II but involves critical regulatory steps as observed in a plethora of evidence from the earliest to most recent studies.
2.1 Evidence suggesting a mode of transcription regulation in eukaryotes occurs early after initiation
2.1.1. Peaks of paused Pol II on proximal promoters
Studies of transcription regulation have for decades focused primarily on how regulatory signals and key transcription factors act at the level of the recruitment of Pol II to promoters or the initiation step of transcription (103). This model gained strong support for all genes that were closely examined in S. cerevisiae. Activation of transcription generally produced a recruitment of general transcription factors (GTFs) and Pol II to promoters, and the recruited Pol II then began transcribing and produced full gene-length transcripts and a relatively even distribution of Pol II across each gene(126).
One set of observations that seemed at least partially at odds with all transcription being controlled at initiation was that genes in chicken (37), Drosophila heat shock genes (41, 106) and human c-Myc and c-Fos (64, 100, 127), appeared to have much higher levels (peaks) of Pol II near their promoters - at levels that dwarf that on gene bodies. Moreover, this promoter-proximal Pol II was transcriptionally engaged, that is, it had already initiated transcription and had progressed beyond the point of known abortive initiation. These Pol II molecules were also shown to associate with melted DNA found in elongation complexes (39) and could elongate efficiently in nuclear run-on reactions if inhibitory factors were removed by high salt or sarkosyl treatments (both of which block new initiation) (111). These results indicated that a ‘paused’ Pol II elongation complex can accumulate to high levels at least on some genes without efficiently entering into productive elongation.
Whereas promoter-proximal pausing of Pol II was first characterized in a handful of genes, it turns out to be a general mechanism for other genes across species. Pol II ChIP experiments using genome-wide methods (44, 85, 151) and GRO-seq (27, 67, 82) have shown that Pol II pausing is widespread near promoters. Furthermore, similar to Hsp70, other genes in Drosophila are sensitive to nuclear run-on with sarkosyl treatment, indicating that the promoter proximal Pol II in other genes are also physically tethered at the paused state by inhibitory factors (28).
Genes that have paused Pol II are also highly regulated and the escape of Pol II from the pause into productive elongation can be activated in the cases of heat shock genes by stress and in c-Myc by serum stimulation (116). Thus, regulation of these genes was not solely at Pol II recruitment or initiation, but rather, a main component appeared to be at the escape of the paused Pol II into productive elongation.
2.1.2. In vitro transcription systems needed pausing factors and a kinase to recapitulate full regulation seen in metazoans in vivo
The early in vitro transcription systems derived from human whole nuclear extracts were shown to initiate properly and elongate on model DNA templates. All that seemed to be required to produce transcripts was the recruitment of GTFs and Pol II to promoters. Therefore, reconstituted systems composed of GTFs and Pol II were used, and many regulatory transcription factors were shown to facilitate the production of transcripts indicating a mode of regulation at the level of recruitment of GTFs and Pol II (110).
Interestingly, these early reconstituted transcription systems were insensitive to an inhibitor of transcription in mammalian cells, DRB (135). DRB is a kinase inhibitor, and was postulated to act at the elongation stage to reduce the production of full length transcripts (24). The search for factors that make a transcription system responsive to DRB inhibition led to the discovery and characterization of protein complexes that act to stabilize paused Pol II and a kinase that overcame this rate limiting pausing by phosphorylating the pausing complexes (79, 135, 144)
Wada et al. purified a complex from HeLa nuclear that conferred DRB sensitivity to the human in vitro transcription system(135). This complex, DSIF, consists of two subunits, and remarkably, they were the homologs of yeast Spt4 and Spt5 genes identified from the early genetic studies by Winston and colleagues (135).
Studies by Price and colleagues helped clarify the mechanism by which DSIF inhibits elongation. Using Drosophila nuclear extract, they hypothesized that Pol II is tightly associated with negative transcription elongation factor (N-TEF), and a positive transcription elongation factor (P-TEF) relieves the effect of N-TEF. (78). A series of biochemical complementation analyses showed that DRB inhibits P-TEF, and that N-TEF is epistatic to P-TEF inhibition (78, 79). Therefore, it was concluded that DSIF plays the role of N-TEF. However, in the presence of recombinant DSIF alone, transcription was not fully sensitive to DRB inhibition. Handa and colleagues purified an additional N-TEF protein factor that conferred complete DRB sensitivity and was named NELF (144). NELF is a multiprotein complex composed of four subunits - A, B, C or D, and E (86) - and has been shown to physically interact with DSIF and Pol II (144).
In summary, the discovery of Pol II pausing near promoters and identifying several pausing factors that are essential for full regulation of transcription has revealed another critical layer of transcription control that occurs during early elongation.
2.2. Sequence of events leading to promoter proximal pausing
Follow-up on these pioneering studies revealed that many factors and promoter features that are part of early elongation control. Additionally, the mechanisms of early elongation were found to be connected both temporally and spatially to that of initiation and the promoter structure.
The promoter is a dynamic structure that can direct transcription initiation and pausing. In Drosophila, promoter DNA sequences by themselves often have a tendency to position a nucleosome at the TSS (40), and these promoter regions are typically packaged by nucleosomes under repressed conditions (35) (Figure 1a). On the other hand, many yeast or human promoters have DNA features such as the A-T richness or CpG islands that can intrinsically favor an accessible chromatin structure (45). In either case, for a gene to be poised for activation, the accessibility of the promoter to transcription factors is critical (35). Transcription activators or other DNA binding proteins such as GAGA factor in Drosophila (132) can recruit chromatin remodelers to produce an open promoter structure (35) (Figure 1b). Importantly, prolonged transcription factor binding (35) and Pol II pausing itself (40) contribute to the maintenance of this open promoter structure.
Figure 1. Structure of the promoter and Pol II before and during early elongation.
(a) The promoter is occupied by a nucleosome in a closed configuration. This conformation represents a Drosophila promoter unbound by Pol II before priming (40). The nucleosome (nuc) positions are the distance between the dyad axes and the TSS in base-pairs (bp), based on the average micrococcal nuclease sequencing profile (40). (b) The promoter is in an open configuration, unbound by nucleosome and occupied by general transcription factors (35). (c) The pre-initiation complex (PIC) is assembled (zoomed in relative to other panels). Part of the TFIID structure is removed to visualize the assembly of other general transcription factors. TBP is a subunit of the TFIID structure but is not removed to illustrate its binding to the promoter DNA and Pol II. (d) Pol II is paused between the promoter and the first nucleosome (+1 nuc). The pausing position is at +40 from TSS, which is typical in Drosophila promoters. Pol II bound NELF, DSIF, and TFIIS are also shown. TFIID may or may not be present at the promoter depending on its residency in a re-initiation scaffold (149). The illustrated molecules are based on crystal (19, 23, 75, 80) and cryo-electron microscope (43, 61) structures except for NELF (structure not available), and are scaled proportional to their actual dimensions. The length of each DNA turn is about 10 bp or 3.4 nm.
General transcription factors (GTFs) are assembled on core promoters (35), and direct the recruitment and transcriptional initiation of Pol II (Figure 1c). The TFIID complex binds provides a strong foundation for binding to the promoter region. The interaction between the TATA box and TATA box binding protein (TBP), a major subunit of TFIID, as well as interactions of TFIID-associated proteins with elements downstream of the TSS contribute to TFIID binding in many promoters. TFIIA and TFIIB also associate with TBP to facilitate TFIID binding and Pol II recruitment (19). Pol II in complex with TFIIF binds to the promoter followed by TFIIE and TFIIH. TFIIH contains the XPB DNA helicase subunit that melts the DNA and generates the open Pol II complex to expose the template strand of DNA and allow initiation of transcription (131).
Another important function of TFIIH is to phosphorylate the C-terminal domain (CTD) of the Pol II Rpb1 subunit (125, 131). Pol II CTD is composed of a species-specific number of 7 amino acid repeats, Tyr-Ser2-Pro-Thr-Ser5-Pro-Ser7, ranging from 26 in yeast up to over 50 in mammals. A cyclin-dependent kinase Cdk7 subunit of TFIIH phosphorylates the fifth residue (Ser5) (105). Ser2 and Ser7 can also be selectively phosphorylated later on, which allows the interaction of accessary elongation factors with Pol II.
After the very early elongation of nascent RNA (up to ~10 nt), Pol II escapes this initiation stage. Some GTFs are dissociated, but part of the initiation complex such as TFIID, TFIIA, and TFIIH may remain associated with the promoter providing a scaffold for re-initiation (149). Moreover, TFIIF can remain associated with the Pol II as an elongation factor (10, 130). During the course of early elongation, Pol II is prone to arrests in a backtracked position, where TFIIS binds to Pol II facilitating the cleavage of protruded nascent RNA and rescue from the arrested state (10, 23).
In higher eukaryotes, after transcribing the initial 20–60nt of RNA, Pol II becomes paused between the promoter and the first (+1) nucleosome (Figure 1d). Pausing factors NELF and DSIF can bind to paused Pol II and stabilize it in vivo (69). However, the mechanistic details for these complexes are still being determined. A possible mechanism by which these pausing factors mediate their function could be related to their RNA binding property. NELF contains the subunit NELF-E which contains an RNA recognition motif (36, 86), and DSIF has been shown to bind shorter nascent RNA as it emerges from the elongating Pol II complex (83). Consistent with this, the crystal structure of DSIF bound to archaeal RNA polymerase (RNAP) shows that DSIF binds near the active center cleft and RNA exit channel (80) where it may interact with the transcription bubble and modulate the processivity of the RNA polymerase.
3. MECHANISMS OF PROMOTER PROXIMAL PAUSING
To better understand proximal pausing and its role as a rate-limiting step of transcriptional regulation, we need to review in detail the mechanics of early Pol II elongation. In addition to what is known about the factors involved, we need to consider how DNA sequence elements and nucleosomes influence Pol II pausing and its escape. In this section, we discuss the mechanisms of how elongating Pol II can become paused near promoters and what the fate of the paused Pol II is. Also, we describe genomic approaches to identify the properties of promoter that determine the extent and the pattern of promoter proximal pausing.
3.1. Molecular models of promoter proximal pausing
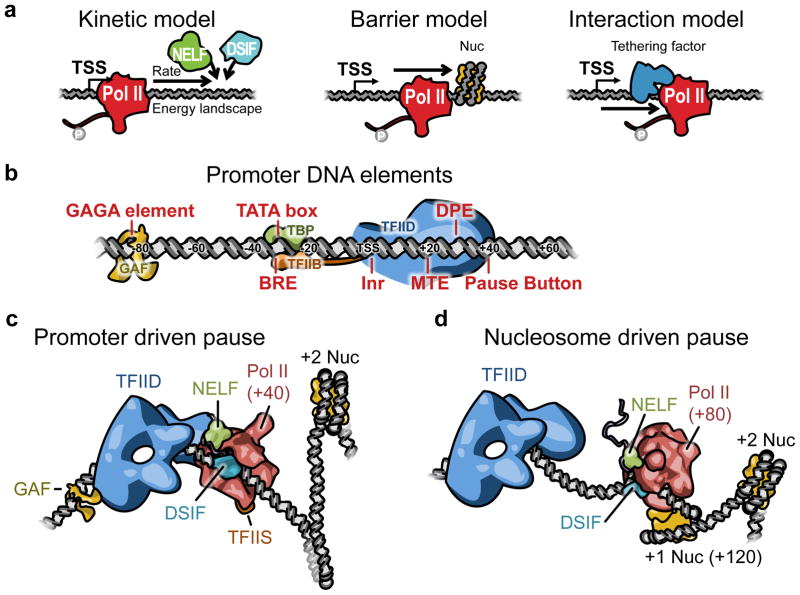
After initiation, the early elongating Pol II meets its first roadblock and pauses. The identity of this first roadblock is very important, because it provides the strongest rate-limiting step in the course of Pol II elongation along the gene. Various in vitro studies have identified many different components that comprise this stop sign. The mechanisms for establishing the pause in vivo can be summarized into three models that are not mutually exclusive (Figure 2a). The actual promoter environment may be a blend of these individual models, and different promoters may use different blends.
Figure 2. Models of promoter proximal Pol II pausing.
(a) The driving force for establishing the position and the extent of Pol II pausing near promoter is summarized in 3 models. (left) In the Kinetic model, the recruitment of the pausing factors competes with the elongation of Pol II. (middle) In the Barrier model, physical barriers such as nucleosomes impede with elongating Pol II. (right) In the Interaction model, a tethering factor that binds to DNA and Pol II at the same time drives Pol II pausing. (b) Promoter DNA elements in Drosophila are shown (18, 92). Protein factors such as GAGA-factor(GAF), TBP, and TFIIB can bind to their DNA elements. TFIID complex can make specific contacts with the DNA elements at multiple positions that are relatively downstream. (c) In promoters with stronger DNA elements, Pol II is more proximal to the TSS within the contact range of the promoter complex (65), and the interaction between the promoter and Pol II can drive the pause. Pausing is stronger in these promoters and the first nucleosome may be absent (40). (d) In promoters with weaker DNA elements, Pol II is closer to the dyad axis of the first nucleosome (65), and is more compatible with pausing driven by nucleosome barriers. The interaction with the promoter complex and the downstream DNA is weaker and may generate less resistance to the Pol II. The illustrations in (c) and (d) are based on actual structures (43, 61, 75, 93)
The first can be described as the kinetic model, where the balance between early elongation and the recruitment of pausing factors determines Pol II pausing. Intrinsic features of Pol II such as the early elongation rate, and DNA and RNA sequence specific features such as the DNA-RNA hybrid energy will affect the kinetics and the energy landscape of elongating Pol II complexes (6). In regions with a higher energy state, Pol II may spontaneously backtrack, and need to be rescued from this backtrack by TFIIS, which is associated with paused Pol II (1). These kinetic processes along with rate of pausing factor binding and the rate and energetics of Pol II will determine the level and the position of promoter proximal pausing (87).
Second is the barrier model where a physical barrier on the chromatin blocks Pol II elongation. Nucleosomes have long been known to provide barriers to Pol II elongation in vitro (52). In Drosophila, it was shown that the first nucleosomes make contacts with paused Pol II and can be cross-linked together in vivo (81). However, in some highly paused genes, the first nucleosome barriers may not be present, for example, the nucleosome free region around the Drosophila Hsp70 promoter extends to +250 bp from TSS (35, 40).
Third is the interaction model where sequence specific binding factors interact with elongating Pol II and determine pausing. This has been an underappreciated model in higher eukaryotes, but for bacteria, it successfully explains the sequence specific pausing by DNA or RNA binding factors that interact with RNAP. For instance, RNA secondary structure at the leader pause site of the E. coli his gene was shown to interact with Pol II (138), and at other genes, the initiation factor σ70 induces pausing by interacting with the downstream DNA elements during its retention within Pol II after the initiation event (109). In higher eukaryotes, there is evidence that pausing may be determined by RNA binding of NELF or DSIF (83, 145). Additionally, a testable hypothesis is that like σ70, the components of the initiation complex may remain bound to the promoter DNA and tether Pol II to induce pausing. Therefore, the general features of E. coli pausing may provide a framework of a molecular model for the promoter proximal pausing of Pol II.
3.2. High-resolution mapping allows integration of pausing models
Advances in the genome-wide approaches have made it possible to define the status of Pol II at the promoter for thousands of genes. In mammalian cells, GRO-seq identified about 40–46% of active genes as paused (27, 82). Likewise in a Drosophila embryonic cell line, 70%–89% of active promoters contained paused Pol II (28). However, although a large fraction of the active genes are paused, the degree of pausing and distribution of pause sites within a promoter varies between genes depending on the features of the promoter (65), and here we consider two types of cis-acting elements: promoter DNA elements and the nucleosome.
The promoter region around a TSS contains several conserved sequence motifs recognized by the general transcription factors (GTFs) and Pol II (Figure 2b). In Drosophila, these include the TATA box, initiator (Inr), TFIIB recognition element (BRE), downstream promoter element (DPE) (18), and motif-ten element (MTE) (92) (Figure 2b). The extent to which promoter DNA elements contribute to GTF binding does not simply rely on true or false type of matching to the consensus motif but on the strength and the quality of the elements (108). The search for the DNA motifs in paused genes in Drosophila embryo have shown the enrichment of some of the core promoter elements such as Inr and DPE, and more upstream GAGA element (48, 139). These elements were also found to be required for pausing in Hsp70 in assays of transgenic mutant promoters (121). A novel GC rich element bearing sequence similarity to DPE was also identified as enriched on paused genes and called the ‘Pause Button’ (48). In addition, the strength of these and other core promoter DNA elements at the consensus position strongly correlated with the extent of pausing (65). Therefore, the strength of a promoter complex as a whole may be a determinant of pausing as well as initiation (40).
The precise position of pausing also relates to the strength of the promoter DNA elements. In Drosophila, promoters with stronger DNA elements tend to have more proximal pausing (+40), while promoters with weaker elements have more distal dispersed pausing (up to +80) (65). Since the average position of the center of the first nucleosome is around +120 from TSS (40), the distal pause position is more consistent with the nucleosome barrier mechanism (25, 52, 65).
These findings lead us to propose an amalgam of molecular models of pausing for different promoters that involves distinct inputs and mechanisms for a proximal, promoter driven pausing (Figure 2c) and for a distal, nucleosome driven pausing (Figure 2d). For the ‘kinetic’ model, the energy landscape and rates are defined not only by the DNA-RNA sequences but also by features of the ‘interaction’ and the ‘barrier’ models. Simply put, the ‘kinetic’ model is an integral part of the ‘interaction’ and ‘barrier’ models. For a promoter with stronger DNA elements, the ‘interaction’ model dominates the energy landscape and the output is a more proximal, promoter driven pausing (Figure 2c). On the other hand, for a promoter with weaker DNA elements, the nucleosome barrier defines the energy landscape more dominantly and the result is a more distal, nucleosome driven pausing (Figure 2d). The interaction and barrier models can even co-exist within a single promoter, and often one may mask the other depending on the balance between the two principal components.
3.3 Fate of paused Pol II
Promoter proximal paused Pol II can resume the NTP catalysis and enter productive elongation. It has been shown that the vast majority of paused genes have detectable levels of elongating Pol II in the gene bodies (28, 82). Measuring the ratio between paused and elongating Pol II will make it possible to estimate how frequently Pol II escapes into productive elongation and assess the activity of the promoter. However, confounding this calculation is the loss of paused Pol II though termination at or near the pause site.
It is also possible the paused Pol II may terminate instead of resuming elongation. Premature termination from the pause or arrest has been extensively documented in the bacterial Rho dependent termination mechanism, but this mechanism usually comes into play downstream of any promoter-associated pause. In eukaryotic cells, stalled Pol II at DNA damage sites can be remove by the ubiquitination mediated degradation pathway during transcription coupled nucleotide excision repair (15, 107). This is not necessarily dependent on the DNA damage itself but is a response to prolonged Pol II stalling (124).
In mammals, mRNA de-capping and 5′→3′ exonuclease Xrn2 mediated termination, similar to the torpedo-like mechanism at the 3′ end of the gene, can take place in promoter proximal region. RNAi depletion of de-capping proteins or the termination factors result in the redistribution of Pol II from the proximal region into the gene bodies (14). Although it is technically challenging to assess the balance between termination and pause escape, it will be critical to quantify the extent of each in order to understand their roles in the regulatory mechanism.
4. ESCAPE INTO PRODUCTIVE ELONGATION
Productive elongation is defined here as the efficient progression of Pol II through the gene body, and the ‘escape into productive elongation’ refers to the transition of the paused Pol II into this productive mode. During Pol II’s residence in the promoter proximal region, two important modifications to the major components of transcription elongation complex take place. One is the 5′ capping of nascent RNA, and the other is the phosphorylation of the paused Pol II complex. The latter is mediated by protein kinase complex P-TEFb that also phosphorylates the pausing factors DSIF and NELF. At this stage, Pol II and its associated nascent RNA are modified so as to allow recruitment of additional elongation and RNA processing factors that help polymerase overcome further barriers in the gene body and couple transcription and RNA processing (116).
4.1. 5′ capping as a checkpoint for productive elongation
Transcription is coordinated with other essential events that lead to proper processing of RNA (97). This requires the loading Pol II with an entourage of proteins that facilitate the RNA processing and help Pol II navigate properly through chromatin. RNA capping has been regarded as a mechanism dependent on early elongation, but accumulating data suggests that it may also have an active role as a control point.
Capping the 5′ end of the nascent RNA requires three enzymatic activities: digestion of the 5′ triphosphate of RNA, addition of a guanine base in 5′ to 5′ linkage, and methylation of the added guanine base (97). In yeast, these activities depend on three different genes, whereas in metazoans, a single Capping Enzyme (CE) performs the first two activities. CE can be recruited to Pol II when the nascent RNA is about 30 nt long through the interaction with phosphorylated Ser5 and DSIF (77). Recruitment of CE can relieve the action of NELF and provide a platform for P-TEFb loading (70, 77).
Examination of the capped state of Pol II-associated RNAs at high resolution in the pause region of heat shock genes under non-heat shock conditions showed that capping occurs progressively as Pol II moves through the pause region – the most proximal paused RNAs are largely uncapped and the more distal (beyond +30) are completely capped (106). Thus, capping could be required for escape into productive elongation, but given the low rate of escape in non-HS conditions, it appears that capping may not be sufficient on its own to drive this escape.
4.2. Major role of P-TEFb in pause escape
4.2.1. Identification of P-TEFb
As described previously, biochemical analysis of the elongation activity in Drosophila Kc nuclear extract showed that the majority of Pol II complexes were only able to produce short, paused transcripts. However, a fraction of the Pol II complexes were able to produce full-length transcripts from the template in a DRB sensitive manner (78). This fraction was postulated to contain positive transcription elongation factor (P-TEF), and later a homogenous fraction was purified that contained P-TEFb (79).
P-TEFb is a heterodimer complex of Cyclin T (CycT) and Cyclin-dependent kinase 9 (Cdk9) subunits (102). Its major role is to phosphorylate Ser2 of Pol II CTD (105), and the pausing factors DSIF (58, 143) and NELF (36). However, there is partial redundancy and non-specificity in Pol II CTD phosphorylation by P-TEFb. P-TEFb can also phosphorylate Ser5 in vitro (105), and Cdk12 (8) or Brd4 (31) can also perform Ser2 phosphorylation. Nevertheless, inhibition of P-TEFb kinase activity with another potent inhibitor flavopiridol resulted in a 5′ to 3′ clearing of Pol II within the gene body in Drosophila Hsp70 (90) and globally in mouse embryonic stem cells (104). Upon this inhibition, Pol II molecules that are already elongating at the time of inhibition continue elongation through the gene body and still maintain a high level of Ser2 phosphorylation level near the 3′ end of Hsp70 (90). Therefore, P-TEFb plays a role in pause escape by phosphorylating the pausing factors and releasing NELF, but it is less clear whether Ser2 phosphorylation itself is required for pause escape.
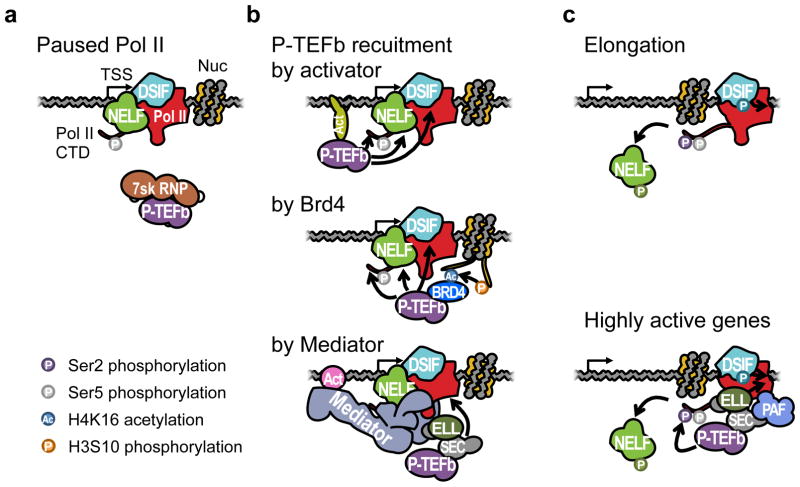
The recruitment of active P-TEFb is a critical step in controlling gene expression, and is regulated at multiple stages. In human cells, the free form of P-TEFb is sequestered by 7SK snRNP in an inactive state (89, 146) (Figure 3a). Upon activation, P-TEFb is released from the sequestration and recruited to the promoter in a variety of ways (Figure 3b). After completing its function, P-TEFb can be sequestered again by 7SK snRNP, but in some highly active genes, it remains bound with the elongating Pol II in Super Elongation Complexes (SECs) (76) (Figure 3c).
Figure 3. Productive transcription elongation complex.
(a) Paused Pol II is bound by NELF and DSIF. The CTD is phosphorylated at Ser5. P-TEFb is held inactive by 7SK RNP. (b) P-TEFb is activated and recruited to the paused Pol II by various mechanisms. The first is directly by activator that binds to the DNA. Recruited P-TEFb can phosphorylate NELF, DSIF, and Ser2 of Pol II CTD. The second is through Brd4 that binds to acetylated histone tails. In human cells, Brd4 can bind to H4K16 acetylation which is dependent on H3S10 phosphorylation through the ‘histone crosstalk’ (153). The third is indirectly through the Mediator complex, which links the activator (Act) and Pol II. A Mediator subunit MED26 can recruit SEC, which also contains P-TEFb (129). (c) Pol II escapes pausing. Phosphorylated NELF is dissociated from Pol II, and DSIF turns into a positive elongation factor after being phosphorylated by P-TEFb. Alternatively, P-TEFb can remain bound to Pol II by SEC that also interacts with PAF in genes with highly active elongation (76). P-TEFb can continuously phosphorylate Ser2, and a SEC component ELL can facilitate elongation. PAF can also recruit additional elongation factors. RNA is not shown for the clarity of viewing the complexes.
4.2.2. Recruitment of P-TEFb by activator, Brd4, and Mediator
There are several ways for P-TEFb to be recruited to paused Pol II. The first is directly by activators. It has been shown that c-Myc can physically interact with the CycT subunit of P-TEFb during transcriptional activation (32, 55). Treating mouse ES cells with the chemical inhibitor of c-Myc/Max complex resulted in a decrease of pause release preferentially in known c-Myc targets (104). Similarly, the RelA subunit of NF-κB also binds to CycT and recruits P-TEFb to TNF-α target genes (7). Finally, the HIV Tat transactivator can bind CycT and directly recruit P-TEFb to the 5′ LTR region through Tat binding to TAR hairpin RNA, adjacent to where Pol II is paused (102).
A second way that P-TEFb is recruited is through the co-activator Brd4 (53, 147). Brd4 is a bromodomain protein that can recognize acetylated histone tails, and can also bind to the CycT subunit of P-TEFb once it is released from 7SK snRNP (53). Recruitment of Brd4 may also require a ‘histone crosstalk’ (153) triggered by PIM1 kinase to phosphorylate H3Ser10 (H3S10ph) (152). 14-3-3 adaptor protein recognizes H3S10ph and recruits histone acetyl-transferase complex MOF to acetylate H4Lys16 which serves as a binding platform for Brd4 (153). Brd4 was also identified as a therapeutic target of acute myeloid leukemia that can control the expression of Myc at the elongation level (154). Thus Brd4-mediated recruitment of P-TEFb may be a key mechanism of gene regulation.
P-TEFb can also be recruited through the Mediator complex. Human Mediator subunit MED26 can serve as a binding platform for SEC, which contains P-TEFb. During initiation and pausing, MED26 is associated with TFIID, but MED26 can be switched to recruit SEC instead (129). Recruitment of P-TEFb through Mediator is believed to be more sensitive in a subpopulation of Pol II that contains a tightly associated factor Gdown1 (50). Gdown1 can compete with TFIIF and therefore might reduce the efficiency of initiation (54). In contrast, after Pol II initiates, Gdown1 stabilizes Pol II in a paused configuration (22). In addition to P-TEFb recruitment, an in vitro study also showed that Mediator helps Pol II overcome the +1 nucleosome barrier on a mononucleosomal template (91), further strengthening its positive function during the early elongation steps of the transcription cycle.
4.3. Factors required for more efficient elongation
There is a set of factors associated with elongating Pol II that is critical for its efficient transcription. Historically, some of these factors were discovered from a class of frequent genetic mutations involving mixed lineage leukemia (MLL) gene in a type of hematopoietic malignancy (112). More surprisingly, all of these factors turned out to be associated with each other to form a single complex, both functionally and physically (76).
The first factor discovered among these is Elongin. Elongin was purified from a fractionation of rat liver nuclear extract that enhanced the in vitro transcription rate of Pol II from the adenovirus 2 major late promoter (AdML) template, composed of three subunits A, B, and C. Elongin’s function in transcription elongation was further confirmed when an Elongin related gene in humans, eleven-nineteen lysine-rich leukemia 1 (ELL1), was found to be a frequent fusion partner MLL in translocation t(11;19)(q23;p13.1) (120). Functional characterization showed that the Elongin and ELL family of proteins increase the net catalytic rate of Pol II, presumably by reducing the transient pauses in gene bodies (76).
ELL can interact with the elongating Pol II as a part of a multi-protein complex named super elongation complex (SEC). SEC consists of AFF1 or AFF4 (AF4/FMR2 family member 1 and 4), ENL (eleven-nineteen leukemia) or AF9 (ALL fused gene from chromosome 9), ELL1 or ELL2, and active P-TEFb (71, 76). The possible combinations of the factors generate diverse subtypes SECs that can activate distinct sets of genes under various physiological contexts (76). In addition, AFF1, AF9, and ENL also are frequent translocation partners of MLL (Rowley, 1998). MLL can bind to Menin and LEDGF (lens epithelial-derived growth factor), and these two factors are responsible for the aberrant tethering of MLL-fused SEC to promoters of Hox genes, resulting in misregulation at the elongation stage and disease progression into leukemia (76).
5. ELONGATION THROUGH THE NUCLEOSOME AND GENE BODY
Even after pause escape, Pol II still has to break further roadblocks in the gene body, most of which are nucleosomes (Figure 4). Genes can use general mechanisms to remove or weaken the nucleosome barriers, such as nucleosome remodeling, exchange of histone variants, and histone tail modifications. Elongating Pol II can make use of an entourage of its associated chaperones to overcome the nucleosome barriers, which can be detected by assaying pausing of Pol II or the loss of the evicted barriers.
Figure 4. Pol II transcribing through gene body nucleosomes.
Pol II may use multiple mechanisms to get through a nucleosome, and not all the steps are used. Step 1, Pol II approaches and makes contact with a nucleosome. Step 2, the outer wrap of nucleosomal DNA can be easily unwrapped (17), and Pol II moves into the nucleosome near the dyad axis. Pol II active site is at around −40 from the dyad axis. The nucleosome binding is strong at this point and Pol II often pauses transiently (25, 52, 65). Step 3, H2A/H2B dimer is dissociated from the DNA and the nucleosome is now a hexamer. A dissociated dimer can still remain through its association with FACT and be re-deposited later (9). Step 4, H3/H4 core nucleosomal particle is evicted from DNA. H3 can remain associated and be re-deposited by Spt6 or Asf1 (13, 118) Step 5, nucleosome hexamer transfers upstream of Pol II while Pol II transcribes into downstream region. A looping intermediate may form during the transfer. Step 6, nucleosome octamer transfers upstream of Pol II, which can be facilitated by histone chaperones. Step 7, Pol II evicts the nucleosome by transcribing through it. Step 8, Pol II transcribes through the nucleosome leaving a hexamer. Step 9, Pol II transcribes through the nucleosome leaving an octamer.
5.1. Overcoming the nucleosome barrier
5.1.1. FACT
Nucleosomes provide a barrier to elongation. In vitro assembled nucleosomal templates strongly inhibit the generation of full-length transcripts by Pol II (52, 94). However, adding HeLa nuclear extract allowed transcription through these chromatinized templates, and FACT was identified from the biochemical fraction that allowed Pol II to overcome the nucleosome barriers (94). FACT has 2 subunits, Spt16 and SSRP1 (95). It functions to disassemble an H2A-H2B dimer from nucleosomes and Pol II can transcribe through the remaining histone hexamer without being displaced (9). Afterwards, the disassembled H2A-H2B dimer can be re-deposited by FACT, leaving the reassembled histone octamer in a more dynamic state (142). FACT’s in vivo association with chromatin was also confirmed on Drosophila Hsp70, and it showed localization and recruitment kinetics that are consistent with elongating Pol II(117).
5.1.2. Spt6
Spt6 was initially identified from the Suppressor of Ty genetic screen in yeast (140, 141). Spt6 can physically interact with histones H3 and H4 directly, and has histone chaperone activity (13). Spt6 travels with Pol II, as it shows an in vivo association with chromatin on Drosophila Hsp70 that is consistent with elongating Pol II (117). Its function is proposed to displace the nucleosome in front of Pol II and reassemble it in the back, allowing Pol II to transcribe through (13). However, Spt6 can also increase the elongation rate of Pol II on naked DNA in vitro (33) and independent of the nucleosomes in vivo(4, 98). Depletion of Spt6 by RNAi decreases the elongation rate on Drosophila Hsp70 under heat-shock conditions (4), when the nucleosomes on Hsp70 are already lost (98, 99). Therefore, Spt6 may both increase the intrinsic elongation rate of Pol II and remove nucleosome barriers at the same time.
5.1.3. PAF complex
PAF complex is another Pol II associated complex that has a role in elongation, and was first identified from the immuno-precipitating yeast whole cell extract using an antibody to the Pol II CTD (119). Like Spt6, PAF also is associated with elongating Pol II in vivo(117), suggesting that it travels with Pol II. PAF complex by itself is not known to contain any enzymatic activity, but serves as a platform for recruiting elongation factor complexes such as SEC (47, 140, 141), FACT (96), and histone modifying enzymes to elongating Pol II (122). In particular, PAF complex was required for H2BK123 monoubiquitination by RNF20/40 (Rad6) and UbcH6 in vitro, and this H2B modification shows a cooperative effect with FACT to allow Pol II to transcribe through nucleosomes (96).
5.1.4. PARP
All the factors above in this section relieve the nucleosome barriers in transcription-dependent manner, associated with elongating Pol II. However, recent studies suggested that PARP (poly(ADP-ribose) polymerase) can achieve this in a transcription-independent way (98, 99). PARP is required for chromatin loosening at transcriptionally active puffs in Drosophila polytene chromosomes (134) and is bound to the promoters of active genes in a pattern that is reciprocal to histone H1 (62). On Drosophila Hsp70, heat shock factor (HSF) response recruits Tip60 histone acetyl-transferase complex to acetylate H2AK5 and subsequently activate PARP. This results in a creation of poly(ADP-ribose) chains and rapid nucleosome loss that precedes the elongation of Pol II through the gene body(99).
5.2. Single molecule studies provide insights to mechanism
Using biochemical experiments, it was shown that SP6 RNAP can transcribe into partially unwrapped DNA that rewraps on the same nucleosome as soon as the polymerase has transcribed through, and the nucleosome ‘steps around’ (~ 50 bp upstream) the polymerase (128). However, Pol II is about 1.7 times larger than SP6 RNAP in dimension, and in other in vitro studies, most Pol II molecules were unable to overcome the nucleosome barrier by itself (128). The precise mechanism could be assessed by examining the Pol II molecules individually to characterize their elongation movements and by defining the conditions under which Pol II can transcribe through the nucleosomes.
Using optical tweezers, it was shown that Pol II could transcribe through the nucleosome under higher ionic strength conditions. The kinetics of Pol II movements were consistent with the model that Pol II transcribes into locally unwrapped DNA (49), similar to SP6 RNAP. Atomic force microscopy made it possible to identify the intermediate structure with locally unwrapped DNA containing Pol II looping back to the same nucleosome. Also, some nucleosomal particles have lost H2A/H2B dimers to become hexamers, and even the whole octamers were lost when the Pol II elongation rates were faster under higher NTP concentrations (12).
When the histones were modified, the kinetics of Pol II through the nucleosomes change. The position of Pol II pausing relative to the nucleosome could be at the entry, central, and exit regions, but reached the maxima at the central region (−35 to +5bp from dyad) (10, 11, 49). This is consistent with in vitro (52) and in vivo (25, 65) positions of Pol II relative to gene body nucleosomes (not the first nucleosome) where the pause is on average ~ 30 bp inside the upstream margin and ~ −40 bp relative to the dyad of the nucleosomes. When the histone tails are acetylated or absent, the pause density at the entry region is decreased, indicating that the unacetylated tails normally interact with the outer turn of the DNA surrounding the nucleosome. When the core histone H3 and H4 are mutated, the pause density in the central region is decreased indicating that the relaxed interaction near the dyad allows Pol II to transcribe through better (11).
One limitation of these methods is that the experiments were carried out under ionic strengths that are higher than the physiological conditions (11, 49). However, by adding reconstituted elongation factors to the system and reproducing in vivo conditions, these sets of single molecule methods will be powerful tools to define precisely how Pol II overcomes the nucleosome barrier and how elongation factors cooperate with this process.
5.3. Elongation rates
The elongation rate of Pol II is an important property of the transcriptional machinery that can direct the level, timing, and the processing of nascent RNA into mature mRNA. It has two components: the pause-free velocity and the frequency/duration of the pauses. The catalytic rate of Pol II sets up the pause-free velocity. It depends on the translocation of the catalytic active site, and this rate is known to be governed by the ‘trigger loop’ structure of Pol II around the active site (68). Therefore, elongation factors that can bind to this region of Pol II may affect the trigger loop structure and alter the translocation rate. The net elongation rate of Pol II would then be a composite of this pause-free velocity and the frequency/duration of pausing or arrests due to underlying sequences and nucleosomes.
The in vitro assays that were used to identify factors related to elongation were basically net elongation rate assays over time courses to determine how long it takes to transcribe the full-length templates. These rates do not represent the true elongation rates in vivo, since rate can be affected by limiting NTP concentrations and the presence of additional elongation factors. To measure the elongation rate in vivo, the traditional approach was to use a rapid induction or shut-down of a gene and measure the time for the “first wave” or the “last wave” of Pol II to reach a certain position on the gene (3). A collection of these rates from yeast to Drosophila and mammals range anywhere between 1–5 kilobases per minute (kb min−1) (3). However these measurements are limited in numbers and types of genes examined, and a systematic comparative analysis between the genes was difficult.
Another approach to measuring the elongation rate is by fluorescence recovery after photobleaching (FRAP). Upon photobleaching fluorescently tagged Pol II, the time for full recovery is the time it takes for the newly initiated polymerase to finish transcribing the whole gene. In Drosophila, salivary gland polytene chromosomes were used to identify the endogenous Hsp70 locus under heat shock condition for the FRAP (148). Similarly, nascent RNA synthesis rate can be measured by FRAP using fluorescently labeled RNA binding proteins. In diploid cells, transgenic genes were used that had structured RNA regions to recruit fluorescently tagged RNA binding proteins (16, 150).
Spt6 was the first among known elongation factors for which the role in elongation rate was robustly confirmed in vivo. Using ChIP, it was shown that depletion of Spt6 in Drosophila embryonic cells delayed the traversal of the first wave of heat shock response in Hsp70 gene (4). In addition, FRAP in Drosophila polytene chromosomes showed that the elongation rates on induced Hsp70 in Spt6 knock-down animals is more than two fold slower than the wild type (4, 148).
More recently, GRO-seq is being used as a tool to measure elongation rates at a large number of genes simultaneously (30). This was made possible because the resolution and the sensitivity of GRO-seq allow the detection elongating Pol II in gene bodies. By stimulating human cells with estrogen or TNFα and measuring the time-course of Pol II induction wave in longer genes, the elongation rates of over 160 responsive genes could be determined. Elongation rates differed between cell types and genes, ranging from 0.4–3.6 kb min−1 in estrogen responsive genes (30). Also genes with higher levels of Pol II in the gene body had faster elongation rates consistent with the in vitro model where cooperative Pol II interactions help rescue arrested Pol II (30, 34, 114).
Although the use of stimulus responsive genes yielded elongation rates in one to two hundred genes, using a more general inhibitor of elongation may greatly expand the number. For example, a DRB washout strategy has been used to induce an artificial first wave of Pol II and pre-mRNA level at various positions were detected to measure the elongation rate in several long human genes (123). Applying genome-wide technology to this type of inhibitor approach will be highly informative in determining the rates and the role of elongation factors in vivo (3).
6. CONCLUSION
Traditional genetic and biochemical studies have revealed that elongation is as critical as initiation in transcriptional regulation of some genes. Recent genome-wide analyses of the distribution of transcription complexes reveal that on many genes in metazoans significant regulated barriers to elongation exist, most prominently near promoters. This promoter-proximal pausing and its regulated escape to productive elongation are being better understood with the advent of high resolution methods for precisely mapping Pol II relative to promoter elements and nucleosome barriers. The pausing intensity and position depend both on core promoter complex interactions with Pol II and the first nucleosome barrier, both appear to contribute to differing extents on different promoters. New single molecule technologies are also providing unprecedented views of individual RNA polymerases during elongation and are providing further insights to the mechanics of elongation and the transit of Pol II through nucleosome barriers during elongation. Further studies in the field will be directed towards testing how mechanistically the promoter-proximal pausing is regulated by transcription factors and the how factors influence the efficiency and rate of elongation in cells.
Sidebar. CO-TRANSCRIPTIONAL RNA SPLICING AND ELONGATION.
The idea that splicing can take place co-transcriptionally is now a general consensus in the field. Splicing can be functionally coupled to transcription and elongation rate can affect splicing efficiency (74). It has been reported that reduction of elongation rate can result in exon inclusion (66) and conversely, Pol II can be paused near 3′ splicing sites (2, 65). The structural coupling between the splicing machinery and elongating Pol II is thought to be mediated through Pol II CTD domain (84). However, the mechanisms of how co-transcriptional splicing machinery recognize features of the DNA and chromatin to determine specific splicing site usage are less known. Recently, DBIRD (DBC1-ZNF326) complex in human was discovered to mediate the physical coupling between splicing machinery and elongating Pol II in alternatively spliced exons, and its depletion led to exon skipping and the reduction of the Pol II accumulation on the exon (26). Future studies involving genome-wide approaches to dissect the association between recruitment of splicing factors and properties of the DNA and chromatin as well as elongating Pol II may help the understanding of how splicing is regulated co-transcriptionally.
SUMMARY POINTS.
Pol II pausing near promoters can control transcriptional outcome by limiting the amount of productively elongating Pol II.
Recruitment of pausing factors such as NELF and DSIF, the barrier provided by nucleosome, and the interaction of Pol II with the promoter complex can all contribute to determining the extent and the pattern of pausing.
Paused Pol II can transition into productive elongation, but may terminate as well.
P-TEFb plays a major role in release of Pol II from pausing, and can be recruited directly by activators or co-activators.
Factors and complexes that facilitate elongation such as SEC, PAF, FACT and Spt6 appear to accompany elongating Pol II.
Nucleosome provides a barrier to productively elongating Pol II in the gene body causing Pol II to pause, and may be displaced and reassembled when Pol II transcribes through.
Elongation rate may be different between genes and cell types, and can both affect and be affected by transcription level and co-transcriptional processing.
FUTURE ISSUES.
The complete molecular structure of a promoter during the course of early elongation and pausing in vivo involving all the factors needs to be defined in sub-nanometer resolution.
The effects of disrupting pausing factors and DNA elements at high resolution and genome-wide need to be identified to allow a more complete understanding of the mechanisms of creating paused Pol II.
How enhancer-associated regulatory factors catalyze efficient escape of paused Pol II into productive elongation should be investigated.
The targets of all the kinases involved in initiation and pausing, and the functional consequence of their activity need to be identified unambiguously.
The mechanisms by which P-TEFb can be recruited/activated to allow Pol II escape into productive elongation in a genome-wide scale should be established.
The mechanisms elongation factors use to facilitate Pol II transcription through nucleosomes should be defined in single molecule experiments.
The rates of elongation and termination needs to be established genome-wide.
Acronyms and Definitions
- Backtracked/backtracking
status of Pol II with upstream displacement from the catalytic position, with active site blocked by protruded nascent RNA
- Brd4
Bromodomain containing protein 4
- ChIP
Chromatin Immunoprecipitation, an experimental method to identify the DNA bound to a protein of interest in vivo
- DRB
5,6-dichloro-1-β-D-ribofuranosylbenzimidazole
- DSIF
DRB Sensitivity Inducing Factor
- ELL
Eleven-nineteen Lysine-rich Leukemia, the gene involved in t(11;19) translocation in mixed lineage leukemia
- FACT
Facilitates Chromatin Transcription
- GRO-seq
Global Run-On sequencing, a recently developed high sensitivity technology to identify engaged RNA polymerase genome-wide
- Histone octamer/hexamer
nucleosome composed of 8 histone molecules (H2A·H2B)2(H32·H42) (octamer) or 6 molecules (H2A·H2B)(H32·H42) (hexamer)
- Mediator
multiprotein coactivator complex composed of nearly 30 subunits that bridges activator to promoter initiation complex and Pol II
- NELF
Negative Elongation Factor
- PAF
RNA polymerase associated factor
- Pol II
RNA polymerase II in eukaryotes, distinguished from RNAP (RNA polymerase) in prokaryotes
- P-TEFb
Positive Transcription Elongation Factor b, composed of CycT (Cyclin T) and Cdk9 (cyclin dependent kinase 9) subunits
- SEC
Super Elongation Complex
- Ser2/Ser5
Specific serine residues in Pol II C-terminal domain (CTD) repeats that can be phosphorylated
- Spt
Suppressor of Ty, a classic genetic screen in transcriptional regulation, or the genes identified from the screen
- TFIIA/B/D/E/F/H
Transcription Factor II A/B/D/E/F/H, general transcription factors of transcription initiation
- TFIIS
Transcription Factor II S, sometimes referred to as SII for the biochemical fraction where it was identified
- TSS
Transcription Start Site
Contributor Information
Hojoong Kwak, Email: hk572@cornell.edu.
John T. Lis, Email: jtl10@cornell.edu.
Literature Cited
- 1.Adelman K, Marr MT, Werner J, Saunders A, Ni Z, Andrulis ED, Lis JT. Efficient release from promoter-proximal stall sites requires transcript cleavage factor TFIIS. Mol Cell. 2005;17(1):103–112. doi: 10.1016/j.molcel.2004.11.028. [DOI] [PubMed] [Google Scholar]
- 2.Alexander RD, Innocente SA, Barrass JD, Beggs JD. Splicing-Dependent RNA Polymerase Pausing in Yeast. Mol Cell. 2010;40(4):12–12. doi: 10.1016/j.molcel.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ardehali MB, Lis JT. Tracking rates of transcription and splicing in vivo. Nat Struct Mol Biol. 2009;16(11):1123–1124. doi: 10.1038/nsmb1109-1123. [DOI] [PubMed] [Google Scholar]
- 4.Ardehali MB, Yao J, Adelman K, Fuda NJ, Petesch SJ, Webb WW, Lis JT. Spt6 enhances the elongation rate of RNA polymerase II in vivo. EMBO J. 2009;28(8):1067–1077. doi: 10.1038/emboj.2009.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aso T, Lane W, Conaway J, Conaway R. Elongin (SIII): a multisubunit regulator of elongation by RNA polymerase II. Science. 1995;269(5229):1439–1443. doi: 10.1126/science.7660129. [DOI] [PubMed] [Google Scholar]
- 6.Bai L, Shundrovsky A, Wang MD. Sequence-dependent kinetic model for transcription elongation by RNA polymerase. J Mol Biol. 2004;344(2):335–349. doi: 10.1016/j.jmb.2004.08.107. [DOI] [PubMed] [Google Scholar]
- 7.Barboric M, Nissen RM, Kanazawa S, Jabrane-Ferrat N, Peterlin BM. NF-kappaB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol Cell. 2001;8(2):327–337. doi: 10.1016/s1097-2765(01)00314-8. [DOI] [PubMed] [Google Scholar]
- 8.Bartkowiak B, Liu P, Phatnani HP, Fuda NJ, Cooper JJ, Price DH, Adelman K, Lis JT, Greenleaf AL. CDK12 is a transcription elongation-associated CTD kinase, the metazoan ortholog of yeast Ctk1. Genes Dev. 2010;24(20):2303–2316. doi: 10.1101/gad.1968210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003;301(5636):1090–1093. doi: 10.1126/science.1085703. [DOI] [PubMed] [Google Scholar]
- 10.Bengal E, Flores O, Krauskopf A, Reinberg D, Aloni Y. Role of the mammalian transcription factors IIF, IIS, and IIX during elongation by RNA polymerase II. Mol Cell Biol. 1991;11(3):1195–1206. doi: 10.1128/mcb.11.3.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bintu L, Ishibashi T, Dangkulwanich M, Wu Y-Y, Lubkowska L, Kashlev M, Bustamante C. Nucleosomal elements that control the topography of the barrier to transcription. Cell. 2012;151(4):738–749. doi: 10.1016/j.cell.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bintu L, Kopaczynska M, Hodges C, Lubkowska L, Kashlev M, Bustamante C. The elongation rate of RNA polymerase determines the fate of transcribed nucleosomes. Nat Struct Mol Biol. 2011;18(12):1394–1399. doi: 10.1038/nsmb.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bortvin A, Winston F. Evidence That Spt6p Controls Chromatin Structure by a Direct Interaction with Histones. Science. 1996;272(5267):1473–1476. doi: 10.1126/science.272.5267.1473. [DOI] [PubMed] [Google Scholar]
- 14.Brannan K, Kim H, Erickson B, Glover-Cutter K, Kim S, Fong N, Kiemele L, et al. mRNA decapping factors and the exonuclease Xrn2 function in widespread premature termination of RNA polymerase II transcription. Mol Cell. 2012;46(3):311–324. doi: 10.1016/j.molcel.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bregman DB, Halaban R, van Gool AJ, Henning KA, Friedberg EC, Warren SL. UV-induced ubiquitination of RNA polymerase II: a novel modification deficient in Cockayne syndrome cells. Proc Natl Acad Sci U S A. 1996;93(21):11586–11590. doi: 10.1073/pnas.93.21.11586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brody Y, Neufeld N, Bieberstein N, Causse SZ, Böhnlein E-M, Neugebauer KM, Darzacq X, Shav-Tal Y. The in vivo kinetics of RNA polymerase II elongation during co-transcriptional splicing. PLoS Biol. 2011;9(1):e1000573. doi: 10.1371/journal.pbio.1000573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brower-Toland B, Wacker DA, Fulbright RM, Lis JT, Kraus WL, Wang MD. Specific contributions of histone tails and their acetylation to the mechanical stability of nucleosomes. J Mol Biol. 2005;346(1):135–146. doi: 10.1016/j.jmb.2004.11.056. [DOI] [PubMed] [Google Scholar]
- 18.Butler JEF, Kadonaga JT. The RNA polymerase II core promoter: a key component in the regulation of gene expression. Genes Dev. 2002;16(20):2583–2592. doi: 10.1101/gad.1026202. [DOI] [PubMed] [Google Scholar]
- 19.Carey M. PICking apart Pol II initiation. Nat Struct Mol Biol. 2012;19(8):737–738. doi: 10.1038/nsmb.2349. [DOI] [PubMed] [Google Scholar]
- 20.Carey M, Li B, Workman JL. RSC exploits histone acetylation to abrogate the nucleosomal block to RNA polymerase II elongation. Mol Cell. 2006;24(3):481–487. doi: 10.1016/j.molcel.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia W-J, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123(4):581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 22.Cheng B, Li T, Rahl PB, Adamson TE, Loudas NB, Guo J, Varzavand K, et al. Functional association of Gdown1 with RNA polymerase II poised on human genes. Mol Cell. 2012;45(1):38–50. doi: 10.1016/j.molcel.2011.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheung ACM, Cramer P. Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature. 2011;471(7337):249–253. doi: 10.1038/nature09785. [DOI] [PubMed] [Google Scholar]
- 24.Chodosh LA, Fire A, Samuels M, Sharp PA. 5,6-Dichloro-1-beta-D-ribofuranosylbenzimidazole inhibits transcription elongation by RNA polymerase II in vitro. J Biol Chem. 1989;264(4):2250–2257. [PubMed] [Google Scholar]
- 25.Churchman LS, Weissman JS. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2011;469(7330):368–373. doi: 10.1038/nature09652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Close P, East P, Dirac-Svejstrup AB, Hartmann H, Heron M, Maslen S, Chariot A, Söding J, Skehel M, Svejstrup JQ. DBIRD complex integrates alternative mRNA splicing with RNA polymerase II transcript elongation. Nature. 2012;484(7394):386–389. doi: 10.1038/nature10925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Core LJ, Waterfall JJ, Lis JT. Nascent RNA Sequencing Reveals Widespread Pausing and Divergent Initiation at Human Promoters. Science. 2008 doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Core LJ, Waterfall JJ, Gilchrist DA, Fargo DC, Kwak H, Adelman K, Lis JT. Defining the status of RNA polymerase at promoters. Cell Rep. 2012;2(4):1025–1035. doi: 10.1016/j.celrep.2012.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cowling VH, Cole MD. The Myc transactivation domain promotes global phosphorylation of the RNA polymerase II carboxy-terminal domain independently of direct DNA binding. Mol Cell Biol. 2007;27(6):2059–2073. doi: 10.1128/MCB.01828-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Danko CG, Hah N, Luo X, Martins AL, Core LJ, Lis JT, Siepel AA, Kraus WL. Signaling Pathways Differentially Affect RNA Polymerase II Initiation, Pausing, and Elongation Rate in Cells. Mol Cell. 2013 doi: 10.1016/j.molcel.2013.02.015. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Devaiah BN, Lewis BA, Cherman N, Hewitt MC, Albrecht BK, Robey PG, Ozato K, Sims RJ, Singer DS. BRD4 is an atypical kinase that phosphorylates serine2 of the RNA polymerase II carboxy-terminal domain. Proc Natl Acad Sci U S A. 2012;109(18):6927–6932. doi: 10.1073/pnas.1120422109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eberhardy SR, Farnham PJ. Myc recruits P-TEFb to mediate the final step in the transcriptional activation of the cad promoter. J Biol Chem. 2002;277(42):40156–40162. doi: 10.1074/jbc.M207441200. [DOI] [PubMed] [Google Scholar]
- 33.Endoh M, Zhu W, Hasegawa J, Watanabe H, Kim D-K, Aida M, Inukai N, et al. Human Spt6 stimulates transcription elongation by RNA polymerase II in vitro. Mol Cell Biol. 2004;24(8):3324–3336. doi: 10.1128/MCB.24.8.3324-3336.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Epshtein V, Nudler E. Cooperation between RNA polymerase molecules in transcription elongation. Science. 2003;300(5620):801–805. doi: 10.1126/science.1083219. [DOI] [PubMed] [Google Scholar]
- 35.Fuda NJ, Ardehali MB, Lis JT. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature. 2009;461(7261):186–192. doi: 10.1038/nature08449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fujinaga K, Irwin D, Huang Y, Taube R, Kurosu T, Peterlin BM. Dynamics of human immunodeficiency virus transcription: P-TEFb phosphorylates RD and dissociates negative effectors from the transactivation response element. Mol Cell Biol. 2004;24(2):787–795. doi: 10.1128/MCB.24.2.787-795.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gariglio P, Bellard M, Chambon P. Clustering of RNA polymerase B molecules in the 5′ moiety of the adult beta-globin gene of hen erythrocytes. Nucleic Acids Res. 1981;9(11):2589–2598. doi: 10.1093/nar/9.11.2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ghavi-Helm Y, Michaut M, Acker J, Aude J-C, Thuriaux P, Werner M, Soutourina J. Genome-wide location analysis reveals a role of TFIIS in RNA polymerase III transcription. Genes Dev. 2008;22(14):1934–1947. doi: 10.1101/gad.471908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Giardina C, Pérez-Riba M, Lis JT. Promoter melting and TFIID complexes on Drosophila genes in vivo. Genes Dev. 1992;6(11):2190–2200. doi: 10.1101/gad.6.11.2190. [DOI] [PubMed] [Google Scholar]
- 40.Gilchrist DA, Santos Dos G, Fargo DC, Xie B, Gao Y, Li L, Adelman K. Pausing of RNA Polymerase II Disrupts DNA-Specified Nucleosome Organization to Enable Precise Gene Regulation. Cell. 2010;143(4):12–12. doi: 10.1016/j.cell.2010.10.004. Describes how Pol II pausing functionally interacts with the promoter nucleosome structure in Drosophila. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gilmour DS, Lis JT. RNA polymerase II interacts with the promoter region of the noninduced hsp70 gene in Drosophila melanogaster cells. Mol Cell Biol. 1986;6(11):3984–3989. doi: 10.1128/mcb.6.11.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gkikopoulos T, Schofield P, Singh V, Pinskaya M, Mellor J, Smolle M, Workman JL, Barton GJ, Owen-Hughes T. A role for Snf2-related nucleosome-spacing enzymes in genome-wide nucleosome organization. Science. 2011;333(6050):1758–1760. doi: 10.1126/science.1206097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grob P, Cruse MJ, Inouye C, Peris M, Penczek PA, Tjian R, Nogales E. Cryo-electron microscopy studies of human TFIID: conformational breathing in the integration of gene regulatory cues. Structure. 2006;14(3):511–520. doi: 10.1016/j.str.2005.11.020. [DOI] [PubMed] [Google Scholar]
- 44.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130(1):77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guertin MJ, Lis JT. Mechanisms by which transcription factors gain access to target sequence elements in chromatin. Curr Opin Genet Dev. 2012 doi: 10.1016/j.gde.2012.11.008. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hartzog GA, Wada T, Handa H, Winston F. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 1998;12(3):357–369. doi: 10.1101/gad.12.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He N, Chan CK, Sobhian B, Chou S, Xue Y, Liu M, Alber T, Benkirane M, Zhou Q. Human Polymerase-Associated Factor complex (PAFc) connects the Super Elongation Complex (SEC) to RNA polymerase II on chromatin. Proc Natl Acad Sci U S A. 2011;108(36):E636–45. doi: 10.1073/pnas.1107107108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hendrix DA, Hong J-W, Zeitlinger J, Rokhsar DS, Levine MS. Promoter elements associated with RNA Pol II stalling in the Drosophila embryo. Proc Natl Acad Sci U S A. 2008;105(22):7762–7767. doi: 10.1073/pnas.0802406105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hodges C, Bintu L, Lubkowska L, Kashlev M, Bustamante C. Nucleosomal fluctuations govern the transcription dynamics of RNA polymerase II. Science. 2009;325(5940):626–628. doi: 10.1126/science.1172926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu X, Malik S, Negroiu CC, Hubbard K, Velalar CN, Hampton B, Grosu D, Catalano J, Roeder RG, Gnatt AA. A Mediator-responsive form of metazoan RNA polymerase II. Proc Natl Acad Sci U S A. 2006;103(25):9506–9511. doi: 10.1073/pnas.0603702103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ishimi Y, Kikuchi A. Identification and molecular cloning of yeast homolog of nucleosome assembly protein I which facilitates nucleosome assembly in vitro. J Biol Chem. 1991;266(11):7025–7029. [PubMed] [Google Scholar]
- 52.Izban MG, Luse DS. Transcription on nucleosomal templates by RNA polymerase II in vitro: inhibition of elongation with enhancement of sequence-specific pausing. Genes Dev. 1991;5(4):683–696. doi: 10.1101/gad.5.4.683. [DOI] [PubMed] [Google Scholar]
- 53.Jang MK, Mochizuki K, Zhou M, Jeong H-S, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell. 2005;19(4):523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 54.Jishage M, Malik S, Wagner U, Uberheide B, Ishihama Y, Hu X, Chait BT, Gnatt A, Ren B, Roeder RG. Transcriptional Regulation by Pol II(G) Involving Mediator and Competitive Interactions of Gdown1 and TFIIF with Pol II. Mol Cell. 2012;45(1):51–63. doi: 10.1016/j.molcel.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kanazawa S, Soucek L, Evan G, Okamoto T, Peterlin BM. c-Myc recruits P-TEFb for transcription, cellular proliferation and apoptosis. Oncogene. 2003;22(36):5707–5711. doi: 10.1038/sj.onc.1206800. [DOI] [PubMed] [Google Scholar]
- 56.Kasten M, Szerlong H, Erdjument-Bromage H, Tempst P, Werner M, Cairns BR. Tandem bromodomains in the chromatin remodeler RSC recognize acetylated histone H3 Lys14. EMBO J. 2004;23(6):1348–1359. doi: 10.1038/sj.emboj.7600143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kettenberger H, Armache K-J, Cramer P. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol Cell. 2004;16(6):955–965. doi: 10.1016/j.molcel.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 58.Kim JB, Sharp PA. Positive transcription elongation factor B phosphorylates hSPT5 and RNA polymerase II carboxyl-terminal domain independently of cyclin-dependent kinase-activating kinase. J Biol Chem. 2001;276(15):12317–12323. doi: 10.1074/jbc.M010908200. [DOI] [PubMed] [Google Scholar]
- 59.Kim J-H, Lane WS, Reinberg D. Human Elongator facilitates RNA polymerase II transcription through chromatin. Proc Natl Acad Sci U S A. 2002;99(3):1241–1246. doi: 10.1073/pnas.251672198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim T, Buratowski S. Dimethylation of H3K4 by Set1 Recruits the Set3 Histone Deacetylase Complex to 5′ Transcribed Regions. Cell. 2009;137(2):259–272. doi: 10.1016/j.cell.2009.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kostek SA, Grob P, De Carlo S, Lipscomb JS, Garczarek F, Nogales E. Molecular architecture and conformational flexibility of human RNA polymerase II. Structure. 2006;14(11):1691–1700. doi: 10.1016/j.str.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 62.Krishnakumar R, Gamble MJ, Frizzell KM, Berrocal JG, Kininis M, Kraus WL. Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science. 2008;319(5864):819–821. doi: 10.1126/science.1149250. [DOI] [PubMed] [Google Scholar]
- 63.Krogan NJ, Kim M, Tong A, Golshani A, Cagney G, Canadien V, Richards DP, et al. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol Cell Biol. 2003;23(12):4207–4218. doi: 10.1128/MCB.23.12.4207-4218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Krumm A, Meulia T, Brunvand M, Groudine M. The block to transcriptional elongation within the human c-myc gene is determined in the promoter-proximal region. Genes Dev. 1992;6(11):2201–2213. doi: 10.1101/gad.6.11.2201. [DOI] [PubMed] [Google Scholar]
- 65.Kwak H, Fuda NJ, Core LJ, Lis JT. Precise Maps of RNA Polymerase Reveal How Promoters Direct Initiation and Pausing. Science. 2013;339(6122):950–953. doi: 10.1126/science.1229386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.la Mata de M, Alonso CR, Kadener S, Fededa JP, Blaustein M, Pelisch F, Cramer P, Bentley D, Kornblihtt AR. A Slow RNA Polymerase II Affects Alternative Splicing In Vivo. Mol Cell. 2003;12(2):8–8. doi: 10.1016/j.molcel.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 67.Larschan E, Bishop EP, Kharchenko PV, Core LJ, Lis JT, Park PJ, Kuroda MI. X chromosome dosage compensation via enhanced transcriptional elongation in Drosophila. Nature. 2011;471(7336):115–118. doi: 10.1038/nature09757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Larson MH, Zhou J, Kaplan CD, Palangat M, Kornberg RD, Landick R, Block SM. Trigger loop dynamics mediate the balance between the transcriptional fidelity and speed of RNA polymerase II. Proc Natl Acad Sci U S A. 2012;109(17):6555–6560. doi: 10.1073/pnas.1200939109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee C, Li X, Hechmer A, Eisen M, Biggin MD, Venters BJ, Jiang C, Li J, Pugh BF, Gilmour DS. NELF and GAGA factor are linked to promoter-proximal pausing at many genes in Drosophila. Mol Cell Biol. 2008;28(10):3290–3300. doi: 10.1128/MCB.02224-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lenasi T, Peterlin BM, Barboric M. Cap-binding protein complex links pre-mRNA capping to transcription elongation and alternative splicing through positive transcription elongation factor b (P-TEFb) J Biol Chem. 2011;286(26):22758–22768. doi: 10.1074/jbc.M111.235077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, Florens L, Washburn MP, Conaway JW, Conaway RC, Shilatifard A. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol Cell. 2010;37(3):429–437. doi: 10.1016/j.molcel.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu X, Bushnell DA, Silva D-A, Huang X, Kornberg RD. Initiation complex structure and promoter proofreading. Science. 2011;333(6042):633–637. doi: 10.1126/science.1206629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lorch Y, Maier-Davis B, Kornberg RD. Chromatin remodeling by nucleosome disassembly in vitro. Proc Natl Acad Sci U S A. 2006;103(9):3090–3093. doi: 10.1073/pnas.0511050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T. Epigenetics in alternative pre-mRNA splicing. Cell. 2011;144(1):16–26. doi: 10.1016/j.cell.2010.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389(6648):251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 76.Luo Z, Lin C, Shilatifard A. The super elongation complex (SEC) family in transcriptional control. Nat Rev Mol Cell Biol. 2012;13(9):543–547. doi: 10.1038/nrm3417. Reviews the recent progresses on the study of SEC. [DOI] [PubMed] [Google Scholar]
- 77.Mandal SS, Chu C, Wada T, Handa H, Shatkin AJ, Reinberg D. Functional interactions of RNA-capping enzyme with factors that positively and negatively regulate promoter escape by RNA polymerase II. Proc Natl Acad Sci U S A. 2004;101(20):7572–7577. doi: 10.1073/pnas.0401493101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Marshall NF, Price DH. Control of formation of two distinct classes of RNA polymerase II elongation complexes. Mol Cell Biol. 1992;12(5):2078–2090. doi: 10.1128/mcb.12.5.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Marshall NF, Price DH. Purification of P-TEFb, a Transcription Factor Required for the Transition into Productive Elongation. J Biol Chem. 1995;270(21):12335–12338. doi: 10.1074/jbc.270.21.12335. [DOI] [PubMed] [Google Scholar]
- 80.Martinez-Rucobo FW, Sainsbury S, Cheung AC, Cramer P. Architecture of the RNA polymerase–Spt4/5 complex and basis of universal transcription processivity. EMBO J. 2011;30(7):1302–1310. doi: 10.1038/emboj.2011.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mavrich TN, Jiang C, Ioshikhes IP, Li X, Venters BJ, Zanton SJ, Tomsho LP, et al. Nucleosome organization in the Drosophila genome. Nature. 2008;453(7193):358–362. doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Min IM, Waterfall JJ, Core LJ, Munroe RJ, Schimenti J, Lis JT. Regulating RNA polymerase pausing and transcription elongation in embryonic stem cells. Genes Dev. 2011;25(7):742–754. doi: 10.1101/gad.2005511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Missra A, Gilmour DS. Interactions between DSIF (DRB sensitivity inducing factor), NELF (negative elongation factor), and the Drosophila RNA polymerase II transcription elongation complex. Proc Natl Acad Sci U S A. 2010;107(25):11301–11306. doi: 10.1073/pnas.1000681107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Muñoz MJ, la Mata de M, Kornblihtt AR. The carboxy terminal domain of RNA polymerase II and alternative splicing. Trends Biochem Sci. 2010;35(9):497–504. doi: 10.1016/j.tibs.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 85.Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39(12):1507–1511. doi: 10.1038/ng.2007.21. Nature Publishing Group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Narita T, Yamaguchi Y, Yano K, Sugimoto S, Chanarat S, Wada T, Kim D-K, et al. Human transcription elongation factor NELF: identification of novel subunits and reconstitution of the functionally active complex. Mol Cell Biol. 2003;23(6):1863–1873. doi: 10.1128/MCB.23.6.1863-1873.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nechaev S, Fargo DC, Santos dos G, Liu L, Gao Y, Adelman K. Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science. 2010;327(5963):335–338. doi: 10.1126/science.1181421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell. 2003;11(3):709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 89.Nguyen VT, Kiss T, Michels AA, Bensaude O. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature. 2001;414(6861):322–325. doi: 10.1038/35104581. [DOI] [PubMed] [Google Scholar]
- 90.Ni Z, Saunders A, Fuda NJ, Yao J, Suarez J-R, Webb WW, Lis JT. P-TEFb is critical for the maturation of RNA polymerase II into productive elongation in vivo. Mol Cell Biol. 2008;28(3):1161–1170. doi: 10.1128/MCB.01859-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nock A, Ascano JM, Barrero MJ, Malik S. Mediator-regulated transcription through the +1 nucleosome. Mol Cell. 2012;48(6):837–848. doi: 10.1016/j.molcel.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ohler U, Liao GC, Niemann H, Rubin GM. Computational analysis of core promoters in the Drosophila genome. Genome Biol. 2002;3(12):research0087.1–0087.12. doi: 10.1186/gb-2002-3-12-research0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Omichinski JG, Pedone P, Felsenfeld G. The solution structure of a specific GAGA factor-DNA complex reveals a modular binding mode. Nat Struct Biol. 1997;4(2):122–132. doi: 10.1038/nsb0297-122. [DOI] [PubMed] [Google Scholar]
- 94.Orphanides G, LeRoy G, Chang CH, Luse DS, Reinberg D. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell. 1998;92(1):105–116. doi: 10.1016/s0092-8674(00)80903-4. [DOI] [PubMed] [Google Scholar]
- 95.Orphanides G, Wu WH, Lane WS, Hampsey M, Reinberg D. The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature. 1999;400(6741):284–288. doi: 10.1038/22350. [DOI] [PubMed] [Google Scholar]
- 96.Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125(4):703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 97.Perales R, Bentley D. “Cotranscriptionality”: the transcription elongation complex as a nexus for nuclear transactions. Mol Cell. 2009;36(2):178–191. doi: 10.1016/j.molcel.2009.09.018. Reviews co-transcriptional processes and how transcription elongation complex plays a central role in nucleus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Petesch SJ, Lis JT. Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell. 2008;134(1):74–84. doi: 10.1016/j.cell.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Petesch SJ, Lis JT. Activator-induced spread of poly(ADP-ribose) polymerase promotes nucleosome loss at Hsp70. Mol Cell. 2012;45(1):64–74. doi: 10.1016/j.molcel.2011.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Plet A, Eick D, Blanchard JM. Elongation and premature termination of transcripts initiated from c-fos and c-myc promoters show dissimilar patterns. Oncogene. 1995;10(2):319–328. [PubMed] [Google Scholar]
- 101.Pokholok DK, Hannett NM, Young RA. Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol Cell. 2002;9(4):799–809. doi: 10.1016/s1097-2765(02)00502-6. [DOI] [PubMed] [Google Scholar]
- 102.Price DH. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol Cell Biol. 2000;20(8):2629–2634. doi: 10.1128/mcb.20.8.2629-2634.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386(6625):569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 104.Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141(3):432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ramanathan Y, Rajpara SM, Reza SM, Lees E, Shuman S, Mathews MB, Pe’ery T. Three RNA polymerase II carboxyl-terminal domain kinases display distinct substrate preferences. J Biol Chem. 2001;276(14):10913–10920. doi: 10.1074/jbc.M010975200. [DOI] [PubMed] [Google Scholar]
- 106.Rasmussen EB, Lis JT. In vivo transcriptional pausing and cap formation on three Drosophila heat shock genes. Proc Natl Acad Sci U S A. 1993;90(17):7923–7927. doi: 10.1073/pnas.90.17.7923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ratner JN. Ultraviolet Radiation-induced Ubiquitination and Proteasomal Degradation of the Large Subunit of RNA Polymerase II. J Biol Chem. 1998;273(9):5184–5189. doi: 10.1074/jbc.273.9.5184. [DOI] [PubMed] [Google Scholar]
- 108.Rhee HS, Pugh BF. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature. 2012;483(7389):295–301. doi: 10.1038/nature10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ring BZ, Yarnell WS, Roberts JW. Function of E. coli RNA polymerase sigma factor sigma 70 in promoter-proximal pausing. Cell. 1996;86(3):485–493. doi: 10.1016/s0092-8674(00)80121-x. [DOI] [PubMed] [Google Scholar]
- 110.Roeder RG. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996;21(9):327–335. [PubMed] [Google Scholar]
- 111.Rougvie AE, Lis JT. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell. 1988;54(6):795–804. doi: 10.1016/s0092-8674(88)91087-2. [DOI] [PubMed] [Google Scholar]
- 112.Rowley JD. The critical role of chromosome translocations in human leukemias. Annu Rev Genet. 1998;32(1):495–519. doi: 10.1146/annurev.genet.32.1.495. [DOI] [PubMed] [Google Scholar]
- 113.Rufiange A, Jacques P-E, Bhat W, Robert F, Nourani A. Genome-wide replication-independent histone H3 exchange occurs predominantly at promoters and implicates H3 K56 acetylation and Asf1. Mol Cell. 2007;27(3):393–405. doi: 10.1016/j.molcel.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 114.Saeki H, Svejstrup JQ. Stability, flexibility, and dynamic interactions of colliding RNA polymerase II elongation complexes. Mol Cell. 2009;35(2):191–205. doi: 10.1016/j.molcel.2009.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sainsbury S, Niesser J, Cramer P. Structure and function of the initially transcribing RNA polymerase II–TFIIB complex. Nature. 2012;493(7432):437–440. doi: 10.1038/nature11715. [DOI] [PubMed] [Google Scholar]
- 116.Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nat Rev Mol Cell Biol. 2006;7(8):557–567. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- 117.Saunders A, Werner J, Andrulis ED, Nakayama T, Hirose S, Reinberg D, Lis JT. Tracking FACT and the RNA polymerase II elongation complex through chromatin in vivo. Science. 2003;301(5636):1094–1096. doi: 10.1126/science.1085712. [DOI] [PubMed] [Google Scholar]
- 118.Schwabish MA, Struhl K. Asf1 mediates histone eviction and deposition during elongation by RNA polymerase II. Mol Cell. 2006;22(3):415–422. doi: 10.1016/j.molcel.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 119.Shi X, Finkelstein A, Wolf AJ, Wade PA, Burton ZF, Jaehning JA. Paf1p, an RNA polymerase II-associated factor in Saccharomyces cerevisiae, may have both positive and negative roles in transcription. Mol Cell Biol. 1996;16(2):669–676. doi: 10.1128/mcb.16.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Shilatifard A, Lane WS, Jackson KW, Conaway RC, Conaway JW. An RNA Polymerase II Elongation Factor Encoded by the Human ELL Gene. Science. 1996;271(5257):1873–1876. doi: 10.1126/science.271.5257.1873. [DOI] [PubMed] [Google Scholar]
- 121.Shopland LS, Hirayoshi K, Fernandes M, Lis JT. HSF access to heat shock elements in vivo depends critically on promoter architecture defined by GAGA factor, TFIID, and RNA polymerase II binding sites. Genes Dev. 1995;9(22):2756–2769. doi: 10.1101/gad.9.22.2756. [DOI] [PubMed] [Google Scholar]
- 122.Sims RJ, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 2004;18(20):2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- 123.Singh J, Padgett RA. Rates of in situ transcription and splicing in large human genes. Nat Struct Mol Biol. 2009;16(11):1128–1133. doi: 10.1038/nsmb.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Somesh BP, Reid J, Liu W-F, Søgaard TMM, Erdjument-Bromage H, Tempst P, Svejstrup JQ. Multiple mechanisms confining RNA polymerase II ubiquitylation to polymerases undergoing transcriptional arrest. Cell. 2005;121(6):913–923. doi: 10.1016/j.cell.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 125.Spangler L, Wang X, Conaway JW, Conaway RC, Dvir A. TFIIH action in transcription initiation and promoter escape requires distinct regions of downstream promoter DNA. Proc Natl Acad Sci U S A. 2001;98(10):5544–5549. doi: 10.1073/pnas.101004498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Stargell LA, Struhl K. Mechanisms of transcriptional activation in vivo: two steps forward. Trends Genet. 1996;12(8):311–315. doi: 10.1016/0168-9525(96)10028-7. [DOI] [PubMed] [Google Scholar]
- 127.Strobl LJ, Eick D. Hold back of RNA polymerase II at the transcription start site mediates down-regulation of c-myc in vivo. EMBO J. 1992;11(9):3307–3314. doi: 10.1002/j.1460-2075.1992.tb05409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Studitsky VM, Clark DJ, Felsenfeld G. A histone octamer can step around a transcribing polymerase without leaving the template. Cell. 1994;76(2):371–382. doi: 10.1016/0092-8674(94)90343-3. [DOI] [PubMed] [Google Scholar]
- 129.Takahashi H, Parmely TJ, Sato S, Tomomori-Sato C, Banks CAS, Kong SE, Szutorisz H, et al. Human mediator subunit MED26 functions as a docking site for transcription elongation factors. Cell. 2011;146(1):92–104. doi: 10.1016/j.cell.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tan S, Conaway RC, Conaway JW. Dissection of transcription factor TFIIF functional domains required for initiation and elongation. Proc Natl Acad Sci U S A. 1995;92(13):6042–6046. doi: 10.1073/pnas.92.13.6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tirode F, Busso D, Coin F, Egly J-M. Reconstitution of the Transcription Factor TFIIH. Mol Cell. 1999;3(1):87–95. doi: 10.1016/s1097-2765(00)80177-x. [DOI] [PubMed] [Google Scholar]
- 132.Tsukiyama T, Becker PB, Wu C. ATP-dependent nucleosome disruption at a heat-shock promoter mediated by binding of GAGA transcription factor. Nature. 1994;367(6463):525–532. doi: 10.1038/367525a0. [DOI] [PubMed] [Google Scholar]
- 133.Tsukiyama T, Wu C. Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell. 1995;83(6):1011–1020. doi: 10.1016/0092-8674(95)90216-3. [DOI] [PubMed] [Google Scholar]
- 134.Tulin A, Spradling A. Chromatin loosening by poly(ADP)-ribose polymerase (PARP) at Drosophila puff loci. Science. 2003;299(5606):560–562. doi: 10.1126/science.1078764. [DOI] [PubMed] [Google Scholar]
- 135.Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, et al. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 1998;12(3):343–356. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wagschal A, Rousset E, Basavarajaiah P, Contreras X, Harwig A, Laurent-Chabalier S, Nakamura M, et al. Microprocessor, Setx, Xrn2, and Rrp6 co-operate to induce premature termination of transcription by RNAPII. Cell. 2012;150(6):1147–1157. doi: 10.1016/j.cell.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Walfridsson J, Khorosjutina O, Matikainen P, Gustafsson CM, Ekwall K. A genome-wide role for CHD remodelling factors and Nap1 in nucleosome disassembly. EMBO J. 2007;26(12):2868–2879. doi: 10.1038/sj.emboj.7601728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wang D, Severinov K, Landick R. Preferential interaction of the his pause RNA hairpin with RNA polymerase beta subunit residues 904–950 correlates with strong transcriptional pausing. Proc Natl Acad Sci U S A. 1997;94(16):8433–8438. doi: 10.1073/pnas.94.16.8433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wang YV, Tang H, Gilmour DS. Identification in vivo of different rate-limiting steps associated with transcriptional activators in the presence and absence of a GAGA element. Mol Cell Biol. 2005;25(9):3543–3552. doi: 10.1128/MCB.25.9.3543-3552.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Winston F, Chaleff DT, Valent B, Fink GR. Mutations affecting Ty-mediated expression of the HIS4 gene of Saccharomyces cerevisiae. Genetics. 1984;107(2):179–197. doi: 10.1093/genetics/107.2.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Winston F. 47 Analysis of SPT Genes: A Genetic Approach toward Analysis of TFIID, Histones, and Other Transcription Factors of Yeast. Cold Spring Harbor Monograph Archive. 1992;22B(0):1271–1293. [Google Scholar]
- 142.Xin H, Takahata S, Blanksma M, McCullough L, Stillman DJ, Formosa T. yFACT induces global accessibility of nucleosomal DNA without H2A-H2B displacement. Mol Cell. 2009;35(3):365–376. doi: 10.1016/j.molcel.2009.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Yamada T, Yamaguchi Y, Inukai N, Okamoto S, Mura T, Handa H. P-TEFb-mediated phosphorylation of hSpt5 C-terminal repeats is critical for processive transcription elongation. Mol Cell. 2006;21(2):227–237. doi: 10.1016/j.molcel.2005.11.024. [DOI] [PubMed] [Google Scholar]
- 144.Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. NELF, a Multisubunit Complex Containing RD, Cooperates with DSIF to Repress RNA Polymerase II Elongation. Cell. 1999;97(1):11–11. doi: 10.1016/s0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- 145.Yamaguchi Y, Inukai N, Narita T, Wada T, Handa H. Evidence that negative elongation factor represses transcription elongation through binding to a DRB sensitivity-inducing factor/RNA polymerase II complex and RNA. Mol Cell Biol. 2002;22(9):2918–2927. doi: 10.1128/MCB.22.9.2918-2927.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Yang Z, Zhu Q, Luo K, Zhou Q. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature. 2001;414(6861):317–322. doi: 10.1038/35104575. [DOI] [PubMed] [Google Scholar]
- 147.Yang Z, Yik JHN, Chen R, He N, Jang MK, Ozato K, Zhou Q. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell. 2005;19(4):535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 148.Yao J, Ardehali MB, Fecko CJ, Webb WW, Lis JT. Intranuclear distribution and local dynamics of RNA polymerase II during transcription activation. Mol Cell. 2007;28(6):978–990. doi: 10.1016/j.molcel.2007.10.017. [DOI] [PubMed] [Google Scholar]
- 149.Yudkovsky N, Ranish JA, Hahn S. A transcription reinitiation intermediate that is stabilized by activator. Nature. 2000;408(6809):225–229. doi: 10.1038/35041603. [DOI] [PubMed] [Google Scholar]
- 150.Yunger S, Rosenfeld L, Garini Y, Shav-Tal Y. Single-allele analysis of transcription kinetics in living mammalian cells. Nat Methods. 2010;7(8):631–633. doi: 10.1038/nmeth.1482. [DOI] [PubMed] [Google Scholar]
- 151.Zeitlinger J, Stark A, Kellis M, Hong J-W, Nechaev S, Adelman K, Levine M, Young RA. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39(12):1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zippo A, De Robertis A, Serafini R, Oliviero S. PIM1-dependent phosphorylation of histone H3 at serine 10 is required for MYC-dependent transcriptional activation and oncogenic transformation. Nat Cell Biol. 2007;9(8):932–944. doi: 10.1038/ncb1618. [DOI] [PubMed] [Google Scholar]
- 153.Zippo A, Serafini R, Rocchigiani M, Pennacchini S, Krepelova A, Oliviero S. Histone crosstalk between H3S10ph and H4K16ac generates a histone code that mediates transcription elongation. Cell. 2009;138(6):1122–1136. doi: 10.1016/j.cell.2009.07.031. [DOI] [PubMed] [Google Scholar]
- 154.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478(7370):524–528. doi: 10.1038/nature10334. Demonstrates how the mechanisms of transcriptional elongation can be directly linked to medicine. [DOI] [PMC free article] [PubMed] [Google Scholar]
Related Resources
- Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet. 2012;13(10):720–731. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermejo R, Lai MS, Foiani M. Preventing replication stress to maintain genome stability: resolving conflicts between replication and transcription. Mol Cell. 2012;45(6):710–718. doi: 10.1016/j.molcel.2012.03.001. [DOI] [PubMed] [Google Scholar]
- Petesch SJ, Lis JT. Overcoming the nucleosome barrier during transcript elongation. Trends Genet. 2012;28(6):285–294. doi: 10.1016/j.tig.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selth LA, Sigurdsson S, Svejstrup JQ. Transcript Elongation by RNA Polymerase II. Annu Rev Biochem. 2010;79:271–293. doi: 10.1146/annurev.biochem.78.062807.091425. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Li T, Price DH. RNA Polymerase II Elongation Control. Annu Rev Biochem. 2012;81(1):119–143. doi: 10.1146/annurev-biochem-052610-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]