SUMMARY
Recruitment of the Legionella pneumophila effector DrrA to the Legionella-containing vacuole, where it activates and AMPylates Rab1, is mediated by a P4M domain that binds phosphatidylinositol 4-phosphate [PI(4)P] with high affinity and specificity. Despite the importance of PI(4)P in Golgi trafficking and its manipulation by pathogens, the structural bases for PI(4)P-dependent membrane recruitment remain poorly defined. Here, we determined the crystal structure of a DrrA fragment including the P4M domain in complex with dibutyl PI(4)P and investigated the determinants of phosphoinositide recognition and membrane targeting. Headgroup recognition involves an elaborate network of direct and water-mediated interactions with basic and polar residues in the context of a deep, constrictive binding pocket. An adjacent hydrophobic helical element packs against the acyl chains and inserts robustly into PI(4)P-containing monolayers. The structural, biochemical, and biophysical data reported here support a detailed structural mechanism for PI(4)P-dependent membrane targeting by DrrA.
INTRODUCTION
Phosphoinositides derived by phosphorylation of D-myo-phosphatidylinositol at the 3-, 4-, and/or 5-positions play major roles in signal transduction, cytoskeletal architecture, cell migration, and membrane dynamics (Di Paolo and De Camilli, 2006). The spatiotemporal distribution of phosphoinositides is tightly regulated by lipid kinases and phosphatases. Together with small GTPases, these lipids define the identity of cellular compartments and constitute essential signals that direct intracellular membrane trafficking (Behnia and Munro, 2005).
Many proteins contain modular domains that play essential roles in recruitment to intracellular membranes through stereospecific recognition of distinct phosphoinositide headgroups (Moravcevic et al., 2012). The best-studied phosphoinositide recognition modules include Pleckstrin homology (PH); Fab1, YOTB, Vac1, EEA1 (FYVE); and phagocyte oxidase homology (PX) domains. Crystal and nuclear magnetic resonance (NMR) structures have been described for several of these domains in complex with short chain phosphoinositides or headgroups, including the phospholipase C δ PH domain with inositol 1,4,5-trisphosphate (Ferguson et al., 1995), the Grp1 PH domain with inositol 1,3,4,5-tetrakisphosphate (Ferguson et al., 2000; Lietzke et al., 2000), the EEA1 FYVE domain with inositol 1,3-bisphosphate (Dumas et al., 2001; Kutateladze and Overduin, 2001), and the p40Phox PX domain with dibutyl phosphatidy-linositol 3-phosphate [PI(3)P] (Bravo et al., 2001). These and other studies have provided detailed insight into the mechanisms for binding and recognition of phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2], phosphatidylinositol (3,4)-bisphosphate, phosphatidylinositol-(3,4,5)-triphosphate [PI(3,4,5)P3], and PI(3)P. Structural mechanisms for recognition of phosphatidylinositol 4-phosphate [PI(4)P], phosphatidylinositol 5-phosphate [PI(5)P], and phosphatidylinositol 3,5-bisphosphate [PI(3,5)P2], however, remain poorly characterized (Moravcevic et al., 2012).
Synthesized by phosphatidylinositol 4-kinases (PI4Ks), PI(4) P is an important lipid component of the Golgi complex and plasma membrane (PM), and a major regulator of conserved eukaryotic cellular processes, including protein and lipid export from the Golgi to the cell surface, vesicle biogenesis and cargo sorting at the trans-Golgi network, and modulation of sphingolipid biosynthesis (Hammond et al., 2012). Two main classes of proteins that target Golgi membranes in eukaryotic cells have been characterized: adaptor and coat complexes (i.e., AP-1, GGA, and EpsinR) and lipid-transfer proteins (OSBP, CERT, and FAPP) (D’Angelo et al., 2008; Graham and Burd, 2011). Recruitment of these proteins to the Golgi is driven by phosphoinositide binding modules that recognize PI(4)P (e.g., PH, PX, epsin N-terminal homology [ENTH], and GGA and Tom1 [GAT] domains), some of these domains also bind the small GTPase Arf1, thereby increasing their specificity for Golgi membranes (D’Angelo et al., 2008; Moravcevic et al., 2012).
The Legionella pneumophila effector DrrA, also known as SidM, contains a novel PI(4)P binding module, the PI(4)P binding module (P4M) domain, that has high affinity for PI(4)P (Brombacher et al., 2009; Schoebel et al., 2010). DrrA is a guanine nucleotide exchange factor (GEF) specific for the Rab1 GTPase (Ingmundson et al., 2007; Machner and Isberg, 2006, 2007; Murata et al., 2006) and also has adenylyl transferase (ATase) activity for several Rab GTPases, including Rab1 (Müller et al., 2010). Rab1 manipulation by DrrA and at least five other L. pneumophila effectors redirects vesicular trafficking to the nascent Legionella-containing vacuole (LCV) within infected host cells (Hilbi and Haas, 2012; Neunuebel and Machner, 2012).
Crystal structures of eukaryotic PI(4)P binding domains and of DrrA constructs that include the P4M domain have been reported previously (He et al., 2011; Schoebel et al., 2010; Stahelin et al., 2007; Wood et al., 2009; Zhu et al., 2010). Although these structures define the fold and locate the PI(4)P binding site, they contain either sulfate ions or no ligand in the binding pocket. Thus, the structural basis for stereospecific recognition of PI(4)P and membrane targeting remains unclear. Here, we determined the crystal structure of a DrrA fragment spanning the GEF and P4M domains in complex with dibutyl PI(4)P. We demonstrate that DrrA binds with unprecedentedly high affinity to PI(4)P-containing membranes. In addition, DrrA exhibits distinctively strong PI(4)P-dependent membrane insertion. With the use of structure-based mutagenesis, we establish that both tight headgroup coordination in a deep binding pocket and insertion of a hydrophobic helical element into the membrane bilayer contribute to the exceptionally high binding affinity and penetrating power of DrrA for PI(4)P-containing membranes.
RESULTS
Phosphoinositide Binding and Recognition
To investigate the phosphoinositide binding specificity of DrrA in a membrane environment, cosedimentation was used to profile the partitioning of a construct spanning the GEF and P4M domains (DrrA321–647) with sucrose-loaded large unilamellar vesicles ([LUVs], hereafter liposomes) containing a neutral 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) background and 3 mol % of each of the seven phosphoinositides. As shown in Figure 1A, >90% of DrrA321–647 partitioned with the pellet after ultracentrifugation with PI(4)P-containing liposomes over the entire range of phospholipid concentrations (0.18–0.9 mM). Substantially lower partitioning, ranging from 25% to 80% depending on the lipid concentration, was observed for other phosphoinositides. For PI(3,4,5)P3 or POPC alone, the fraction in the pellet was similar to that in the absence of phospholipids. These results show that DrrA321–647 exhibits high affinity and specificity for PI(4)P present at a low mole fraction in neutral POPC membranes, implying a stereoselective mode of headgroup recognition.
Figure 1. Specificity and Affinity of DrrA Binding to PI(4)P.
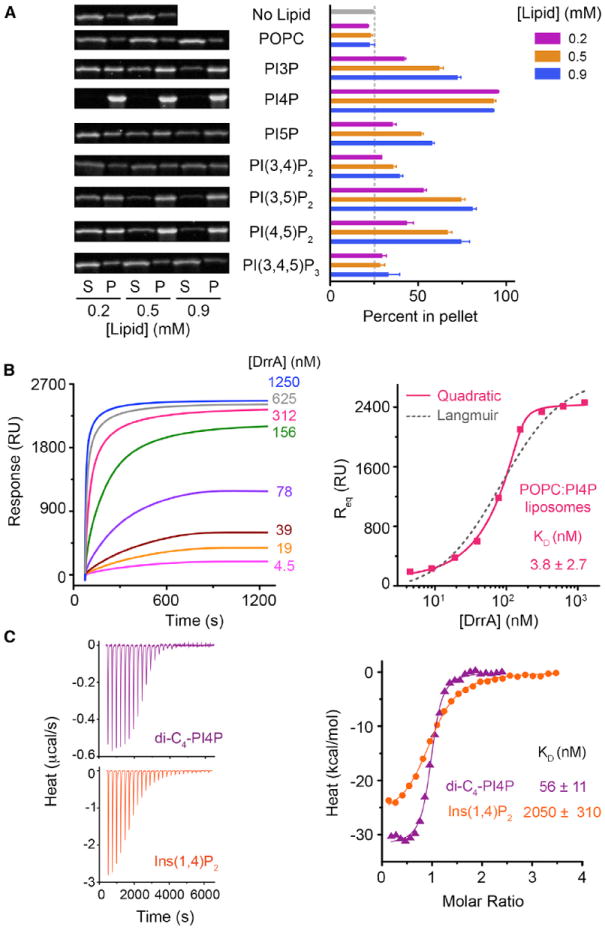
(A) DrrA321–647 partitioning with sucrose-loaded POPC LUVs containing 3% of the indicated phosphoinositides at three lipid concentrations. Values and error bars are mean and SD (n = 3). Gels show representative supernatant (S) and pellet (P) fractions at the indicated lipid concentrations.
(B) SPR analysis of DrrA321–647 association with POPC LUVs containing 3% PI(4)P. Left: representative sensorgrams after curve alignment and reference subtraction. Right: equilibrium responses (Req) from the data on the left as a function of [DrrA321–647]. Solid and dotted lines are fitted quadratic and Langmuir binding models, respectively. RU, resonance units. See also Figure S1 and Table S1.
(C) Binding of WT DrrA321–647 to dibutyl PI(4)P (purple) and Ins(1,4)P2 (orange) determined by ITC at 20°C. Left: baseline-corrected titration data. Right, fitted 1:1 binding models. KD values are mean ± SD (n = 3 or 4).
Binding of DrrA321–647 to PI(4)P-containing liposomes was quantified by surface plasmon resonance (SPR) and showed concentration-dependent association that approached equilibrium on the time scale of the injection. The equilibrium response (Req) obtained from the average response at the end of the injection saturated at protein concentrations in the nanomolar range (Figure 1B). Interestingly, the concentration dependence of Req deviated markedly from the Langmuir model; however, the data were well described by a quadratic binding model, a model that is the exact analytical solution for fraction bound for the standard two-state binding model (see Experimental Procedures). Consistent with the cosedimentation data, where full partitioning was observed at the lowest phospholipid concentration, the fitted dissociation constant (KD = 3.8 ± 2.7 nM) is 40-fold lower than the fitted PI(4)P concentration (150 ± 52 nM). An equivalent KD value of 3.7 nM was obtained when KD was treated as a global parameter in a simultaneous fit (Figure S1A available online). The KD for PI(4)P-containing liposomes is exceptionally low compared with that of other PI(4)P binding domains (Table S1). For example, the binding of the FAPP1 PH domain under similar conditions follows a Langmuir isotherm with KD values 100- to 1,000-fold higher than those observed for DrrA, depending on the composition of the liposomes and the mole fraction of PI(4)P (Figures S1C and S1D).
Binding to soluble dibutyl PI(4)P and the corresponding head-group was compared by isothermal titration calorimetry ([ITC]; Figure 1C). Although nearly identical to the value reported previously (Schoebel et al., 2010), the KD of 56 ± 11 nM for dibutyl PI(4) P is 13-fold higher than that determined by SPR yet 36-fold lower than the KD for inositol 1,4-bisphosphate [Ins(1,4)P2]. These surprisingly large differences suggest that PI(4)P binding involves interactions with both the headgroup and diacylglycerol (DAG) moiety as well as distinct protein-lipid interactions dependent on an intact membrane bilayer.
Crystal Structure of a GEF-P4M Construct of DrrA in Complex with Dibutyl PI(4)P
To investigate the structural basis for PI(4)P recognition, the crystal structure of DrrA330–647 in complex with dibutyl PI(4)P was determined at 1.83 Å resolution (Figure 2; Table 1). The structure comprises the GEF (α1–α9) and P4M (α10–α15) domains of DrrA (Figures 2A and 2B). As expected, the overall domain architecture and tertiary structures of the individual domains are similar to those of the unliganded forms (Schoebel et al., 2010; Zhu et al., 2010).
Figure 2. Crystal Structure of DrrA330–647 in Complex with Dibutyl PI(4)P.
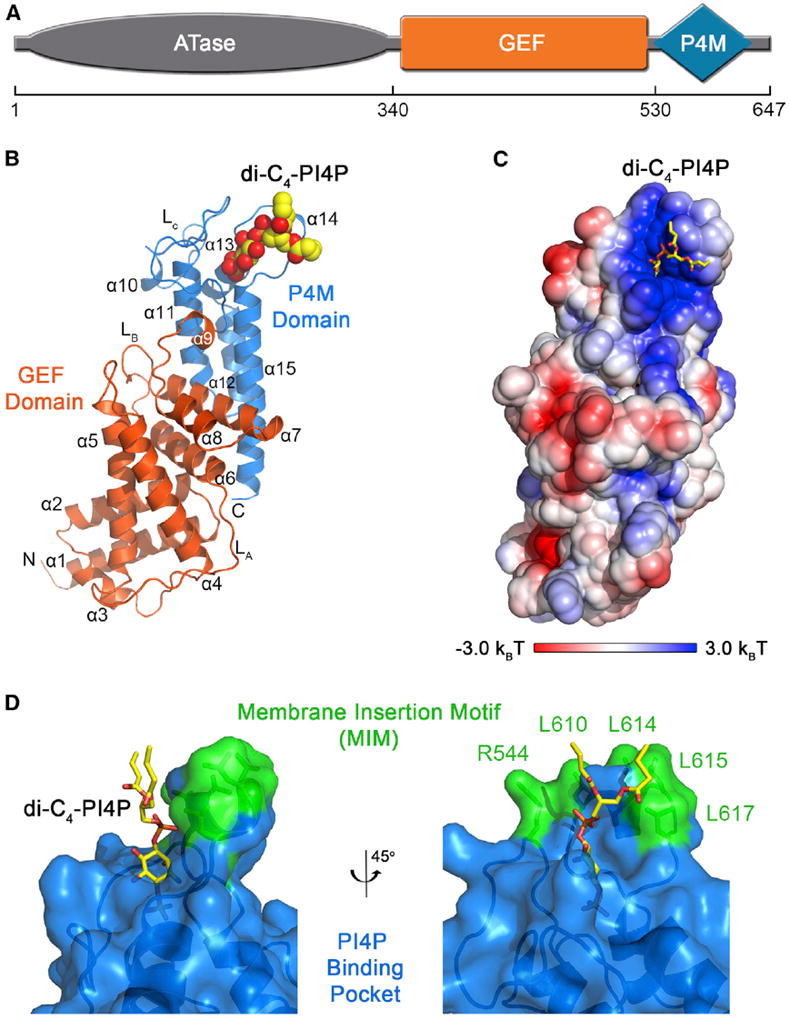
(A) Domain architecture of DrrA.
(B) Overall view with the GEF and P4M domains colored as indicated and dibutyl PI(4)P depicted as spheres. Secondary structural elements are numbered starting with the first helix of the GEF domain. di-C4-PI4P, dibutyl PI(4)P.
(C) Surface representation of the P4M domain colored according to electrostatic potential calculated with APBS (Baker et al., 2001). Dibutyl PI(4)P is shown as sticks.
(D) View of the PI(4)P binding pocket with DrrA rendered as ribbons with a semitransparent surface. Dibutyl PI(4)P and side chains in the putative membrane insertion motif are shown as sticks.
Table 1.
Data Collection and Refinement Statistics
| DrrA330–647-di-C4-PI4P Complex (NSLS X12B)a | |
|---|---|
| Data Collection | |
| Space group | I41 |
| Cell dimensions | |
| a, b, c (Å) | 128.55, 128.55, 53.22 |
| α, β, γ (°) | 90, 90, 90 |
| Resolution (Å) | 1.83 |
| Rsym (%) | 7.9 (54.3) |
| I/σ(I) | 26.3 (2.9) |
| Completeness (%) | 99.98 (99.97) |
| Redundancy | 3.7 (3.5) |
| Refinement | |
| Resolution (Å) | 26.90–1.83 |
| No. of unique reflections | 38,272 |
| Rwork/Rfree (%) | 18.9/20.8 |
| No. of atoms | |
| Protein | 2,465 |
| Ligand | 35 |
| Solvent | 196 |
| Average B-factors (Å2) | |
| Protein | 26.65 |
| Ligand | 22.63 |
| Solvent | 32.61 |
| Root mean square deviations | |
| Bond lengths (Å) | 0.009 |
| Bond angles (°) | 1.18 |
Numbers in parentheses represent values from the highest resolution shell.
The P4M domain is composed of six helices and an ordered loop (LC) connecting to the GEF domain (Figure 2B). Three parallel helices (α11, α12, and α15) form a pillar structure that supports the base of the electropositive binding pocket (Figures 2B and 2C). Near the top of the pillar, residues from helices α10, α13, and α14, together with LC, envelope most of the head-group and pack against one surface of the DAG moiety. Helix α14 extends well above the binding pocket and contains several leucines that are exposed or lie in van der Waals contact with the acyl chains (Figure 2D). Given the high potential for leucine side chains to penetrate into the hydrocarbon core of bilayer membranes (White and Wimley, 1999), these observations suggest that α14 likely functions as a “membrane insertion motif” (MIM).
Structural Basis for High-Affinity PI(4)P Binding and Recognition
Clear electron density for dibutyl PI(4)P allowed a detailed analysis of the interactions involved in PI(4)P recognition (Figure 3A). The 4-phosphate is coordinated through an intricate polar network involving the side chains of two basic (Arg541 and Lys568) and three polar residues (Tyr532, Gln608, and Ser621) in addition to the backbone NH groups of Ser620 and Ser621 (Figure 3B). The 1-phosphate is secured by direct and water-mediated interactions with the backbone NH and side chain hydroxyl groups of Thr611 and Thr612 from α14. The 2- and 3-hydroxyls and the 4-phosphate accept hydrogen bonds from the backbone NH groups of Gly609, Gln608, and Ser620, respectively, whereas the 5-hydroxyl forms hydrogen bonds with the side chains of Ser620 and Arg541 (Figure 3B). Extending the direct interactions, a chain of four water molecules interconnects the DAG moiety with the upper part of the headgroup and His543. In addition to exploiting the hydrogen-bonding potential of the ligand, the polar residues lining the binding pocket define a deep, narrow cavity complementary to the headgroup of PI(4)P, suggesting that other phosphoinositides must bind in suboptimal alternative orientations to avoid steric clashes with residues in α14 and α15.
Figure 3. Structural Basis for PI(4)P Recognition and High-Affinity Binding.
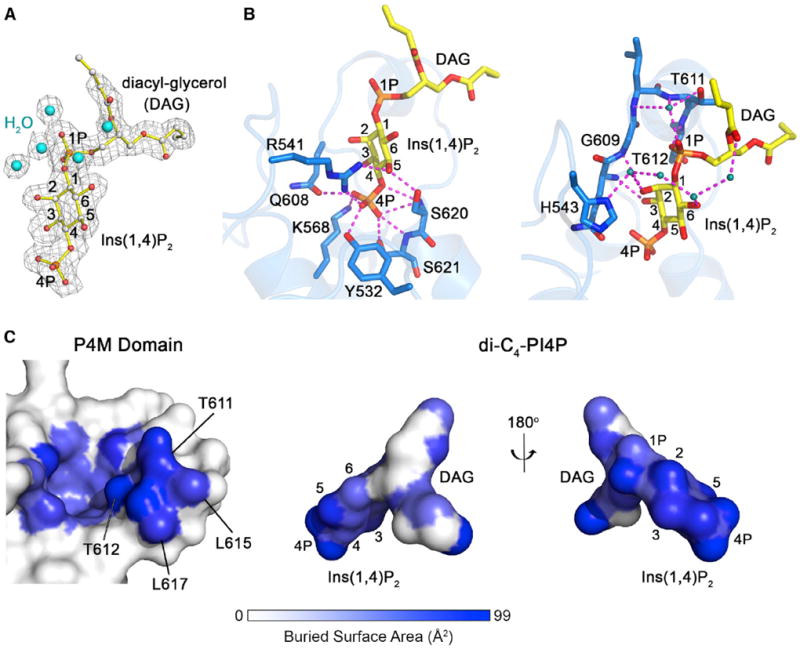
(A) Dibutyl PI(4)P and waters from the final refined model overlaid with electron density from the σA-weighted 2|Fo| - |Fc| map calculated before addition of the ligand/solvent and contoured at 1.0 σ.
(B) Polar contacts between DrrA and the lower (left) and upper (right) regions of dibutyl PI(4)P.
(C) View of the P4M domain and dibutyl PI(4)P colored according to buried surface area upon complex formation.
An accessible surface area of 658 Å2 is buried upon dibutyl PI(4)P binding, with the largest changes involving Thr611, Thr612, and Leu617 in the putative MIM. These residues contact the upper region of the headgroup and proximal surface of the acyl chains (Figure 3C). Thus, van der Waals packing and solvent exclusion from nonpolar surfaces appear to contribute substantially to headgroup specificity as well as the considerably higher affinity for dibutyl PI(4)P compared with Ins(1,4)P2.
In the unliganded structures (Schoebel et al., 2010; Zhu et al., 2010), a common sulfate from the crystallization solution occupies the 4-phosphate pocket, whereas a second sulfate in one structure (Schoebel et al., 2010) occupies a location distinct from the 1-phosphate in the dibutyl PI(4)P complex. Small-to-moderate tertiary structural variations between the various structures appear to reflect crystal packing and suggest that the MIM loop may have some flexibility, at least in the unliganded state. Nevertheless, the structural differences are not obviously related to ligand binding, because the conformation of the MIM in the dibutyl PI(4)P complex is similar to that in one of the unliganded structures (Zhu et al., 2010).
Determinants of PI(4)P Recognition and Membrane Targeting
To further explore the determinants of PI(4)P recognition and membrane targeting, residues in the binding pocket and putative MIM were mutated individually or in combination, and the effects were analyzed by SPR, ITC, or both. A 1:1 quadratic binding model was used to fit the SPR equilibrium responses of high-affinity mutants (KD < 50 nM), whereas the Langmuir binding model was used for weaker binding mutants (KD > 200 nM) (Figure S1B; Table S2).
Alanine substitution of Lys568, which engages the 4-phosphate, caused the largest reduction in affinity for PI(4)P-containing liposomes (2,600-fold; Figure 4; Table S2). Substitution of other 4-phosphate-interacting residues (Y532A, R541A, and Q608R) reduced affinity 150- to 280-fold, whereas substitution of the tandem threonines that contact the 1-phosphate (T611A/T612A) or the tandem serines that contact the 4-phosphate and 5-hydroxyl (S620A/S621A) reduced affinity by 31- and 10-fold, respectively. Mutation of headgroup-coordinating residues also prevented or strongly impaired binding to dibutyl PI(4)P, Ins(1,4)P2, or both (Figure 4; Table S3).
Figure 4. Determinants of PI(4)P Recognition and Membrane Targeting.
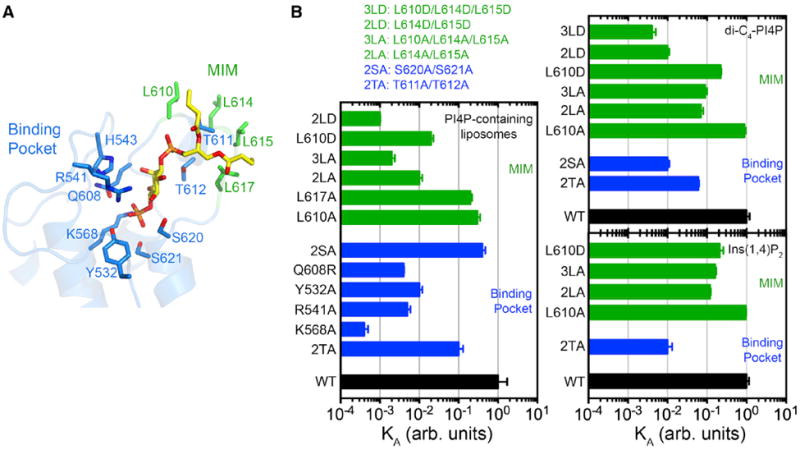
(A) Location of mutated residues.
(B) Affinities (KA) of WT and mutant DrrA321–647 for PI(4)P-containing LUVs measured by SPR (left) and soluble PI(4)P analogs measured by ITC (right). Values and error bars are mean and SD (n = 2–4). arb. units, arbitrary units.
See also Figure S1B and Tables S2 and S3.
The affinity for PI(4)P in liposomes decreased with increasing substitution of exposed leucines in the MIM (Figure 4). Single mutations (L610A and L617A) reduced affinity 4- to 6-fold, whereas double (L614A/L615A; 2LA) and triple (L610A/L614A/L615A; 3LA) mutations reduced affinity by 91- and 440-fold, respectively (Figure 4; Table S2). The effects of these mutations on binding to dibutyl PI(4)P or Ins(1,4)P2 were less dramatic; a single mutation (L610A) maintained wild-type (WT) affinity and double and triple mutations reduced affinity 6-to 14-fold (2LA and 3LA). Stronger effects in both SPR and ITC were observed for aspartic acid substitutions.
These results demonstrate that the majority of residues contacting the head-group contribute substantially to the affinity for PI(4)P and suggest that the exposed leucines in the MIM play an important role in high-affinity binding to PI(4)P-containing membranes, likely by partitioning into the hydrocarbon core.
Biphasic Responses of DrrA in Monolayer Insertion
To explore the ability of DrrA to penetrate phospholipid membranes and assess the contribution of the MIM, monolayer insertion experiments were performed with WT DrrA and the K568A and 3LA variants. Insertion of DrrA into DOPC lipid monolayers containing 20% PI(4)P shows two phases dictated by the initial surface pressure (Πi) (Figure 5A). At low Πi and therefore low packing densities, the equilibrium surface pressure (Πe) is independent of Πi, suggesting little effect of the lipid monolayer on the insertion of DrrA at the air-liquid interface. The surface pressure response at a low Πi is designated as phase I of insertion (Figure 5A). At higher pressures (phase II) the slope changes abruptly, indicating a positive synergistic response of the lipid monolayer in promoting protein insertion that might be explained by a change in anionic charge density and therefore surface potential of the lipid monolayer containing PI(4)P or by an increasing protein-protein interaction when inserted at high enough densities with constrained protein orientation (Calvez et al., 2011).
Figure 5. Biphasic Surface Pressure Responses of DrrA in Monolayer Insertion.
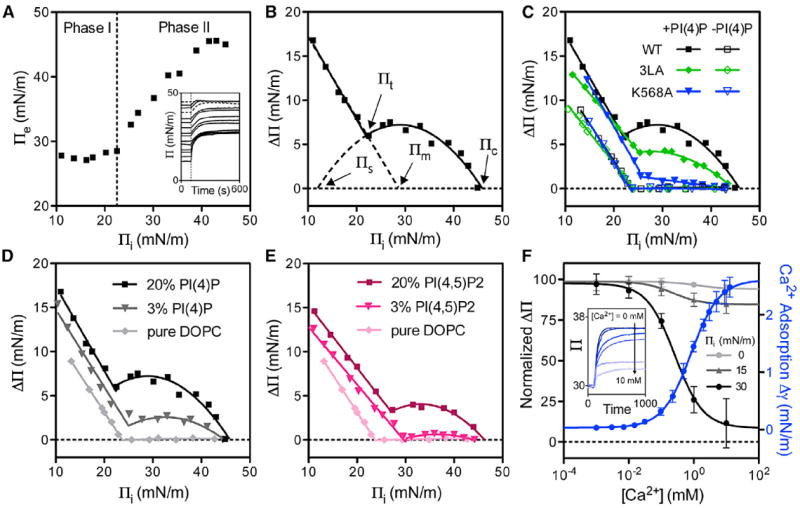
(A) Biphasic insertion of DrrA is indicated by a slope change in a Πe versus Πi plot. Inset: insertion time courses at different Πis; dotted lines are baseline corrected.
(B) The same data set was recast into a ΔΠ versus Πi plot and fitted with a biphasic insertion model (see text).
(C) Monolayer insertion of WT DrrA and the K568A and L610A/614A/615A (3LA) mutants with and without 20 mol % PI(4)P in the lipid monolayer.
(D and E) Monolayer insertion of DrrA at the indicated mole fractions of PI(4)P (D) and PI(4,5)P2 (E).
(F) Normalized Ca2+-inhibited DrrA insertion at different Πis overlaid with Ca2+ affinity measurements determined by surface pressure titration. Inset: insertion time courses of DrrA at different Ca2+ concentrations. Values and errors bars represent mean and SD for two to four independent measurements.
The change in surface pressure (ΔΠ) calculated from the data in Figure 5A is plotted as a function of Πi in Figure 5B. The biphasic behavior can be described by the following equation with all parameters labeled in Figure 5B:
where k1 is a coefficient of phase I insertion that depends on the packing geometry of DrrA at the air-water interface (Zhang et al., 2002); k2 is a phase II insertion coefficient with a unit of (mN/m)−1 and values ranging from 0.01 to 0.03, depending on both the binding affinity and the mechanical work done by DrrA; Πt is the transition surface pressure between phase I and II; Πm is the maximum penetration pressure of phase I insertion; Πc is the critical surface pressure of the system; and Πs is the extrapolated surface pressure threshold at which the phase II insertion starts to occur. A value of Πs smaller than Πt suggests that the insertion mode in phase II happens before the Πt is met, although below Πt it cannot be distinguished from the other insertion mode at Πi < Πt. This model for insertion in phase I is the same as that used in most monolayer insertion studies, but it also describes the surface pressure response in phase II once a threshold Πs is reached. The pressure increase declines as Πi approaches Πc because more free energy is needed to insert the protein into the monolayer.
Insertion of DrrA WT, K568A, and 3LA into DOPC monolayers was compared in the presence or absence of 20% PI(4)P (Figure 5C). In the presence of PI(4)P, the three traces are similar in phase I with similar Πt. In phase II, the 3LA mutation lowers the magnitude of ΔΠ and the K568A mutation almost abolishes insertion. When PI(4)P is absent, the magnitude of ΔΠ in phase I decreases for WT and mutants, and phase II insertion is eliminated. These results show that powerful insertion of DrrA into PI(4)P-containing membranes at physiological surface pressures is PI(4)P-dependent and requires both 4-phosphate recognition by Lys568 and the presence of an intact MIM.
Insertion of DrrA into PI(4)P-containing monolayers depends on the density of PI(4)P in the membrane (Figure 5D). In phase I, an increase in the mol % of PI(4)P has no effect on the slope (k1) but induces an increase in Πm. In phase II, ΔΠmax increases with the PI(4)P mol %, yet DrrA insertion at both 3 and 20 mol % PI(4)P has the same Πc (~46 mN/m), suggesting that this upper limit results from a common generic factor, such as monolayer packing frustration.
To determine whether the biphasic insertion of DrrA is PI(4)P specific, insertion into PI(4,5)P2-containing monolayers was measured at two different PI(4,5)P2 densities. Insertion is dependent on the mol % of PI(4,5)P2 analogous to what was observed with PI(4)P-containing monolayers (Figure 5E). The surface pressure responses in phase I are very similar between the two different lipid compositions; in phase II however, ΔΠmax decreases when PI(4,5)P2 substitutes for PI(4)P (Figure 5E). This difference is consistent with binding specificity to phosphoinositide-containing liposomes and further supports the hypothesis that the insertion of DrrA into lipid membranes depends on stereospecific PI(4)P recognition and not solely on electrostatic attraction, which would be stronger for PI(4,5)P2.
To test whether the biphasic feature of DrrA insertion into membranes is due to a change in physical state of the lipids as suggested previously (Boisselier et al., 2012; Calvez et al., 2009), fluorescence microscopy and compressional elastic modulus analysis of a compression-area isotherm were used in PI(4)P-containing monolayers. The phase transition at Πt was not detected with either of these methods (data not shown). Thus, the Πt in the DrrA monolayer insertion model represents the surface pressure at which phase I and phase II of insertion have equivalent effects on pressure.
Because Ca2+ is likely to perturb the lateral distribution (Ellenbroek et al., 2011; Flanagan et al., 1997; Levental et al., 2009; Wang et al., 2012b) or the electrostatic charge (Wang et al., 2012a) of PI(4)P, and changes in intracellular Ca2+ are often involved in vesicle trafficking, the effect of Ca2+ on DrrA insertion was examined. Surface pressure changes at 15 and 30 mN/m (±0.1 mN/m) were selected to represent DrrA insertion in phase I and phase II, respectively. Membrane partitioning of DrrA by itself without lipid monolayers was also studied. Only DrrA insertion in phase II appears to be strongly Ca2+ sensitive (Figure 5F). Ca2+-inhibited monolayer insertion at 30 mN/m is consistent with the apparent Ca2+ affinity measurement (KD,Ca = 0.82 ± 0.06 mM) determined by surface pressure titration at the same lipid composition and surface pressure, suggesting that Ca2+ inhibition of insertion mainly results from changes in lipid configurations, but not protein conformation (Figure 5F). Although Ca2+ at its physiological concentration (<10 μM) does not affect DrrA insertion in vitro in this simplified system, an effect in vivo cannot be excluded given that Ca2+ affinity to a PI(4)P-containing membrane depends on the surface potential of the membrane that is governed by the local concentrations of PI(4)P and other anionic lipids (Hammond et al., 2012; Toner et al., 1988; Wang et al., 2012b)
In summary, our results suggest that the insertion of DrrA into membranes at low lipid densities results from lipid-excluding penetration of the protein to the air-water interface that is relatively nonspecific but still weakly responsive to the addition of PI(4)P or PI(4,5)P2. In contrast, insertion at high lipid densities characteristic of cellular membranes requires PI(4)P headgroup binding as well as residues in the MIM and is sensitive to the Ca2+-induced change in PI(4)P lateral organization. In contrast to monolayer penetration studies of many other phosphoinositide binding proteins, phosphoinositide-specific and -nonspecific membrane penetration in the case of DrrA can be isolated as two separate phases.
DISCUSSION
In this study, we determined the crystal structure of a PI(4)P-specific binding domain in complex with dibutyl PI(4)P and investigated in detail the mechanism for PI(4)P-dependent membrane association. Three main structural features underlie the specificity and high affinity for PI(4)P-containing membranes: (1) the constricted shape of the binding pocket, which excludes optimal binding modalities for other phosphoinositides; (2) the polar and basic residues in the binding pocket, which extensively coordinate the Ins(1,4)P2 headgroup; and (3) the hydrophobic residues in the MIM, which pack against the DAG moiety, contribute to unusually high penetrating power, and likely partition into the hydrocarbon core.
Similar deep binding pockets have been reported for PH and PX domains that bind phosphoinositides with high affinity and selectivity (Lemmon, 2008). The stereochemistry of interactions with the 1- and 4-phosphates is highly reminiscent of PH domains that use a basic and polar motif with lysine, arginine, histidine, and tyrosine residues for stereospecific phosphate coordination (Lemmon, 2008). Despite lacking sequence or structural homology, the P4M domain deploys a similar combination of basic and polar residues for 4-phosphate recognition, suggesting that the basic/histidine/tyrosine motif in these structurally diverse phosphoinositide-binding domains is a consequence of convergent evolution.
Structural studies of other PI(4)P binding domains have provided insight into the mechanisms of PI(4)P recognition by eukaryotic proteins. A combined crystallographic, NMR, and mutational analysis of PI(4)P binding by the FAPP1 PH domain implicated a cluster of critical basic residues in the canonical pocket that overlay with equivalent residues in the Grp1 PH domain required for phosphoinositide binding (He et al., 2011). Similarly, a putative PI(4)P binding pocket and a cationic patch with strong positive electrostatic potential were identified in the crystal structure of the Bem1p PX domain. An arginine in the binding pocket was found to be essential for binding to PI(4)P-containing liposomes, whereas mutation of other residues that could mediate additional polar interactions caused moderate reductions in affinity (Stahelin et al., 2007). In the structure of GOLPH3, a sulfate ion is located in a positively charged pocket and stabilized by electrostatic interactions with an arginine that together with other basic residues and a conserved tryptophan predicted to coordinate PI(4)P is critical for Golgi targeting (Wood et al., 2009). In each case, at least one basic residue extends from the protein core to a location where the 4-phosphate is expected to dock and thereby resembles Lys568 in DrrA that mediates a key interaction with the 4-phosphate of the bound ligand. Thus, ionic interactions in a solvent-excluded environment appear to be a common feature of PI(4)P recognition.
Crystallographic studies of unliganded DrrA fragments containing the P4M domain located the headgroup binding pocket and identified Lys568 as an important residue for PI(4)P binding (Schoebel et al., 2010; Zhu et al., 2010). The structure of the dibutyl PI(4)P complex provides a complete, detailed description of the binding modality, including all of the elements involved in an elaborate protein-lipid interface. The ITC and SPR measurements reveal a strong dependence on the DAG moiety as well as the context in which the phosphoinositide ligand is recognized. The effects of substitutions demonstrate that high-affinity PI(4)P binding requires not only polar and ionic interactions with the headgroup but also several exposed hydrophobic residues in the MIM. Finally, monolayer insertion studies establish that the MIM is partially responsible for the outstanding PI(4)P-dependent penetrating power at physiological phospholipid densities.
The DrrA MIM represents an elaborated version of insertion elements found in eukaryotic membrane-associating domains, such as FYVE, PX, and C2 (Cho and Stahelin, 2005; Hurley, 2006). In FYVE domains, exposed residues at the tip of a membrane insertion/turret loop (Leu-Leu in Vps27 or Val-Thr in EEA1) penetrate into the hydrocarbon core (Vps27) or interfacial region (EEA1) of the bilayer (Dumas et al., 2001; Misra and Hurley, 1999). The PX domain of p40phox includes a membrane attachment loop that contributes to the binding pocket and contains exposed residues that lie in van der Waals contact with the DAG moiety of the bound dibutyl PI(3)P (Bravo et al., 2001).
Monolayer insertion experiments typically show a linear behavior of ΔΠ as a function of Πi (Lumb et al., 2011; Stahelin et al., 2002, 2007). Strikingly, the experiments presented here show unprecedented biphasic membrane insertion, in which PI(4)P-specific binding occurs at or above physiological lipid densities (Πi > 25 mN/m), with a parabolic behavior of ΔΠ as a function of Πi that differs drastically from the linear shape of nonspecific membrane penetration at low lipid densities (Πi < 25 mN/m). The abrupt change in membrane insertion might be explained by a robust PI(4)P-induced electrostatic switch in the binding pocket, followed by an extraordinary synergistic response to accommodate the numerous hydrophobic side chains of the MIM in the hydrocarbon core of the lipid monolayer (Cho and Stahelin, 2005).
DrrA is the L. pneumophila effector essential for recruitment, activation, and AMPylation of Rab1 at the LCV, initiating a cascade of trafficking processes that remodel the LCV from a PM-derived phagosome into an endoplasmic reticulum (ER)-like replicative organelle. Considering the role of DrrA in the manipulation of the early secretory pathway of host cells, it is perhaps not surprising that the affinity, specificity, and membrane-penetrating power of the P4M domain is exceptional compared with that of eukaryotic PI(4)P binding domains. Indeed, the affinity exceeds that of the OSBP PH domain by an order of magnitude and is at least 50-fold higher than other eukaryotic PI(4)P binding domains (Table S1).
The elaborated structural features that determine the affinity and specificity of the DrrA P4M domain may be explained by selective pressure for localization and function in competition with host PI(4)P binding domains. Recent evidence implicates host phosphoinositide-metabolizing enzymes, such as OCRL1/Dd5P4 and PI4KIIIβ, as well as the L. pneumophila-secreted PI phosphatase SidF, in the production and subsequent turnover of PI(4)P at the LCV (Hilbi et al., 2011; Hsu et al., 2012). Transient accumulation of PI(4)P combined with exquisite PI(4)P-dependent membrane partitioning by the P4M domain likely ensures that DrrA and potentially other L. pneumophila effectors are selectively localized to the LCV during host cell invasion, a process analogous to the recruitment of host factors to membrane compartments in response to extracellular or internal stimuli.
The crystal structure of the DrrA GEF-P4M domain complex with PI(4)P provides the basis for a plausible membrane docking model (Figure 6). Aligning the dibutyl glycerol 1-phosphate moiety of the bound dibutyl PI(4)P with the corresponding elements of a simulated fluid phase phospholipid bilayer places α14 in the interfacial region such that the exposed leucines in the MIM penetrate into the hydrocarbon core. In this orientation, the helical axis is roughly parallel to the plane of the bilayer. Although this docking model is based solely on the disposition of the bound dibutyl PI(4)P, the predicted deep insertion of the MIM is consistent with the higher affinity binding to PI(4)P-containing liposomes, the extraordinary PI(4)P-dependent monolayer penetration power, and the differing effects of mutations in the MIM on the affinity for PI(4)P in a membrane context and in solution. Moreover, superposition with the GEF domain from the structure of a nucleotide-free Rab1-DrrA complex (Schoebel et al., 2009) orients the C terminus of the Rab1 GTPase domain toward the bilayer at a distance readily spanned by many potential configurations of the doubly prenylated hypervariable C-terminal region. A composite model for full-length DrrA based on published structures, small angle X-ray scattering, and other observations (Muller et al., 2012), places the active site of the N-terminal ATase domain at a distance that could be spanned by more extended conformations of the hypervariable C-terminal region (Figure S2). Thus, the membrane docking model appears to be compatible with the function of DrrA in activating as well as AMPylating Rab1 anchored on the same membrane via its doubly prenylated C terminus.
Figure 6. Structure-Based Model for PI(4)P-Dependent Membrane Targeting of DrrA.
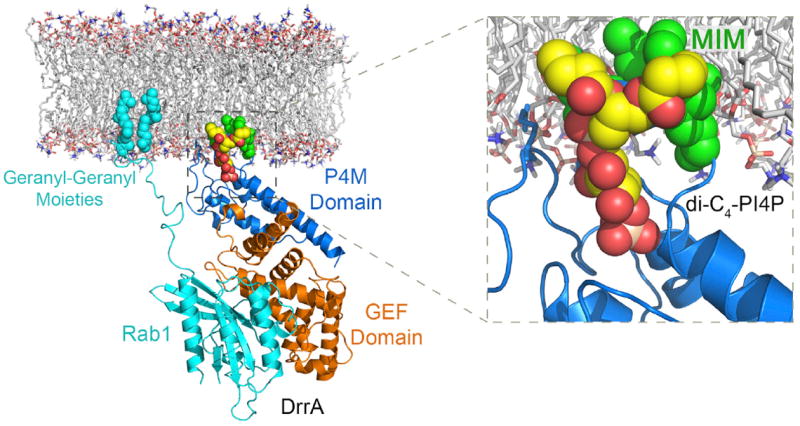
A composite model including Rab1 bound to the GEF domain was generated and docked with a simulated POPC bilayer (see Discussion). The hypervariable C terminus and prenyl groups of Rab1 are modeled in arbitrary but physically plausible conformations.
See also Figure S2.
In addition to PI(4)P-dependent targeting to the LCV, the DrrA P4M domain interacts with PM-derived syntaxins (Arasaki et al., 2012). This interaction may function in concert with Rab1 activation and AMPylation by DrrA to facilitate tethering and fusion of the LCV with ER-derived vesicles. The details of how PM soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs) interact with DrrA and the relationship to PI(4)P binding remain to be determined. Nevertheless, it is likely the membrane-targeting mechanism described here plays an important role in orienting the DrrA-P4M domain for association with SNAREs as well as catalytic interactions with Rab1, thereby contributing to LCV maturation.
EXPERIMENTAL PROCEDURES
Materials, constructs, and methods for expression and purification are described in the Supplemental Experimental Procedures.
Crystallization, Data Collection, and Structure Determination
DrrA330–647 at 10 mg/ml was mixed with a 1.2-fold molar excess of dibutyl PI(4) P in 10 mM Tris (pH 8.0) and 100 mM NaCl and incubated for 1 hr at 4°C. Crystals were grown by vapor diffusion in microseeded hanging drops over 4%–6% polyethylene glycol (PEG) 3350, 100 mM Tris (pH 8.0), and 0.5–1 M NaCl at 18°C. After drop-wise addition of cryostabilizer (20% PEG 3350, 20% glycerol, 0.1 M Tris [pH 8.0], and 0.2 M NaCl), crystals were transferred to fresh drops of cryostabilizer and flash-frozen in liquid nitrogen. A diffraction data set complete to 1.83 Å was collected at the National Synchrotron Light Source (NSLS) X12B beamline, and the structure was solved by molecular replacement using PHASER with a search model derived from the unliganded structure of DrrA317–647 (Protein Data Bank [PDB] code: 3L0M). The model was rebuilt with BUCCANEER (Cowtan, 2006) and refined by simulated annealing with PHENIX (Adams et al., 2010) followed by iterative atom updating with ARP-wARP (Langer et al., 2008), manual rebuilding with COOT (Emsley et al., 2010), and positional/B-factor refinement with REFMAC5 (Murshudov et al., 2011). Dibutyl PI(4)P and solvent molecules were added to the model that was then further improved by iterative refinement with REFMAC5 and manual rebuilding with COOT. Electrostatic potential was calculated with APBS (Baker et al., 2001). Structural figures were rendered with pyMOL (Schrodinger, 2010).
Liposome Partitioning Assays
Partitioning of DrrA321–647 with sucrose-loaded LUVs containing phosphoinositides was assessed by ultracentrifugation (Buser and McLaughlin, 1998). Lipids were mixed in 2:1 chloroform:methanol, dried under argon followed by vacuum for 1 hr, hydrated with buffer (20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES] [pH 7.5] and 170 mM sucrose), disrupted by five freeze-thaw cycles, and extruded through 100 nm filters (Avanti Polar Lipids) to generate LUVs with a mole composition of 97% POPC and 3% dipalmitoyl phosphoinositide. Purified protein (0.6 μM) was incubated with LUVs at total lipid concentrations of 0.18, 0.5, or 0.9 mM for 15 min at 25°C in 20 mM HEPES (pH 7.5) and 0.1 M KCl. Incubation was followed by centrifugation at 100,000 × g for 30 min at 25°C. Normalized volumes of supernatant and resuspended pellet fractions were analyzed by SDS-PAGE. Bands were visualized with Krypton protein stain (Thermo Scientific) using a fluorescent imager (Kodak Image Station 4000MM; excitation wavelength, 520 nm; emission wavelength, 600 nm), and integrated, after background correction (rolling circle, 200 pixel radius), using ImageJ (National Institutes of Health).
Isothermal Titration Calorimetry
Dibutyl PI(4)P and Ins(1,4)P2 were dissolved in ITC buffer (50 mM 3-(N-morpholino)propanesulfonic acid [pH 7.5] and 150 mM NaCl). Proteins were dialyzed and degassed before ITC experiments that were performed at 20°C using a VP-ITC calorimeter (MicroCal) with injections of 10 μl over time intervals of 4 min. For WT DrrA, 5 or 20 μM protein was titrated with 60 or 360 μM dibutyl PI(4)P or Ins(1,4)P2, respectively. For mutants, 20 μM protein was titrated with 360 μM dibutyl PI(4)P or Ins(1,4)P2. After baseline correction, peaks were integrated and fit with a 1:1 binding model (Cronin et al., 2004).
SPR Experiments
SPR experiments were performed at 25°C in HEPES-buffered saline (HBS: 25 mM HEPES [pH 7.5] and 150 mM NaCl) using a BIAcore T100 or 3000 system (GE Healthcare). LUVs prepared in HBS were immobilized on an L1 chip (Narayan and Lemmon, 2006). The chip was primed by consecutive washes with 1% octyl glucoside, 0.5% SDS, and 30% ethanol. POPC LUVs with or without 3% PI(4)P were loaded on the sample and reference channels, respectively. Proteins in HBS were injected at a flow rate of 10 μl/min for 10–20 min, allowing sufficient time for the association reaction to approach Req values. LUVs were washed with 20 μl of 0.1 M NaOH between sample injections. Sensorgrams were obtained for seven to nine concentrations from 10-fold below to 10-fold above the KD value and corrected by subtracting the response of the reference channel. Req values extracted by averaging a region at the end of the association phase were plotted versus protein concentration, and the apparent KD value was determined by nonlinear least-squares fitting with a Langmuir model
or, for the WT protein and high-affinity mutants, a 1:1 quadratic model
KD, Rmax, and [PI4P] were treated as adjustable parameters.
Monolayer Insertion Studies
Monolayer insertion measurements were performed on a DeltaPi tensiometer using a Teflon-coated multiwell plate (Kibron) controlled by the FilmWare 3.57 software package (Kibron). Monolayer subphase composition was 10 mM HEPES, 1 μM EDTA, and 150 mM KCl at pH 6.8 dissolved in 18.2 MΩ double distilled water unless noted otherwise. The system was calibrated without a stir plate but was zeroed with constant stirring using a siliconized miniaturized stir bar (Chrono-log) before lipid deposition. The stir speed and the position of the plate were not changed after zeroing because the probe is sensitive to the change in magnetic field. Premixed lipids were deposited on a 1 ml buffered solution on a multiwell plate. The surface pressure was monitored with a surface probe using the Wilhelmy method (Kates, 1986), after allowing the monolayer to sit for at least 10 min for organic solvent to evaporate. After reaching the target surface pressure, 1–3 μl of concentrated (~10 mg/ml) DrrA stock solution (<0.3% of subphase volume fraction) was injected into the subphase with constant stirring. A final concentration of >500 nM DrrA was used. Multiple injections were performed to make sure the protein concentration reached saturation in terms of surface pressure change. The time course of surface pressure change was recorded over the span of 20–30 min because the surface pressure usually reached a plateau within the first 10 min. Baseline correction was needed in some cases (~10%), mostly when the protein was injected at high Πi (Πi > 38 mN/m) and the protein-induced surface pressure change was relatively small. A baseline was determined by both the point when the protein was injected and the slope of a decaying time course after a plateau was reached.
Ca2+ Binding Affinity Measurements
Similarly to the cation binding affinity studies performed by Ohki (1982) and Ohshima and Ohki (1985), a simplified Ca2+ binding affinity assay was carried out on a MicroTrough XS Langmuir trough (Kibron), and the Ca2+-induced surface pressure drop was analyzed using a Langmuir adsorption model (Levental et al., 2009; Wang et al., 2012b).
Statistical Analysis
Values and errors represent mean ± SD for two to four independent measurements.
Supplementary Material
Acknowledgments
We thank Prof. Bert Van Den Berg and Dr. Elif Eren (University of Massachusetts [UMass] Medical School) and the staff at the NSLS beamline X12B (Brookhaven National Laboratories) for data collection. We also thank Andrew Malaby for critically reading the manuscript and Professors Silvia Corvera and Lawrence Stern (both of UMass Medical School) for sharing lab equipment. This research was supported by National Institutes of Health grants R01 GM056324 (to D.G.L.), R37-AI041699 (to C.R.R.), and R01 DK083592 (to P.A.J. and Y.-H.W.) and National Science Foundation grant DMR-1120901 (to P.A.J. and Y.-H.W.).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, two figures, and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.str.2013.12.018.
ACCESSION NUMBERS
Coordinates and structure factors have been deposited with the PDB under the ID code 4MXP.
References
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arasaki K, Toomre DK, Roy CR. The Legionella pneumophila effector DrrA is sufficient to stimulate SNARE-dependent membrane fusion. Cell Host Microbe. 2012;11:46–57. doi: 10.1016/j.chom.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnia R, Munro S. Organelle identity and the signposts for membrane traffic. Nature. 2005;438:597–604. doi: 10.1038/nature04397. [DOI] [PubMed] [Google Scholar]
- Boisselier E, Calvez P, Demers E, Cantin L, Salesse C. Influence of the physical state of phospholipid monolayers on protein binding. Langmuir. 2012;28:9680–9688. doi: 10.1021/la301135z. [DOI] [PubMed] [Google Scholar]
- Bravo J, Karathanassis D, Pacold CM, Pacold ME, Ellson CD, Anderson KE, Butler PJ, Lavenir I, Perisic O, Hawkins PT, et al. The crystal structure of the PX domain from p40(phox) bound to phosphatidylinositol 3-phosphate. Mol Cell. 2001;8:829–839. doi: 10.1016/s1097-2765(01)00372-0. [DOI] [PubMed] [Google Scholar]
- Brombacher E, Urwyler S, Ragaz C, Weber SS, Kami K, Overduin M, Hilbi H. Rab1 guanine nucleotide exchange factor SidM is a major phosphatidylinositol 4-phosphate-binding effector protein of Legionella pneumophila. J Biol Chem. 2009;284:4846–4856. doi: 10.1074/jbc.M807505200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buser CA, McLaughlin S. Ultracentrifugation technique for measuring the binding of peptides and proteins to sucrose-loaded phospholipid vesicles. Methods Mol Biol. 1998;84:267–281. doi: 10.1385/0-89603-488-7:267. [DOI] [PubMed] [Google Scholar]
- Calvez P, Bussières S, Eric Demers, Salesse C. Parameters modulating the maximum insertion pressure of proteins and peptides in lipid monolayers. Biochimie. 2009;91:718–733. doi: 10.1016/j.biochi.2009.03.018. [DOI] [PubMed] [Google Scholar]
- Calvez P, Demers E, Boisselier E, Salesse C. Analysis of the contribution of saturated and polyunsaturated phospholipid monolayers to the binding of proteins. Langmuir. 2011;27:1373–1379. doi: 10.1021/la104097n. [DOI] [PubMed] [Google Scholar]
- Cho W, Stahelin RV. Membrane-protein interactions in cell signaling and membrane trafficking. Annu Rev Biophys Biomol Struct. 2005;34:119–151. doi: 10.1146/annurev.biophys.33.110502.133337. [DOI] [PubMed] [Google Scholar]
- Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr D Biol Crystallogr. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- Cronin TC, DiNitto JP, Czech MP, Lambright DG. Structural determinants of phosphoinositide selectivity in splice variants of Grp1 family PH domains. EMBO J. 2004;23:3711–3720. doi: 10.1038/sj.emboj.7600388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angelo G, Vicinanza M, Di Campli A, De Matteis MA. The multiple roles of PtdIns(4)P — not just the precursor of PtdIns(4,5)P2. J Cell Sci. 2008;121:1955–1963. doi: 10.1242/jcs.023630. [DOI] [PubMed] [Google Scholar]
- Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- Dumas JJ, Merithew E, Sudharshan E, Rajamani D, Hayes S, Lawe D, Corvera S, Lambright DG. Multivalent endosome targeting by homodimeric EEA1. Mol Cell. 2001;8:947–958. doi: 10.1016/s1097-2765(01)00385-9. [DOI] [PubMed] [Google Scholar]
- Ellenbroek WG, Wang YH, Christian DA, Discher DE, Janmey PA, Liu AJ. Divalent cation-dependent formation of electrostatic PIP2 clusters in lipid monolayers. Biophys J. 2011;101:2178–2184. doi: 10.1016/j.bpj.2011.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson KM, Lemmon MA, Sigler PB, Schlessinger J. Scratching the surface with the PH domain. Nat Struct Biol. 1995;2:715–718. doi: 10.1038/nsb0995-715. [DOI] [PubMed] [Google Scholar]
- Ferguson KM, Kavran JM, Sankaran VG, Fournier E, Isakoff SJ, Skolnik EY, Lemmon MA. Structural basis for discrimination of 3-phosphoinositides by pleckstrin homology domains. Mol Cell. 2000;6:373–384. doi: 10.1016/s1097-2765(00)00037-x. [DOI] [PubMed] [Google Scholar]
- Flanagan LA, Cunningham CC, Chen J, Prestwich GD, Kosik KS, Janmey PA. The structure of divalent cation-induced aggregates of PIP2 and their alteration by gelsolin and tau. Biophys J. 1997;73:1440–1447. doi: 10.1016/S0006-3495(97)78176-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham TR, Burd CG. Coordination of Golgi functions by phosphatidylinositol 4-kinases. Trends Cell Biol. 2011;21:113–121. doi: 10.1016/j.tcb.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond GRV, Fischer MJ, Anderson KE, Holdich J, Koteci A, Balla T, Irvine RF. PI4P and PI(4,5)P2 are essential but independent lipid determinants of membrane identity. Science. 2012;337:727–730. doi: 10.1126/science.1222483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Scott JL, Heroux A, Roy S, Lenoir M, Overduin M, Stahelin RV, Kutateladze TG. Molecular basis of phosphatidylinositol 4-phosphate and ARF1 GTPase recognition by the FAPP1 pleckstrin homology (PH) domain. J Biol Chem. 2011;286:18650–18657. doi: 10.1074/jbc.M111.233015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilbi H, Haas A. Secretive bacterial pathogens and the secretory pathway. Traffic. 2012;13:1187–1197. doi: 10.1111/j.1600-0854.2012.01344.x. [DOI] [PubMed] [Google Scholar]
- Hilbi H, Weber S, Finsel I. Anchors for effectors: subversion of phosphoinositide lipids by Legionella. Front Microbiol. 2011;2:91. doi: 10.3389/fmicb.2011.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu F, Zhu W, Brennan L, Tao L, Luo ZQ, Mao Y. Structural basis for substrate recognition by a unique Legionella phosphoinositide phosphatase. Proc Natl Acad Sci USA. 2012;109:13567–13572. doi: 10.1073/pnas.1207903109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH. Membrane binding domains. Biochim Biophys Acta. 2006;1761:805–811. doi: 10.1016/j.bbalip.2006.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingmundson A, Delprato A, Lambright DG, Roy CR. Legionella pneumophila proteins that regulate Rab1 membrane cycling. Nature. 2007;450:365–369. doi: 10.1038/nature06336. [DOI] [PubMed] [Google Scholar]
- Kates M. Techniques of Lipidology. Second Edition. Amsterdam: Elsevier Science Publishers B. V.; 1986. [Google Scholar]
- Kutateladze T, Overduin M. Structural mechanism of endosome docking by the FYVE domain. Science. 2001;291:1793–1796. doi: 10.1126/science.291.5509.1793. [DOI] [PubMed] [Google Scholar]
- Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protoc. 2008;3:1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- Levental I, Christian DA, Wang YH, Madara JJ, Discher DE, Janmey PA. Calcium-dependent lateral organization in phosphatidylinositol 4,5-bisphosphate (PIP2)- and cholesterol-containing monolayers. Biochemistry. 2009;48:8241–8248. doi: 10.1021/bi9007879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lietzke SE, Bose S, Cronin T, Klarlund J, Chawla A, Czech MP, Lambright DG. Structural basis of 3-phosphoinositide recognition by pleckstrin homology domains. Mol Cell. 2000;6:385–394. doi: 10.1016/s1097-2765(00)00038-1. [DOI] [PubMed] [Google Scholar]
- Lumb CN, He J, Xue Y, Stansfeld PJ, Stahelin RV, Kutateladze TG, Sansom MS. Biophysical and computational studies of membrane penetration by the GRP1 pleckstrin homology domain. Structure. 2011;19:1338–1346. doi: 10.1016/j.str.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machner MP, Isberg RR. Targeting of host Rab GTPase function by the intravacuolar pathogen Legionella pneumophila. Dev Cell. 2006;11:47–56. doi: 10.1016/j.devcel.2006.05.013. [DOI] [PubMed] [Google Scholar]
- Machner MP, Isberg RR. A bifunctional bacterial protein links GDI displacement to Rab1 activation. Science. 2007;318:974–977. doi: 10.1126/science.1149121. [DOI] [PubMed] [Google Scholar]
- Misra S, Hurley JH. Crystal structure of a phosphatidylinositol 3-phosphate-specific membrane-targeting motif, the FYVE domain of Vps27p. Cell. 1999;97:657–666. doi: 10.1016/s0092-8674(00)80776-x. [DOI] [PubMed] [Google Scholar]
- Moravcevic K, Oxley CL, Lemmon MA. Conditional peripheral membrane proteins: facing up to limited specificity. Structure. 2012;20:15–27. doi: 10.1016/j.str.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller MP, Peters H, Blümer J, Blankenfeldt W, Goody RS, Itzen A. The Legionella effector protein DrrA AMPylates the membrane traffic regulator Rab1b. Science. 2010;329:946–949. doi: 10.1126/science.1192276. [DOI] [PubMed] [Google Scholar]
- Muller MP, Shkumatov AV, Oesterlin LK, Schoebel S, Goody PR, Goody RS, Itzen A. Characterization of enzymes from Legionella pneumophila involved in reversible adenylylation of Rab1. J Biol Chem. 2012 doi: 10.1074/jbc.M112.396861. Published online August 7, 2012 http://dx.doi.org/10.1074/jbc.M112.396861. [DOI] [PMC free article] [PubMed]
- Murata T, Delprato A, Ingmundson A, Toomre DK, Lambright DG, Roy CR. The Legionella pneumophila effector protein DrrA is a Rab1 guanine nucleotide-exchange factor. Nat Cell Biol. 2006;8:971–977. doi: 10.1038/ncb1463. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Skubák P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan K, Lemmon MA. Determining selectivity of phosphoinositide-binding domains. Methods. 2006;39:122–133. doi: 10.1016/j.ymeth.2006.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neunuebel MR, Machner MP. The taming of a Rab GTPase by Legionella pneumophila. Small GTPases. 2012;3:28–33. doi: 10.4161/sgtp.18704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohki S. A mechanism of divalent ion-induced phosphatidylserine membrane fusion. Biochim Biophys Acta. 1982;689:1–11. doi: 10.1016/0005-2736(82)90182-1. [DOI] [PubMed] [Google Scholar]
- Ohshima H, Ohki S. Effects of divalent-cations on the surface-tension of a lipid monolayer coated air/water interface. J Colloid Interface Sci. 1985;103:85–94. [Google Scholar]
- Schoebel S, Oesterlin LK, Blankenfeldt W, Goody RS, Itzen A. RabGDI displacement by DrrA from Legionella is a consequence of its guanine nucleotide exchange activity. Mol Cell. 2009;36:1060–1072. doi: 10.1016/j.molcel.2009.11.014. [DOI] [PubMed] [Google Scholar]
- Schoebel S, Blankenfeldt W, Goody RS, Itzen A. High-affinity binding of phosphatidylinositol 4-phosphate by Legionella pneumophila DrrA. EMBO Rep. 2010;11:598–604. doi: 10.1038/embor.2010.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrodinger LLC. The PyMOL Molecular Graphics System. Version 1.3r1 2010 [Google Scholar]
- Stahelin RV, Long F, Diraviyam K, Bruzik KS, Murray D, Cho W. Phosphatidylinositol 3-phosphate induces the membrane penetration of the FYVE domains of Vps27p and Hrs. J Biol Chem. 2002;277:26379–26388. doi: 10.1074/jbc.M201106200. [DOI] [PubMed] [Google Scholar]
- Stahelin RV, Karathanassis D, Murray D, Williams RL, Cho W. Structural and membrane binding analysis of the Phox homology domain of Bem1p: basis of phosphatidylinositol 4-phosphate specificity. J Biol Chem. 2007;282:25737–25747. doi: 10.1074/jbc.M702861200. [DOI] [PubMed] [Google Scholar]
- Toner M, Vaio G, McLaughlin A, McLaughlin S. Adsorption of cations to phosphatidylinositol 4,5-bisphosphate. Biochemistry. 1988;27:7435–7443. doi: 10.1021/bi00419a039. [DOI] [PubMed] [Google Scholar]
- Wang WJ, Anderson NA, Travesset A, Vaknin D. Regulation of the electric charge in phosphatidic acid domains. J Phys Chem B. 2012a;116:7213–7220. doi: 10.1021/jp303840a. [DOI] [PubMed] [Google Scholar]
- Wang YH, Collins A, Guo L, Smith-Dupont KB, Gai F, Svitkina T, Janmey PA. Divalent cation-induced cluster formation by polyphosphoinositides in model membranes. J Am Chem Soc. 2012b;134:3387–3395. doi: 10.1021/ja208640t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White SH, Wimley WC. Membrane protein folding and stability: physical principles. Annu Rev Biophys Biomol Struct. 1999;28:319–365. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]
- Wood CS, Schmitz KR, Bessman NJ, Setty TG, Ferguson KM, Burd CG. PtdIns4P recognition by Vps74/GOLPH3 links PtdIns 4-kinase signaling to retrograde Golgi trafficking. J Cell Biol. 2009;187:967–975. doi: 10.1083/jcb.200909063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Lu YJ, Wang SX, Sui SF. The effects of protein hydrophobic exposure on the slope of the line in the deltapi vs pi(i) plot. IUBMB Life. 2002;54:73–79. doi: 10.1080/15216540214311. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Hu L, Zhou Y, Yao Q, Liu L, Shao F. Structural mechanism of host Rab1 activation by the bifunctional Legionella type IV effector SidM/DrrA. Proc Natl Acad Sci USA. 2010;107:4699–4704. doi: 10.1073/pnas.0914231107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


