Abstract
Transcription regulation occurs frequently through promoter-associated pausing of RNA polymerase II (Pol II). We developed a Precision nuclear Run-On and sequencing assay (PRO-seq) to map the genome-wide distribution of transcriptionally-engaged Pol II at base-pair resolution. Pol II accumulates immediately downstream of promoters, at intron-exon junctions that are efficiently used for splicing, and over 3' poly-adenylation sites. Focused analyses of promoters reveal that pausing is not fixed relative to initiation sites nor is it specified directly by the position of a particular core promoter element or the first nucleosome. Core promoter elements function beyond initiation, and when optimally positioned they act collectively to dictate the position and strength of pausing. We test this ‘Complex Interaction’ model with insertional mutagenesis of the Drosophila Hsp70 core promoter.
Tracking the accumulation of Pol II along genes reveals potential points of regulation(1). For example, a rate limiting step in early elongation, known as promoter-proximal pausing, has revealed a major regulatory block in the transition to productive elongation in Drosophila and mammals(2–8). Also, less extensive, but significant accumulation of Pol II over the 3’ cleavage/polyA region of genes is proposed to facilitate 3’ processing and transcription termination(9,10). Finally, the interplay of transcription rate and splicing efficiency(11) might be reflected in the selective accumulation of Pol II at splice junctions.
Promoter-associated Pol II pausing is a culmination of intrinsic interactions between Pol II and the underlying DNA, as well as extrinsic stabilization by protein complexes(12). Protein factors such as Negative Elongation Factor (NELF) and DRB Sensitivity Inducing Factor (DSIF)(3,13), DNA elements(14,15), DNA sequence composition(16), nascent RNA processing(16), and nucleosomes(17) can influence pausing. Understanding how these elements and factors function mechanistically requires a high resolution view of their spatial relationship. Current tools for precise tracking of the location and status of Pol II in vivo have distinct limitations(18). ChIP-based methods that collect Pol II or associated RNAs do not distinguish paused Pol II from other Pol II-RNA complexes(16,18,19). The genome-wide nuclear run-on approach (GRO-seq method)(6–8) circumvents these issues by enriching nascent transcripts only associated with actively engaged polymerase with high sensitivity, but it has a resolution of only 30–50 bases(18).
We developed a genome-wide, nuclear run-on assay called PRO-seq that has the sensitivity of GRO-seq, but maps Pol II with base-pair resolution. PRO-seq uses biotin-labeled ribonucleotide triphosphate analogs (biotin-NTP) for nuclear run-on reactions, allowing the efficient affinity purification of nascent RNAs for high throughput sequencing from their 3’ ends (Figs. 1A, S1A). Supplying only one of the four biotin-A/C/G/UTP restricts Pol II to incorporate a single or at most a few identical bases, resulting in sequence reads that have the same 3’ end base within each library (table S1). Moreover, the incorporation of the first biotin-base inhibits further transcript elongation, ensuring base-pair resolution (fig. S2).
Fig. 1.
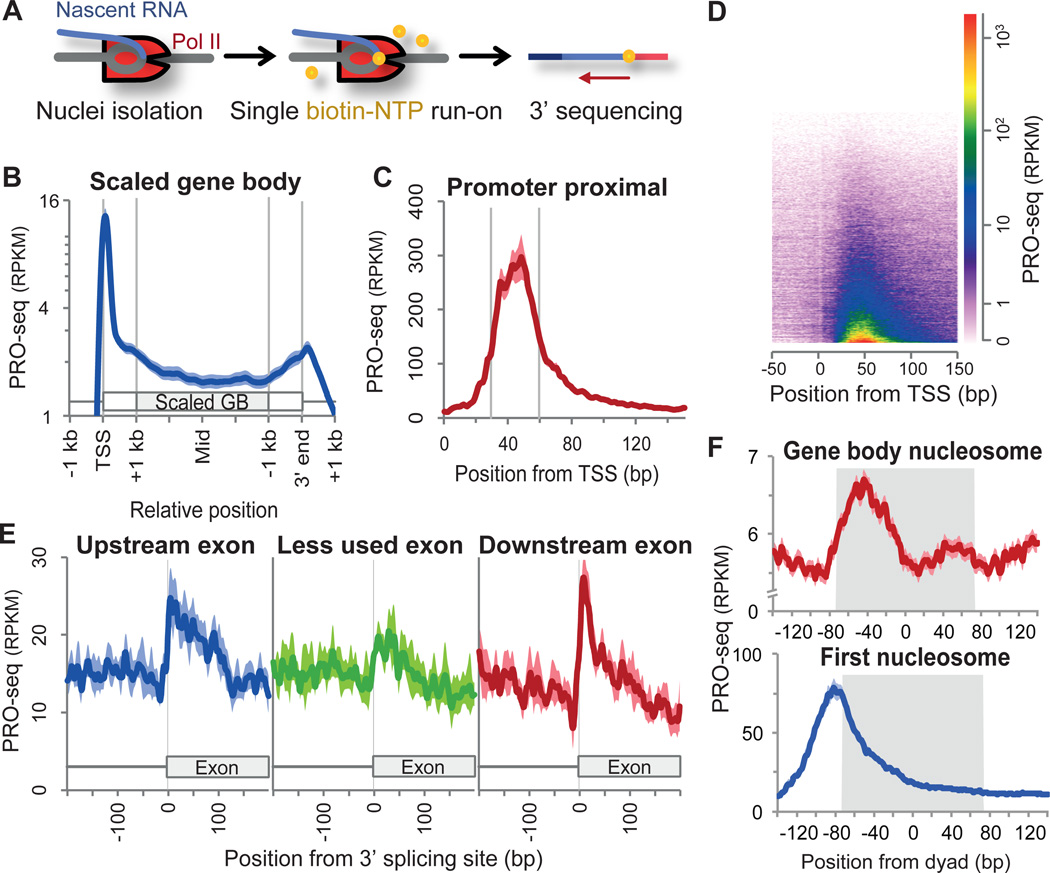
Accumulation of Pol II at promoters, 3’ ends, 3’ splice sites, and nucleosomes. (A) Schematic of PRO-seq. (B) Average PRO-seq profile of non-overlapping genes (n=6,309) for the sense strand. Gene body regions (+1 kb from the 5’ end to −1 kb from the 3’ end) are scaled to 4 kb. Read counts are adjusted to per kilobase per million mapped (RPKM). Shaded margins surrounding the average plot show standard errors of the means. (C) High resolution PRO-seq profile from the TSS to +150 bp (n=16,746). (D) Heatmap visualization of PRO-seq profile of the annotated genes. Genes are arranged by their increasing PRO-seq density. (E) Average PRO-seq profile at 3’ splicing sites of less used and their flanking exons (n=242 each). Less used exons have RNA-seq densities less than 5% of their flanking exons (fig. S5B). (F) Average PRO-seq profile relative to the dyad centers of gene body and first nucleosomes. Region occupied by nucleosome is in grey shades.
The average profile of PRO-seq density (Fig. 1B) reveals pausing of Pol II immediately downstream of the transcription start site (TSS) (Figs. 1C, 1D), and accumulation of Pol II at 3’ cleavage/PolyA sites, consistent with previous GRO-seq studies (Fig. S3)(20, 21). Interestingly, Pol II also accumulates near 3’ splicing sites at spliced exons, but less often at skipped exons (Figs. 1E, S4), suggesting that splicing decisions are connected to differential rates of Pol II elongation through splice junctions(11). Although we have insufficient sequencing coverage to quantify Pol II accumulation at particular 3’ splice sites, our composite analyses support a functional coupling between elongation and splicing.
The highest density of PRO-seq reads map within +30 to +60 from the TSS (Figs. 1C, D), providing a higher resolution view of paused Pol II mapped by GRO-seq(21). Moreover, the pattern of pausing by PRO-seq is consistent with the positions and levels of short nuclear-capped RNAs (scRNAs, fig. S3)(16). Additionally, we demonstrate that PRO-seq maps correspond precisely to positions of engaged Pol II observed in intact cells seen by previous permanganate footprints of transcription bubbles (fig S2G).
Nucleosomes are known to act as barriers to Pol II(12). In the bodies of genes, the average PRO-seq density shows a relative increase at ~ −40 from the previously mapped(22) nucleosome centers (Fig. 1F, S5A). This is consistent with measurements of strong DNA-nucleosome interactions(23) and measured impediments to Pol II transcription through nucleosomes measured in vitro and in yeast(24, 20). However, the PRO-seq density relative to the first (+1) nucleosome is different (Fig. 1F), with the average PRO-seq density at a maximum ~ −80 from the nucleosome centers. Thus, the bulk of promoter-proximal pausing is inconsistent with a standard nucleosome barrier model at least for Drosophila, and is more consistent with tethering of polymerases near the promoter(21).
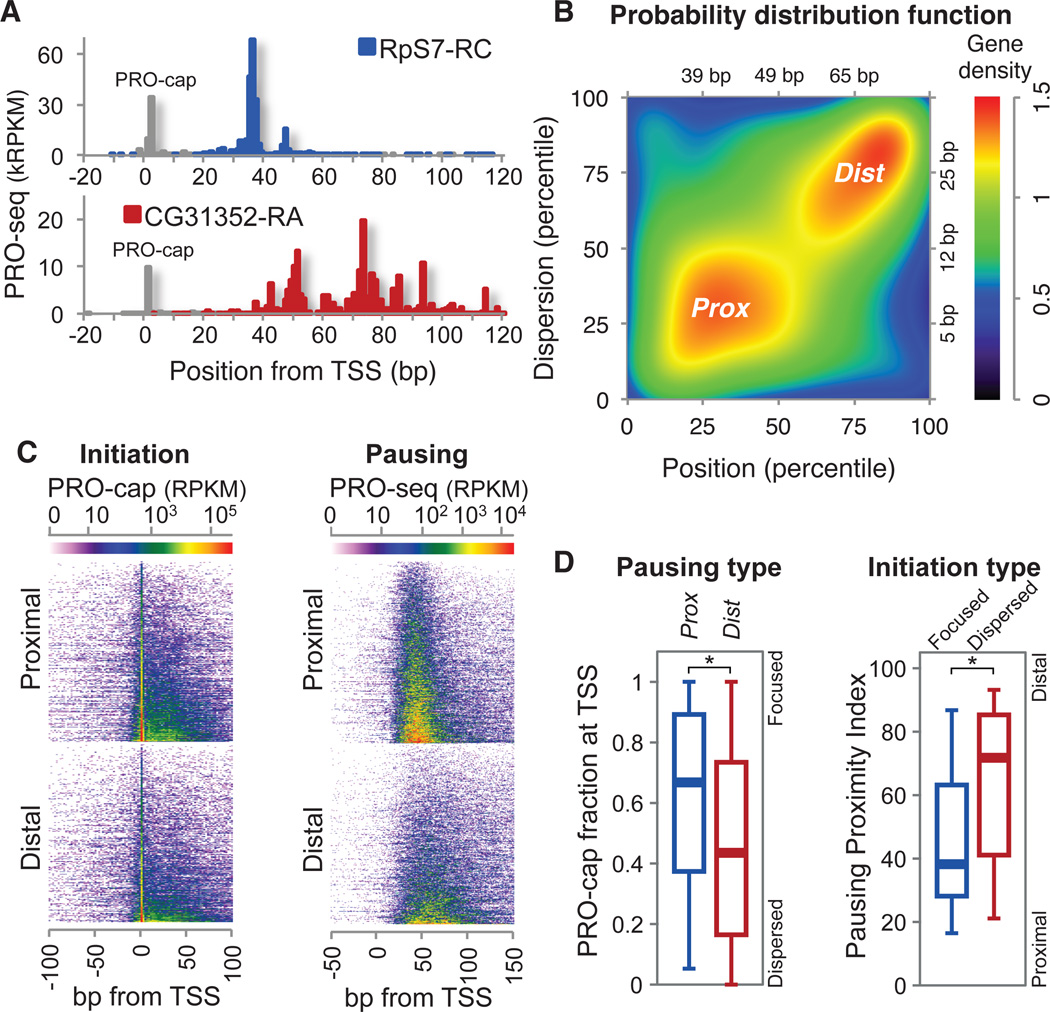
Whereas the average promoter-associated pause location is approximately at +40 from the TSS, pausing is far from uniform. Some genes have more proximal and focused pausing, while others have distal and dispersed pausing (Fig. 2A). We systematically assessed genome-wide pausing positions relative to the TSS and their dispersion to identify two characteristic groups of promoters: focused-proximal (Prox), and dispersed-distal (Dist) promoters (Figs. 2B, S6). The Prox and Dist pausing patterns could arise from a fixed length of elongation from initiation sites that have the same dispersion, or from variable lengths of elongation from more focused initiation sites. Distinguishing between these possibilities requires precise mapping of the initiation sites using the same pool of Pol II-engaged nascent RNAs. Therefore, we modified the PRO-seq method to detect initiation sites (PRO-cap, fig. S1B) and compared the degree of variation in the initiation and the pause sites. We observed that both Prox and Dist genes have relatively focused initiation in general (Figs. 2A, 2E, S2G), and that pausing is overall more dispersed than initiation (fig. S7C). Nonetheless, the degree of the focused initiation, the fraction of initiation arising exactly at a single TSS, is higher for Prox genes, and genes with more focused initiation also have more proximal pausing (Fig. 2D). These findings indicate that although pausing is not fixed to initiation, the mechanisms that produce focused initiation affect the resulting pattern of pausing.
Fig. 2.
Variations of the pause sites and TSSs. (A) Examples of highly paused genes with different pausing patterns. Initiation sites from PRO-cap mapping are shown in grey. (B) Distribution of paused genes (n=3,225) by the pausing position and dispersion percentiles. Focused-proximal (Prox, n=848) and dispersed-distal (Dist, n=846) groups are indicated and axis units in base-pairs are also shown. (C) Heat map of initiation (PRO-cap) and pausing (PRO-seq) for Prox and Dist genes. (D) Association between initiation and pausing patterns. ‘TSS focusing’ in Prox vs Dist genes (left) and ‘Pausing proximity’ in focused vs dispersed initiation genes (right). The fraction of PRO-cap reads at (±1 bp) the TSS over the sum of reads around (±50 bp) the TSS represents the ‘TSS focusing’(22). Focused and dispersed initiation genes are the quartiles of the paused genes with the highest and the lowest TSS focusing respectively. Pausing Proximity Index is defined by the average of pausing position and dispersion percentiles (fig. S6A). Boxes represent 25th, 50th(median), and 75th percentiles; whiskers are 5th and 95th percentiles. Asterisks (*) indicate p<0.001 by Kolmogorov-Smirnov(KS) test.
In an effort to otherwise explain the differential patterns of pausing, we first compared the nucleosome occupancy around Prox and Dist promoters. Prox promoters have less nucleosome occupancy than Dist promoters (fig. S5B), and some Pol II at Dist promoters appear to have more intimate contact with the first nucleosome (Fig S5C). These results (and Fig. 1F) support a nucleosome independent mechanism of pausing for Prox promoters, whereas a subset of Dist promoters could have a component of pausing that is established by direct nucleosome barriers. Because nucleosome position and occupancy does not explain the bulk of Pol II pausing, we investigated the underlying DNA elements around promoters.
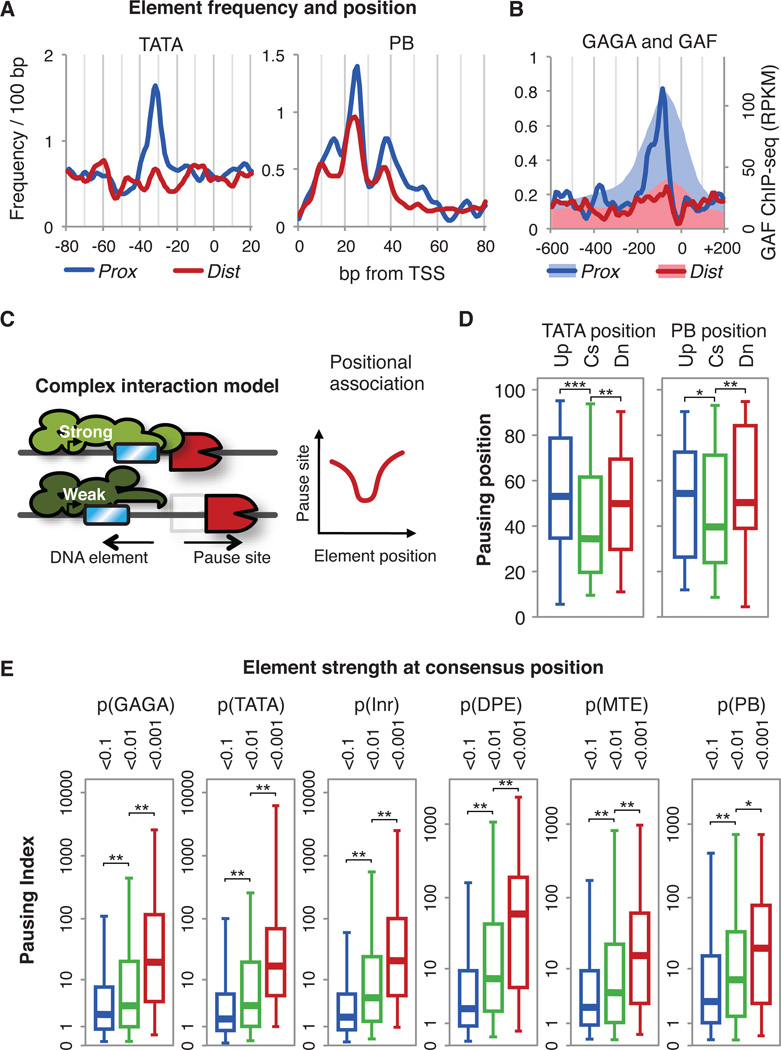
Critical DNA sequence elements within the core promoter direct the position, direction and efficiency of transcription initiation(25). These include the TATA box, Initiator (Inr), Motif Ten Element (MTE), the Downstream Promoter Element (DPE)(25) and a recently discovered element implicated in pausing, the Pause Button(15) (fig. S8A). Core promoter elements are more enriched on Prox than on Dist promoters (Fig. 3A, fig. S8B-D). Additionally, when we searched within the extended promoter regions of Prox and Dist groups for the presence of 232 additional functional DNA elements(26) (fig. S8E), only the GAGA element, residing ~80 bp upstream of the TSS(3,15) shows strong associations with Prox genes (Fig. 3B), as does the level of GAGA-factor binding(27). Thus, core promoter elements and GAGA-factor appear to play a significant role in the mechanism of pausing.
Fig. 3.
Relationship between promoter DNA elements and Pol II pausing. (A) Frequency of TATA box and Pause Button (PB) in Prox and Dist subsets. The average frequency per gene is shown. (B) Frequency of GAGA element (lines) and GAF binding (shades) [27] in Prox and Dist subsets. (C) ‘Complex interaction model’ between DNA elements and paused Pol II. The DNA elements (blue) are at their consensus (Strong) or slightly upstream (Weak) positions and the expected changes of the pausing positions are plotted. (D) Pattern of the positional association between DNA elements and pausing positions. Pausing position percentiles are shown for gene subsets by the element positions (Cs: optimal consensus position, Up: upstream, Dn: downstream; subset information in table S2). Asterisks indicate p-values for the KS test(***: p<0.002, **: p<0.06, *: p<0.14). (E) Association of promoter DNA element strength at consensus positions with Pausing Index(6). Active genes (n=5,471) are divided into three subsets according to the distance-weighted p-values of the DNA elements to the consensus positions (table S3). Asterisks indicate p-values for the KS test (**:p<0.001,*:p<0.01)
Pausing positions could be determined through direct tethering of elongating Pol II to promoter elements. Alternatively, in a ‘Complex Interaction’ model, pausing could be mediated through protein complexes that function best when cognate elements are located at specific positions in the core promoter. Thus, if we examine the association of the positions of the DNA elements and the pausing sites in this model, we expect a ‘V’-shaped plot of association rather than a simple linear correlation: displacement of the element from the optimal position will weaken the interactions within the core complex, resulting in downstream scattering and reduced level of pausing (Fig. 3C). To test this, we examined genes where a particular promoter DNA element occurs only once, and divided genes into three subsets: the optimal consensus position, upstream and downstream. Genes with the DNA elements nearest to the consensus positions have more proximal pausing. Genes with TATA near −30 have more proximal pausing than the genes with TATA at positions of −40 or more, showing a ‘V’-shaped association (Fig. 3D). This ‘V’ pattern was observed in both the upstream elements TATA and Inr (fig. S9A), and the downstream elements PB (Fig. 3D) and MTE (fig. S9B). Also, pausing tends to be stronger in genes with the elements at the optimal positions (fig. S9D). Furthermore, the extent of pausing shows strong dependency on the match of the DNA elements to their consensus sequence and consensus positions (Fig. 3E). Together, these association patterns between core promoter elements and pausing support the ‘Complex Interaction’ model, and explain the strong and focused pausing on Prox promoters .
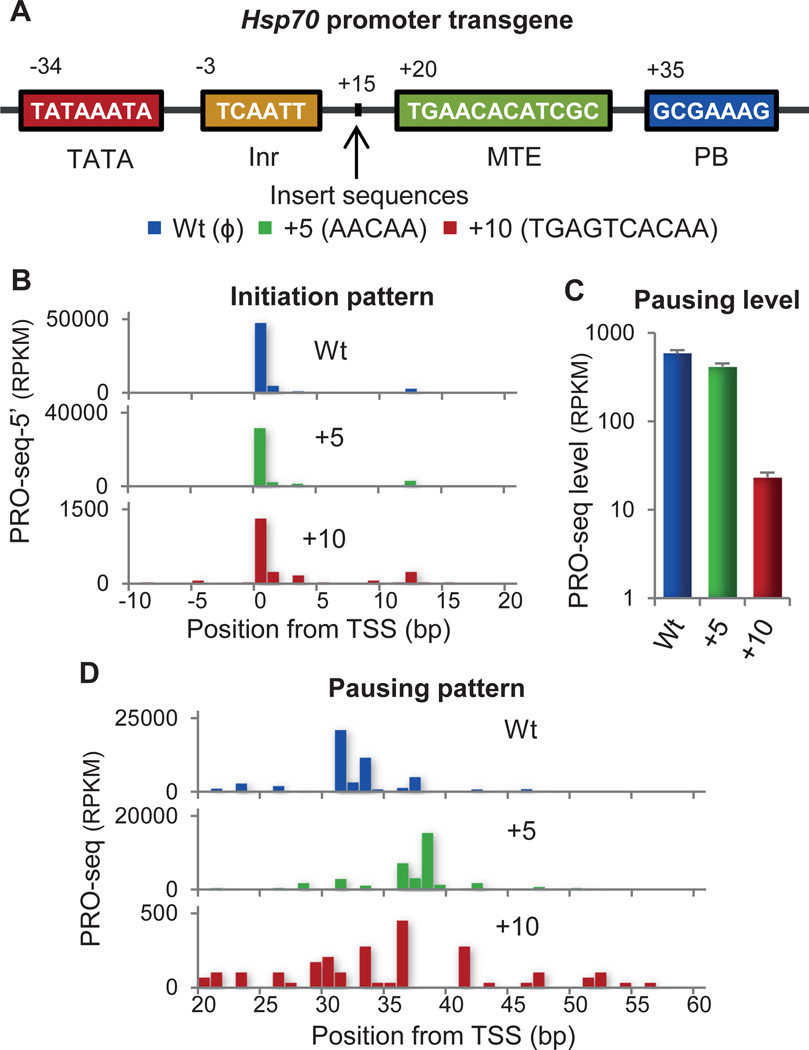
The ‘Complex Interaction’ model depends on both the presence and the correct positioning of core promoter elements. We disrupted the positional relationship of core elements in the well-studied Drosophila gene Hsp70(1). Transgenic fly lines were generated that carry mutant Hsp70 promoters with spacers inserted at the +15 position between the upstream and downstream promoter elements (Fig. 4A), and analyzed by PRO-seq (fig. S10). The initiation sites remain constant in these mutant promoters, indicated by the 5’ ends of the PRO-seq reads (Fig. 4B). The transgenic Hsp70 without spacers inserted shows a strong pause peak mainly at +31 (Fig. 4C). When 5 bp are inserted, the pause peak is shifted 5–7 bp downstream from the original site. The additional bases transcribed before pausing again demonstrates that the position of pausing is not predetermined by elongation distance. When 10 bp are inserted, pausing sites become scattered between +20 to +60 (Fig. 4D) and have fewer reads (Fig 4C). Collectively, these results support the core interaction model and suggest that the interaction complex can accommodate a small change (5 bp) in the positional context of the DNA sequences, but a larger change (10 bp) results in reduced and dispersed pausing.
Fig. 4.
Disruption in the position of downstream DNA elements in Hsp70 promoter. (A) Structure of the Drosophila Hsp70 promoter. Mutant promoters have different length inserts at +15. (B) Initiation patterns at transgenic Hsp70 promoters. 5’ end counts of PRO-seq reads in each transgenic adult fly line are shown to compare the position and pattern of initiation. (C) Level of pausing in transgenic Hsp70 promoters. The sum of the read counts within the pausing region is normalized to the total mapped reads. (D) Positions of pausing in transgenic Hsp70 promoters.
The advances in resolution provided by PRO-seq enabled the precise and genome-wide assessment of the relationship between promoter-proximal pausing and the core promoter structure. For the strong and tightly clustered pausing of the Prox genes, we provide support for a ‘Complex Interaction’ model involving the promoter initiation complex which can extend up to 30 bp from the TSS(28), physically contacting and tethering the pausing complexes. This may share a kinship with bacterial initiation factor σ that is retained within the early elongation complex and interacts with promoter proximal DNA during transcription pausing in E. coli(29). Interestingly the Prox genes are expressed on average at a lower level but show a broader range of expression (fig. S6D), and the Dist genes are enriched in constitutively active genes (table S5). These results suggest that the mechanistic distinctions have regulatory consequences. A well-structured core promoter may strongly recruit Pol II; however, it can also effectively retain Pol II in a paused configuration close to the TSS, until activation signals allow its escape into productive elongation.
Supplementary Material
Acknowledgements
This research was supported by NIH (GM25232 and HG004845 to JTL) and a fellowship from the Howard Hughes Medical Institute (to HK). Sequence data are in Gene Expression Omnibus (GEO) database under accession number GSE42117. Part of this work is included in a broader patent application US patent App. 12/554,472 “Genome-wide Method for Mapping of Engaged RNA Polymerases Quantitatively and at High Resolution”.
References and Notes
- 1.Fuda NJ, Ardehali MB, Lis JT. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature. 2009;461:186. doi: 10.1038/nature08449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rasmussen EB, Lis JT. In vivo transcriptional pausing and cap formation on three Drosophila heat shock genes. Proc. Natl. Acad. Sci. U. S. A. 1993;90:7923. doi: 10.1073/pnas.90.17.7923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee C, et al. NELF and GAGA factor are linked to promoter-proximal pausing at many genes in Drosophila . Mol. Cell. Biol. 2008;28:3290. doi: 10.1128/MCB.02224-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muse GW, et al. RNA polymerase is poised for activation across the genome. Nat. Genet. 2007;39:1507. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zeitlinger J, et al. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat. Genet. 2007;39:1512. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hah N, et al. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell. 2011;145:622. doi: 10.1016/j.cell.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Min IM, et al. Regulating RNA polymerase pausing and transcription elongation in embryonic stem cells. Genes Dev. 2011;25:742. doi: 10.1101/gad.2005511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Core LJ, Lis JT. Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science. 2008;319:1791. doi: 10.1126/science.1150843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glover-Cutter K, et al. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat. Struct. Mol. Biol. 2008;15:71. doi: 10.1038/nsmb1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kornblihtt AR. Chromatin, transcript elongation and alternative splicing. Nat. Struct. Mol. Biol. 2006;13:5. doi: 10.1038/nsmb0106-5. [DOI] [PubMed] [Google Scholar]
- 12.Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nat. Rev. Mol. Cell Biol. 2006;7:557. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- 13.Wu CH, et al. NELF and DSIF cause promoter proximal pausing on the hsp70 promoter in Drosophila . Genes Dev. 2003;17:1402. doi: 10.1101/gad.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu CH, et al. Analysis of core promoter sequences located downstream from the TATA element in the hsp70 promoter from Drosophila melanogaster . Mol. Cell. Biol. 2001;21:1593. doi: 10.1128/MCB.21.5.1593-1602.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hendrix DA, Hong JW, Zeitlinger J, Rokhsar DS, Levine MS. Promoter elements associated with RNA Pol II stalling in the Drosophila embryo. Proc. Natl. Acad. Sci. U. S. A. 2008;105:7762. doi: 10.1073/pnas.0802406105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nechaev S, et al. Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila . Science. 2010;327:335. doi: 10.1126/science.1181421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mavrich TN, et al. Nucleosome organization in the Drosophila genome. Nature. 2008;453:358. doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nechaev S, Adelman K. Pol II waiting in the starting gates: Regulating the transition from transcription initiation into productive elongation. Biochim. Biophys. Acta. 2011;1809:34. doi: 10.1016/j.bbagrm.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Churchman LS, Weissman JS. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2011;469:368. doi: 10.1038/nature09652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Larschan E, et al. X chromosome dosage compensation via enhanced transcriptional elongation in Drosophila . Nature. 2011;471:115. doi: 10.1038/nature09757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Core LJ, et al. Defining the Status of RNA Polymerase at Promoters. Cell Rep. 2012;2:1025. doi: 10.1016/j.celrep.2012.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gilchrist DA, et al. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell. 2010;143:540. doi: 10.1016/j.cell.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hall MA, et al. High-resolution dynamic mapping of histone-DNA interactions in a nucleosome. Nat. Struct. Mol. Biol. 2009;16:124. doi: 10.1038/nsmb.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bondarenko VA, et al. Nucleosomes Can Form a Polar Barrier to Transcript Elongation by RNA Polymerase II. Mol. Cell. 2006;24:469. doi: 10.1016/j.molcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 25.Juven-Gershon T, Hsu JY, Theisen JW, Kadonaga JT. The RNA polymerase II core promoter - the gateway to transcription. Curr. Opin. Cell Biol. 2008;20:253. doi: 10.1016/j.ceb.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stark A, et al. Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature. 2007;450:219. doi: 10.1038/nature06340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharma S, Guertin MJ, Lis JT. The Genomic Binding Profile of GAGA Element Associated Factor (GAF) in Drosophila S2 cells. thesis, Cornell University. 2012 [Google Scholar]
- 28.Emanuel PA, Gilmour DS. Transcription factor TFIID recognizes DNA sequences downstream of the TATA element in the Hsp70 heat shock gene. Proc. Natl. Acad. Sci. U. S. A. 1993;90:8449. doi: 10.1073/pnas.90.18.8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ring BZ, et al. Function of E. coli RNA Polymerase σ Factor σ70 in Promoter-Proximal Pausing. Cell. 1996;86:485. doi: 10.1016/s0092-8674(00)80121-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.