Abstract
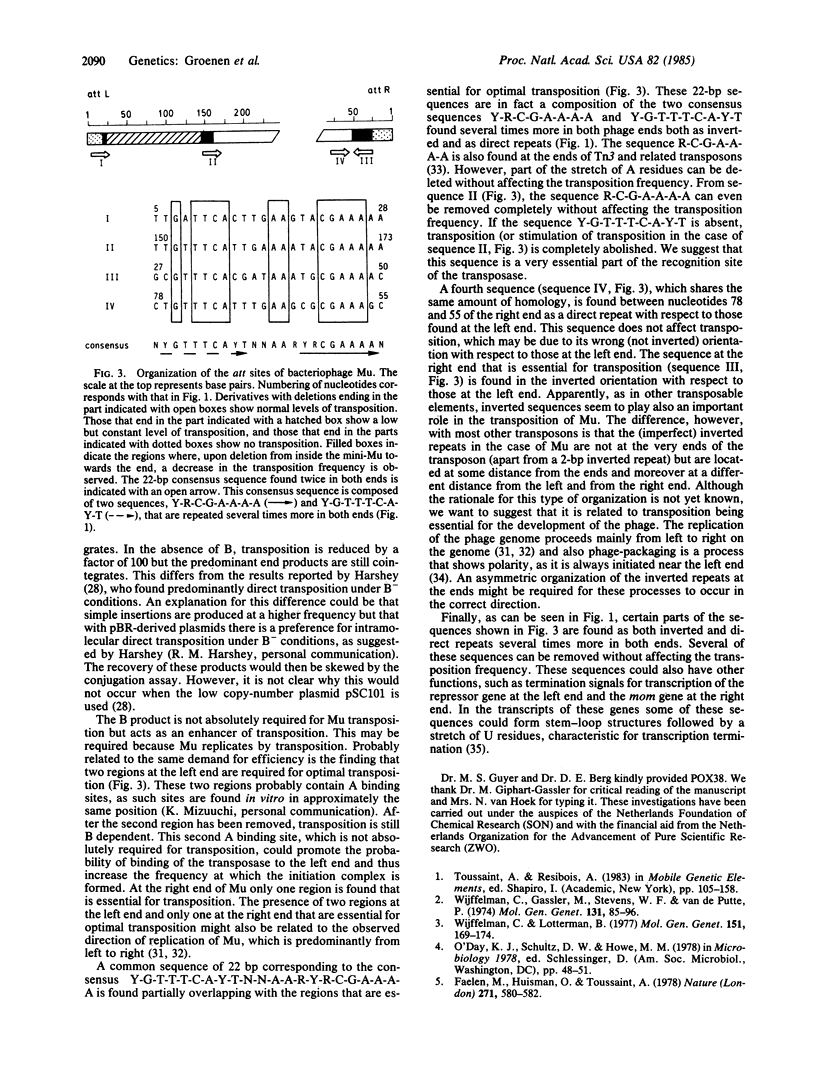
We have determined the minimal DNA sequences at the ends of the genome of bacteriophage Mu that are required for its transposition. A mini-Mu was constructed on a multicopy plasmid that enabled the manipulation of the DNA sequences at its ends without affecting the genes essential for transposition. The genes A and B, which were cloned outside the ends of the mini-Mu on the same plasmid, were both needed for optimal transposition. In our experimental system the predominant end products of the transposition are cointegrates both in the presence and in the absence of B. Two regions ending approximately 25 and 160 bp from the left end and one ending approximately 50 bp from the right end appear to be essential for optimal transposition. Overlapping with these regions, a 22-base-pair sequence was recognized with the consensus Y-G-T-T-C-A-Y-T-N-N-A-A-R-Y-R-C-G-A-A-A-A, where Y and R represent any pyrimidine and purine, respectively. At the left end these sequences occur as direct repeats; at the right end this sequence is inverted with respect to those at the left end.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Biel S. W., Adelt G., Berg D. E. Transcriptional control of IS1 transposition in Escherichia coli. J Mol Biol. 1984 Apr 5;174(2):251–264. doi: 10.1016/0022-2836(84)90337-1. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukhari A. I., Taylor A. L. Influence of insertions on packaging of host sequences covalently linked to bacteriophage Mu DNA. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4399–4403. doi: 10.1073/pnas.72.11.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calos M. P., Miller J. H. Transposable elements. Cell. 1980 Jul;20(3):579–595. doi: 10.1016/0092-8674(80)90305-0. [DOI] [PubMed] [Google Scholar]
- Casadaban M. J., Chou J. In vivo formation of gene fusions encoding hybrid beta-galactosidase proteins in one step with a transposable Mu-lac transducing phage. Proc Natl Acad Sci U S A. 1984 Jan;81(2):535–539. doi: 10.1073/pnas.81.2.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaconas G., Harshey R. M., Sarvetnick N., Bukhari A. I. Predominant end-products of prophage Mu DNA transposition during the lytic cycle are replicon fusions. J Mol Biol. 1981 Aug 15;150(3):341–359. doi: 10.1016/0022-2836(81)90551-9. [DOI] [PubMed] [Google Scholar]
- Chandler M., Galas D. J. Cointegrate formation mediated by Tn9. II. Activity of IS1 is modulated by external DNA sequences. J Mol Biol. 1983 Oct 15;170(1):61–91. doi: 10.1016/s0022-2836(83)80227-7. [DOI] [PubMed] [Google Scholar]
- Faelen M., Huisman O., Toussaint A. Involvement of phage Mu-1 early functions in Mu-mediated chromosomal rearrangements. Nature. 1978 Feb 9;271(5645):580–582. doi: 10.1038/271580a0. [DOI] [PubMed] [Google Scholar]
- Giphart-Gassler M., Reeve J., van de Putte P. Polypeptides encoded by the early region of bacteriophage Mu synthesized in minicells of Escherichia coli. J Mol Biol. 1981 Jan 5;145(1):165–191. doi: 10.1016/0022-2836(81)90339-9. [DOI] [PubMed] [Google Scholar]
- Goosen T., Giphart-Gassler M., Van de Putte P. Bacteriophage Mu DNA replication is stimulated by non-essential early functions. Mol Gen Genet. 1982;186(1):135–139. doi: 10.1007/BF00422925. [DOI] [PubMed] [Google Scholar]
- Guyer M. S. The gamma delta sequence of F is an insertion sequence. J Mol Biol. 1978 Dec 15;126(3):347–365. doi: 10.1016/0022-2836(78)90045-1. [DOI] [PubMed] [Google Scholar]
- Harshey R. M. Switch in the transposition products of Mu DNA mediated by proteins: Cointegrates versus simple insertions. Proc Natl Acad Sci U S A. 1983 Apr;80(7):2012–2016. doi: 10.1073/pnas.80.7.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes W. M., Platt T., Rosenberg M. Termination of transcription in E. coli. Cell. 1983 Apr;32(4):1029–1032. doi: 10.1016/0092-8674(83)90287-8. [DOI] [PubMed] [Google Scholar]
- Kahmann R., Kamp D. Nucleotide sequences of the attachment sites of bacteriophage Mu DNA. Nature. 1979 Jul 19;280(5719):247–250. doi: 10.1038/280247a0. [DOI] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Mizuuchi K. In vitro transposition of bacteriophage Mu: a biochemical approach to a novel replication reaction. Cell. 1983 Dec;35(3 Pt 2):785–794. doi: 10.1016/0092-8674(83)90111-3. [DOI] [PubMed] [Google Scholar]
- Plasterk R. H., Ilmer T. A., Van de Putte P. Site-specific recombination by Gin of bacteriophage Mu: inversions and deletions. Virology. 1983 May;127(1):24–36. doi: 10.1016/0042-6822(83)90367-7. [DOI] [PubMed] [Google Scholar]
- Plasterk R. H., Vollering M., Brinkman A., Van de Putte P. Analysis of the methylation-regulated Mu mom transcript. Cell. 1984 Jan;36(1):189–196. doi: 10.1016/0092-8674(84)90088-6. [DOI] [PubMed] [Google Scholar]
- Priess H., Kamp D., Kahmann R., Bräuer B., Delius H. Nucleotide sequence of the immunity region of bacteriophage Mu. Mol Gen Genet. 1982;186(3):315–321. doi: 10.1007/BF00729448. [DOI] [PubMed] [Google Scholar]
- Reed R. R., Young R. A., Steitz J. A., Grindley N. D., Guyer M. S. Transposition of the Escherichia coli insertion element gamma generates a five-base-pair repeat. Proc Natl Acad Sci U S A. 1979 Oct;76(10):4882–4886. doi: 10.1073/pnas.76.10.4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remaut E., Stanssens P., Fiers W. Plasmid vectors for high-efficiency expression controlled by the PL promoter of coliphage lambda. Gene. 1981 Oct;15(1):81–93. doi: 10.1016/0378-1119(81)90106-2. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro L. J., Mohandas T. DNA methylation and the control of gene expression on the human X chromosome. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):631–637. doi: 10.1101/sqb.1983.047.01.074. [DOI] [PubMed] [Google Scholar]
- Van Leerdam E., Karreman C., van de Putte P. Ner, a cro-like function of bacteriophage Mu. Virology. 1982 Nov;123(1):19–28. doi: 10.1016/0042-6822(82)90291-4. [DOI] [PubMed] [Google Scholar]
- Waggoner B., Pato M., Toussaint A., Faelen M. Replication of mini-Mu prophage DNA. Virology. 1981 Aug;113(1):379–387. doi: 10.1016/0042-6822(81)90163-x. [DOI] [PubMed] [Google Scholar]
- Wijffelman C. A., Westmaas G. C., van de Putte P. Vegetative recombination of bacteriophage Mu-1 in Escherichia coli. Mol Gen Genet. 1972;116(1):40–46. doi: 10.1007/BF00334258. [DOI] [PubMed] [Google Scholar]
- Wijffelman C., Gassler M., Stevens W. F., van de Putte P. On the control of transcription of bacteriophage Mu. Mol Gen Genet. 1974;131(2):85–96. doi: 10.1007/BF00266145. [DOI] [PubMed] [Google Scholar]
- Wijffelman C., Lotterman B. Kinetics of Mu DNA synthesis. Mol Gen Genet. 1977 Mar 7;151(2):169–174. doi: 10.1007/BF00338691. [DOI] [PubMed] [Google Scholar]



