Abstract
Considerable information is known about the extracellular signals that drive pluripotent stem cells through a stepwise program to become cardiomyocytes. A paper by Ohtani et al. in this issue now links extrinsic signals to histone demethylation needed to form mesoderm, the first step in cardiogenesis.
Extrinsic signals drive the stepwise differentiation of pluripotent stem cells (PSCs) into cardiomyocytes, and highly efficient protocols exist for producing large numbers of >90% pure cardiomyocytes by judiciously manipulating the timing and concentration of inducing factors that are provided to the cultures1. As elegantly shown recently by Murry, Bruneau and their colleagues2, 3, the activities of extrinsic, growth factor-like molecules such as TGFβ, and Wnt family members and their inhibitors, become transduced into changes in the 3-dimensional structure of chromatin that locally influences expression of cardiogenic genes. Moreover, rules are emerging for how epigenetic modifications to histones (acetylation and methylation) and DNA (methylation) influence chromatin structure and transcription, making it possible to predict whether a gene is active, inactive, or poised on the basis of the patterns of histone marks2. Despite these advances, we have only a rudimentary understanding of how extrinsic signals elicit specific chromatin modifications and control cell fate.
In this issue, Ohtani et al.4 take a first stab at understanding how functional regulation of histone methylation determines whether PSCs become mesoderm (and hence cardiac). The work focuses on the role of Jmjd3, also known as Kdm6b, a DNA demethylase (DM), which removes repressive marks at H3K27 sites, typically placed by the Polycomb complex5. Their finding that genetic deletion of Jmjd3 selectively affects certain mesodermal genes raises the interesting question of how histone demethylases recognize particular sites in the genome and dictate cell fate choice.
Histone demethylation is regulated genome-wide by LSD and JmjC DMs, which together profoundly influence the transcript profile of cells6. While there are only two LSD family members, at least 18 of 30 JmjC-domain proteins are known to be capable of histone demethylation5. All histones can be methylated, but the most common modifications occur on K4, -9, -27, -36 and -79 of histone H3 and K20 of histone H4. Jmjd3 is selective for H3K27 (the only other DM that can demethylate H3K27 is UTX), and removes methyl groups placed by the Polycomb Repressor Complex 2 (PRC2)5. While DMs are selective for the particular methylated residue, on their own, they do not recognize the surrounding sequence. Considering that Jmjd3 is also involved in endoderm differentiation7, it is intriguing to postulate that extrinsic signaling provides contextual cues that dictate the sites in chromatin occupied by Jmjd3 and other DMs.
Pursuing this idea, Ohtani et al. found that Jmjd3 demethylation of H3K27 residues is needed for Wnt-stimulated nuclear β-catenin to activate certain mesoderm genes. Several recent studies in a variety of biological systems have similarly identified additional extracellular cues that link Jmjd3 to other genes (Figure 1)8-14. While selectivity seems unequivocally under extracellular control, the precise mechanism for recruitment is less evident. Although Wnt/β-catenin signaling and Jmjd3 are both needed to induce mesoderm genes4, 10, 15, neither β-catenin nor Jmjd3 can recognize specific DNA sequences, thus sequence-specific transcription factors probably aid in the selective recruitment of Jmjd3 to particular sites in chromatin.
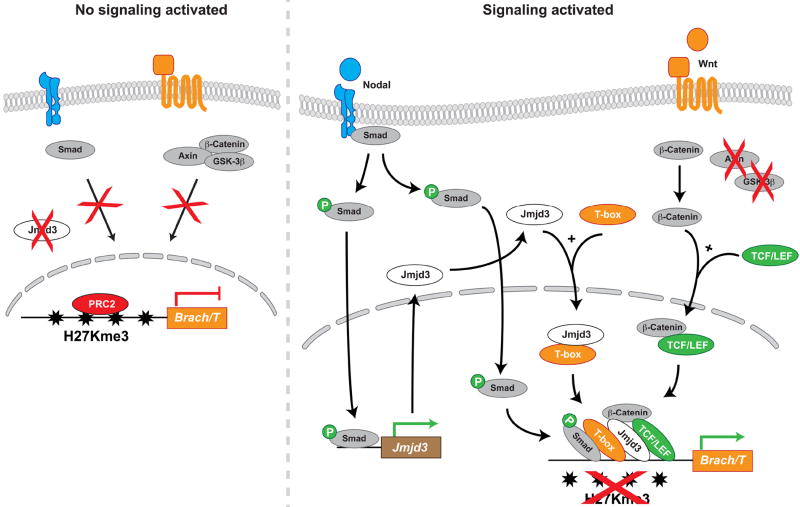
Figure 1. Combinatorial assembly of Jmjd3 histone demethylase with transcription factors directs cell fate.
Context dependent extracellular signals (retinoic acid, Nodal and Wnt, color coded to reflect cell lineages) acting on multipotent progenitors induce specific transcription factors and the histone demethylase Jmjd3. These transcription factors selectively recruit Jmjd3 to particular promoters to activate lineage-specific genes, causing the progenitors adopt particular fates. P, phosphorylation; black arrows, positive effect; green arrows, activation of transcription.
An elegant study by Michael Kuehn and his colleagues10 suggest that Smad2 provides the critical DNA sequence recognition needed to recruit Jmjd3 to T/Brachyury, a key and stereotypical mesodermal gene. They showed that the divergent TGFβ family member Nodal, which phosphorylates Smad2 and -3 and thereby triggers nuclear localization, is needed to bring Jmjd3 to the T/Brachyury locus during mesoderm formation in PSCs. Nodal signaling, in addition, also induces Jmjd3 transcription. Thus, Nodal provides the key input that induces Jmjd3 and provides phosphorylated Smad2/3 proteins that direct it to genes such as T/Brachyury.
Why then are Wnt and β-catenin so important? Ohtani et al. showed that Jmjd3 binds β-catenin directly (and thus associates indirectly with Tcf transcription factors that also recognize DNA). However, Dahle et al.10 found that Wnt signaling alone is unable to remove repressing H3K27 methylation and activate T/Brachyury in the absence of Nodal signaling. Thus, we propose that Jmjd3 participates in a larger complex on the T/Brachyury promoter, and possibly on the promoters of other mesodermal genes, involving both Smad2/3 and β-catenin (Figure 1).
There are a few additional examples of extrinsic cues that direct Jmjd3 to lineage-specific genes. Treating PSCs with high levels of Activin A (to mimic endogenous Nodal) drives Jmjd3 and Smad2/3 to the promoters of endodermal genes7, 9. In this context, however, the high level of Nodal/Activin A signaling, in the absence of Wnt, induces the T-box transcription factors Tbx3 or Eomes depending on the timing of differentiation, and physical association of these T-box proteins presumably provides sequence selectivity for Jmjd3 (Figure 1)7. Similarly, Jmjd3 co-operates with Hes1 to promote transcription of Mash-1 downstream of retinoic acid signaling to drive neural fate in P19 cells13 (Figure 1). In each case, a selective signaling pathway directs the recruitment of a transcription factor (Tbx3, Eomes or Hes1), which then recruits Jmjd3 to specific genomic loci to drive a particular fate.
Interestingly, there is a plethora of lineage-specific T-box transcription factors and it is tempting to speculate that mesodermal T-box proteins recruit Jmjd3 to T/Brachyury and potentially other mesodermal genes. Examining the T/Brachyury promoter by rVista motif analysis, we found that T-box consensus binding sites are interspersed between the Smad and Tcf sites about 1kb away from the start site of transcription. While consensus binding sites are only circumstantial evidence of binding, Tbx6 in the mouse or VegT in Xenopus are candidate factors that might recruit Jmjd3 to the T/Brachyury promoter since they are expressed or active in nascent mesoderm and involved in induction16, 17,18.
Key questions remain about the role of Jmjd3. Does it instruct cell fate or only create conducive conditions by making chromatin accessible to inducing signals? To what extent does it act redundantly or coordinately with other DMs and other chromatin modifying machinery? In the Ohtani work, knocking out Jmjd3 blocked mesoderm formation, but did not trigger a compensatory switch to another lineage. In the early stages of PSC differentiation, a fate switch or ying-yang between mesoderm and ectoderm is well documented19, 20. Both Wnt and Nodal drive mesoderm and ectoderm forms at the expense of mesoderm in their absence19, 20. One explanation might be that the cells are stuck in an epiblast-like stage and simply fail to differentiate when deprived of Jmjd3.
We anticipate that exploring two directions will bring the function of Jmjd3 into sharper focus. The first is to understand Jmjd3’s role in cell fate determination. Even though mesoderm was impaired by the Jmjd3 knockout, 20% of the embryoid bodies still exhibited contracting areas, suggesting that mesoderm differentiation did not fail completely. Functional redundancy could be one explanation for this observation, and indeed, UTX is also needed for mesoderm and ectoderm differentiation and is partially rescued by Jmjd321. Moreover, unlike endoderm, wherein Jmjd3 regulates many of the essential endodermal differentiation genes9, only few mesoderm differentiation genes were targeted by Jmjd34, consistent with the idea that other mechanisms may be required to drive mesoderm differentiation completely. This might explain why removing Jmjd3 does not divert cells to epiblast or ectoderm.
The second area we think would be informative is to define how Jmjd3 and DMs are recruited to specific sites in chromatin. We discussed how association with transcription factors might provide selectivity, but the details of specific factors, and how they are activated or induced by extrinsic signaling, such as from the progenitor cell niche, remain vague. Dahle et al. suggested that occupancy of the T/Brachyury promoter by β-catenin alone is insufficient for Jmjd3 recruitment or transcription. β-catenin/TCF therefore might function as a pioneer complex that binds first and then recruits Jmjd3 and other factors. Alternatively, P-Smad2/3 or even a T-box protein might be the pioneers, and/or some or all of the complex might assemble in the cytoplasm prior to engaging chromatin.
Clearly, the role of Jmjd3 in mesoderm induction is the tip of the iceberg in understanding how chromatin modification directs cardiac cell fate. In addition to defining Jmjd3’s role and basis for gene selectivity, we should also ask whether the many other JmjC DMs are involved in mesoderm and cardiac fate. Furthermore, it will be of particular importance to understand chromatin structural changes that underlie the subsequent steps from mesoderm to terminally differentiated cardiomyocytes, each of which might involve DMs to enable distinct programs of gene expression. We predict that physical interactions between DMs and well-known cardiac transcription factors might be responsible for activating discrete, stage-specific programs of gene expression during cardiogenesis. Furthermore, these mechanisms might be re-employed during compensatory or decompensatory events in heart disease.
Acknowledgments
Funding
NIH (R01HL113601 and R01HL108176)
California Institute for Regenerative Medicine Training Grant (TG2-01162)
Fondation Leducq
Footnotes
Disclosures: None
References
- 1.Burridge PW, Keller G, Gold JD, Wu JC. Production of de novo cardiomyocytes: Human pluripotent stem cell differentiation and direct reprogramming. Cell Stem Cell. 2012;10:16–28. doi: 10.1016/j.stem.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wamstad JA, Alexander JM, Truty RM, Shrikumar A, Li F, Eilertson KE, Ding H, Wylie JN, Pico AR, Capra JA, Erwin G, Kattman SJ, Keller GM, Srivastava D, Levine SS, Pollard KS, Holloway AK, Boyer LA, Bruneau BG. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell. 2012;151:206–220. doi: 10.1016/j.cell.2012.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paige SL, Thomas S, Stoick-Cooper CL, Wang H, Maves L, Sandstrom R, Pabon L, Reinecke H, Pratt G, Keller G, Moon RT, Stamatoyannopoulos J, Murry CE. A temporal chromatin signature in human embryonic stem cells identifies regulators of cardiac development. Cell. 2012;151:221–232. doi: 10.1016/j.cell.2012.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ohtani K, Zhao C, Dobreva G, Manavski Y, Kluge B, Braun T, Rieger MA, Zeiher AM, Dimmeler S. Jmjd3 controls mesodermal and cardiovascular differentiation of embryonic stem cells. Circ Res. 2013 doi: 10.1161/CIRCRESAHA.113.302035. [DOI] [PubMed] [Google Scholar]
- 5.Hubner MR, Spector DL. Role of h3k27 demethylases jmjd3 and utx in transcriptional regulation. Cold Spring Harb Symp Quant Biol. 2011 doi: 10.1101/sqb.2010.75.020. [DOI] [PubMed] [Google Scholar]
- 6.Kooistra SM, Helin K. Molecular mechanisms and potential functions of histone demethylases. Nat Rev Mol Cell Biol. 2012;13:297–311. doi: 10.1038/nrm3327. [DOI] [PubMed] [Google Scholar]
- 7.Kartikasari AE, Zhou JX, Kanji MS, Chan DN, Sinha A, Grapin-Botton A, Magnuson MA, Lowry WE, Bhushan A. The histone demethylase jmjd3 sequentially associates with the transcription factors tbx3 and eomes to drive endoderm differentiation. Embo J. 2013;32:1393–1408. doi: 10.1038/emboj.2013.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramadoss S, Chen X, Wang CY. Histone demethylase kdm6b promotes epithelial-mesenchymal transition. J Biol Chem. 2012;287:44508–44517. doi: 10.1074/jbc.M112.424903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim SW, Yoon SJ, Chuong E, Oyolu C, Wills AE, Gupta R, Baker J. Chromatin and transcriptional signatures for nodal signaling during endoderm formation in hescs. Dev Biol. 2011;357:492–504. doi: 10.1016/j.ydbio.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 10.Dahle O, Kumar A, Kuehn MR. Nodal signaling recruits the histone demethylase jmjd3 to counteract polycomb-mediated repression at target genes. Sci Signal. 2010;3:ra48. doi: 10.1126/scisignal.2000841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ye L, Fan Z, Yu B, Chang J, Al Hezaimi K, Zhou X, Park NH, Wang CY. Histone demethylases kdm4b and kdm6b promotes osteogenic differentiation of human mscs. Cell Stem Cell. 2012;11:50–61. doi: 10.1016/j.stem.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Svotelis A, Bianco S, Madore J, Huppe G, Nordell-Markovits A, Mes-Masson AM, Gevry N. H3k27 demethylation by jmjd3 at a poised enhancer of anti-apoptotic gene bcl2 determines eralpha ligand dependency. Embo J. 2011;30:3947–3961. doi: 10.1038/emboj.2011.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dai JP, Lu JY, Zhang Y, Shen YF. Jmjd3 activates mash1 gene in ra-induced neuronal differentiation of p19 cells. J Cell Biochem. 2010;110:1457–1463. doi: 10.1002/jcb.22703. [DOI] [PubMed] [Google Scholar]
- 14.Fei T, Xia K, Li Z, Zhou B, Zhu S, Chen H, Zhang J, Chen Z, Xiao H, Han JD, Chen YG. Genome-wide mapping of smad target genes reveals the role of bmp signaling in embryonic stem cell fate determination. Genome Res. 2010;20:36–44. doi: 10.1101/gr.092114.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gadue P, Huber TL, Paddison PJ, Keller GM. Wnt and tgf-{beta} signaling are required for the induction of an in vitro model of primitive streak formation using embryonic stem cells. Proc Natl Acad Sci U S A. 2006;103:16806–16811. doi: 10.1073/pnas.0603916103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chapman DL, Agulnik I, Hancock S, Silver LM, Papaioannou VE. Tbx6, a mouse t-box gene implicated in paraxial mesoderm formation at gastrulation. Dev Biol. 1996;180:534–542. doi: 10.1006/dbio.1996.0326. [DOI] [PubMed] [Google Scholar]
- 17.Wardle FC, Papaioannou VE. Teasing out t-box targets in early mesoderm. Curr Opin Genet Dev. 2008;18:418–425. doi: 10.1016/j.gde.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vonica A, Gumbiner BM. Zygotic wnt activity is required for brachyury expression in the early xenopus laevis embryo. Dev Biol. 2002;250:112–127. doi: 10.1006/dbio.2002.0786. [DOI] [PubMed] [Google Scholar]
- 19.Willems E, Leyns L. Patterning of mouse embryonic stem cell-derived pan-mesoderm by activin a/nodal and bmp4 signaling requires fibroblast growth factor activity. Differentiation. 2008;76:745–759. doi: 10.1111/j.1432-0436.2007.00257.x. [DOI] [PubMed] [Google Scholar]
- 20.Lindsley RC, Gill JG, Kyba M, Murphy TL, Murphy KM. Canonical wnt signaling is required for development of embryonic stem cell-derived mesoderm. Development (Cambridge, England) 2006;133:3787–3796. doi: 10.1242/dev.02551. [DOI] [PubMed] [Google Scholar]
- 21.Morales Torres C, Laugesen A, Helin K. Utx is required for proper induction of ectoderm and mesoderm during differentiation of embryonic stem cells. PLoS One. 2013;8:e60020. doi: 10.1371/journal.pone.0060020. [DOI] [PMC free article] [PubMed] [Google Scholar]