Background: Despite the high diversity of glycolipids found in many organisms, only a few glycosyltransferases have been isolated.
Results: A bifunctional glycosyltransferase, synthesizing glucuronosyl or monoglucosyl diacylglycerol, was isolated from Agrobacterium.
Conclusion: Glycolipids and other nonphospholipids can mutually replace each other, enhancing the ability to adapt to changing environments.
Significance: This is the first report on the isolation of a glucuronosyl diacylglycerol synthase.
Keywords: Bacteria, Chromatography, Glycolipids, Glycosyltransferases, Membrane Bilayer, Gram-negative, Q-TOF, Bifunctional, Glucuronic Acid
Abstract
Glycolipids are mainly found in phototrophic organisms (like plants and cyanobacteria), in Gram-positive bacteria, and a few other bacterial phyla. Besides the function as bulk membrane lipids, they often play a role under phosphate deprivation as surrogates for phospholipids. The Gram-negative Agrobacterium tumefaciens accumulates four different glycolipids under phosphate deficiency, including digalactosyl diacylglycerol and glucosylgalactosyl diacylglycerol synthesized by a processive glycosyltransferase. The other two glycolipids have now been identified by mass spectrometry and nuclear magnetic resonance spectroscopy as monoglucosyl diacylglycerol and glucuronosyl diacylglycerol. These two lipids are synthesized by a single promiscuous glycosyltransferase encoded by the ORF atu2297, with UDP-glucose or UDP-glucuronic acid as sugar donors. The transfer of sugars differing in their chemistry is a novel feature not observed before for lipid glycosyltransferases. Furthermore, this enzyme is the first glucuronosyl diacylglycerol synthase isolated. Deletion mutants of Agrobacterium lacking monoglucosyl diacylglycerol and glucuronosyl diacylglycerol or all glycolipids are not impaired in growth or virulence during infection of tobacco leaf discs. Our data suggest that the four glycolipids and the nonphospholipid diacylglyceryl trimethylhomoserine can mutually replace each other during phosphate deprivation. This redundancy of different nonphospholipids may represent an adaptation mechanism to enhance the competitiveness in nature.
Introduction
Although phospholipids are widespread constituents in biological membranes, the occurrence of glycolipids is mainly restricted to plants, cyanobacteria, Gram-positive bacteria, and a few other bacterial phyla (1). Glycolipids are characterized by a high headgroup diversity, which is determined by the number and type of sugars (glucose, galactose, mannose, or charged sugars like glucuronic acid or sulfoquinovose) with different anomeric configurations (α and β) and linkages to each other (1→2, 1→3, 1→4, and 1→6). In Gram-positive bacteria, glycolipids represent building blocks for membranes or serve as membrane anchors for lipoteichoic acids (1, 2). Glycolipids also play an important role in several members of Gram-negative bacteria (Proteobacteria) under phosphate deprivation, similar to plants and cyanobacteria, where they replace phospholipids to save phosphate required for the synthesis of phosphate-containing metabolites (1, 3, 4). Besides glycolipids, further phosphate-free lipids are often involved in this physiological stress response in bacteria (5). Two representatives of such nonphospholipids are diacylglyceryl trimethylhomoserine (DGTS)2 or the glycerol-free ornithine lipid (OL).
These two lipids accumulate in different Proteobacteria, like in the nodule-forming bacterium Sinorhizobium meliloti (Rhizobiaceae and Rhizobiales) when grown under phosphate deprivation. Sinorhizobium further contains the charged glycolipid sulfoquinovosyl diacylglycerol (SQD), which is also increased under these conditions (6). The plant pathogen Agrobacterium tumefaciens (Rhizobiaceae) or the nodule-forming bacterium Mesorhizobium loti (Phyllobacteriaceae and Rhizobiales) were recently shown to synthesize a series of further glycolipids grown under phosphate starvation, i.e. digalactosyl diacylglycerol (DGD), glucosylgalactosyl diacylglycerol (GGD), different triglycosyl diacylglycerols, with all sugars bound in β-anomeric configuration, and two unidentified glycolipids (U1 and U2) (3, 4).
Irrespective of the high variety of bacterial glycolipids, only a few lipid glycosyltransferases have been cloned so far (1, 2). The best studied glycosyltransferases synthesizing monoglucosyl diacylglycerol (MGlcD) and diglucosyl diacylglycerol (DGlcD), with all sugars bound in α-anomeric configuration, were isolated from cell wall-less bacterium Acholeplasma laidlawii (7). The two enzymes MGlcD synthase (alMGS) and DGlcD synthase belong to the glycosyltransferase family 4 in the CAZy database (8). This database classifies glycosyltransferases into different families based on protein sequence similarities. Further glycosyltransferases belonging to glycosyltransferase family 4 are the MGlcD synthases from Deinococcus radiodurans and Thermotoga maritima and the monogalactosyl diacylglycerol (MGD) synthase from Borrelia burgdorferi, as the first cloned galactosyltransferase forming MGD with α-anomeric configuration of the sugar (9, 10). The two glycosyltransferases synthesizing the agrobacterial or mesorhizobial glycolipids DGD, GGD, and triglycosyl diacylglycerols were characterized as processive glycosyltransferases, designated Pgt (3, 4). They transfer both glucose and galactose, with diacylglycerol (DAG) as primary acceptor. The two enzymes with high sequence similarity are members of GT21 (11). The identification and characterization of the enzyme(s) responsible for the synthesis of the two unknown glycolipids U1 and U2 in Agrobacterium and the elucidation of the glycolipid headgroup structures are part of this study.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Plasmids, and Growth Conditions
A. tumefaciens strain C58C1 (pGV2260) was grown at 28 °C in YEP medium in the presence of rifampicin (60 mg/liter) (4). Gentamicin (25 mg/liter) was used for selection of the single mutant Δagt; gentamicin and kanamycin (50 mg/liter) were used for the selection of the double mutant Δagt Δpgt. Escherichia coli strains ElectroSHOX (Bioline) and BL21 (DE3) (Novagen) were used as expression hosts for atu2297. Growth curve experiments of Agrobacterium cells were performed as described (4) starting with an initial A600 of 0.05 in AB minimal medium (12) with high (25 mm) or low (20 μm) phosphate. The A600 was determined for 96 h.
Virulence Tests with Tobacco Leaf Disc Transformation
Leaf disc transformation was performed with Agrobacterium wild type, Δagt, or Δagt Δpgt with each strain in a separate experiment, as described (4).
Cloning of the ORF atu2297 from A. tumefaciens
The ORF atu2297 was amplified using the primers bn799 (CCTAGGTATGACGAGAATCACGATTGTC) and bn800 (GGATCCTCATGCAAGCCGCGAGCGC) containing XmaJI and BamHI restriction sites (as underlined). The PCR product was subcloned in the pGEM-T Easy vector (Promega) and released with XmaJI/BamHI. The vector pTnVagro (11) was used as an expression vector, linearized with XmaJI and BamHI, and ligated with the released ORF atu2297 (pTnV-atu2297). The empty vector (pTnV) was created by blunting the linearized pTnVagro vector (with the Klenow fragment, Thermo Scientific) and subsequent circularization.
Construction of A. tumefaciens Deletion Mutants Δagt and Δagt Δpgt by Disruption of the atu2297 Locus by Homologous Recombination Using Gentamicin and Kanamycin Resistance Cassettes
The mutant line Δagt was generated using the Agrobacterium strain C58C1 (pGV2260). The primers used for the amplification of the homologous sequences were bn934 (TCATCGGCCATCATGGCGC) and bn935 (TATACCATGGTTGCCGCCATTGTGGAACCA) to generate the 5′-flankingsequence with a 3′-NcoI restriction site, whereas bn936 (ATATACGCGTCATTCTGGAAGCGCTGGCCAG) and bn937 (CGTTATCATCTCGCCATCACG) were used for the amplification of the 3′-flanking sequence with a 5′-MluI restriction site. The gentamicin resistance cassette was cloned as described (4), containing 5′-MluI and 3′-NcoI restriction sites. All PCR products were subcloned in pGEM-T easy vectors. MluI, NcoI, and other restriction sites on the cloning vectors were used for further cloning. The two flanking sequences and the gentamicin resistance cassette were cloned in one step in pGEM-T Easy with the gentamicin resistance cassette inserted in antisense orientation relative to the flanking sequences. The double knock-out mutant Δagt Δpgt was generated in the Δpgt background by disruption of the atu2297 locus by insertion of a kanamycin resistance cassette (4, 11). For this purpose, the gentamicin resistance cassette in the deletion construct described above was replaced with a kanamycin resistance cassette. The cloning strategy was similar with the exception of using a PscI restriction site instead of NcoI for cloning of the kanamycin resistance cassette. NcoI and PscI produce compatible ends. The primers used for amplification of the kanamycin resistance cassette were bn1116 (ATATACGCGTCACGCTGCCGCAAGCACTCA) and bn1149 (TCATGACATGTTCAGAAGAACTCGTCAAGAAG). The Δagt single mutant was identified by PCR using the following primer pairs: bn938 (TTGCCCGTTACGTCACCGGA) and bn939 (GCCTCAAATACAGGTCGAGAT); bn249 (AGTGGCTCTCTATACAAAGTTG) and bn938 (TTGCCCGTTACGTCACCGGA); and bn250 (TTCGGTCAAGGTTCTGGACC) and bn939 (GCCTCAAATACAGGTCGAGAT). The Δagt Δpgt double mutant was identified using the following primer pairs: bn938 (TTGCCCGTTACGTCACCGGA) and bn939 (GCCTCAAATACAGGTCGAGAT); bn224 (GCGGACTGGCTTTCTACGTG) and bn938 (TTGCCCGTTACGTCACCGGA) and bn225 (TGCTCGACGTTGTCACTGAAG) and bn939 (GCCTCAAATACAGGTCGAGAT).
Lipid Isolation, Separation, and Analysis by GC-MS and Quadrupole-Time-of-Flight Mass Spectrometry (Q-TOF MS)
Lipids and fatty acids were analyzed as described (3, 4). The fragmentation energy for MGlcD/U1 and GlcAD/U2 analyzed with Q-TOF MS/MS was 12 and 20 V, respectively. The solvent used for one-dimensional TLC was acetone/toluene/water (91:30:8). For two-dimensional TLC, chloroform/methanol/water (65:25:4) was used for the first dimension and chloroform/methanol/acetic acid/water (90:15:10:4) for the second dimension. For NMR spectroscopy, glycolipids from Agrobacterium and E. coli expressing atu2297 were separated by preparative one-dimensional TLC. Purification of U2 from Agrobacterium required two steps because of co-migrating lipids. In the first step, total lipid extracts from Agrobacterium were separated on TLC plates pretreated with ammonium sulfate (0.15 m) and activated by heat (120 °C, 2.5 h) prior to use. In this system, U2 is protonated and migrates clearly above MGD (4). U2 and co-migrating lipids were scraped off and extracted from the silica material with chloroform/methanol (2:1) in an ultrasonic bath for 30 min. The extracted lipid mixture was subjected to a second TLC step using nontreated plates to obtain pure U2 lipid, which migrates similar to DGD in this TLC system (this study).
Enzyme Assays
For enzyme assays, E. coli (ElectroSHOX) cells containing pTnV-atu2297 were grown at 37 °C to an A600 of 0.6, induced with 500 μm isopropyl 1-thio-β-d-galactopyranoside, and incubated for 24 h at 16 °C. The culture was harvested by centrifugation, and the pellet was resuspended in 1 ml of buffer 1 (11) and disrupted with glass beads with the Precellys homogenizer (Peqlab). Cell debris was removed by centrifugation at 70 × g for 1 min. The assays were performed in a final volume of 205 μl with 100 μl of buffer 2 (15 mm Tricine/KOH, pH 7.2, 30 mm MgCl2, 3 mm DTT), 50 μl of UDP-glucuronic acid (UDP-GlcUA) or UDP-glucose (UDP-Glc) (40 pmol/μl), 50 μl of E. coli protein extract, and 5 μl of DAG-14:0/14:0 (10 nmol/μl in ethanol). After incubation for 60 min at 28 °C, the assays were terminated by the addition of 3 ml of chloroform/methanol (2:1) and 0.5 ml of NaCl solution (0.9%). The lipids were extracted as described (3, 4) and separated by TLC. MGD and DGD were used as reference lipids to identify the positions of MGlcD and GlcAD on the TLC plate. Corresponding bands were scraped off the silica plate and extracted for 30 min with chloroform/methanol (2:1) in an ultrasonic bath. The extracted reaction products were analyzed with the Q-TOF mass spectrometer (Agilent) by direct nanospray infusion in the positive mode as described (3, 4) using fragmentation energies of 12 V for MGlcD and 20 V for GlcAD.
Compositional Analyses
To confirm the nature of hexose and hexuronic acid, samples were hydrolyzed with 2 m HCl/MeOH at 85 °C for 2 h, followed by acetylation (85 °C, 10 min) and detection by GC-MS (Hewlett-Packard HP 5890 (series II) gas chromatograph equipped with a fused silica SPB-5 column (Supelco, 30 m × 0.25 mm × 0.25-μm film thickness), flame ionization detector, and MS 5989A mass spectrometer with vacuum gauge controller 59827A). The temperature program was 150 °C for 3 min and then 5 °C/min to 330 °C. The sugars were identified by comparison with the authentic standards. The determination of the absolute configuration of the constituents was performed as described (13).
NMR Spectroscopy
NMR spectroscopy experiments were carried out in CDCl3 with tetramethylsilane (δH 0.00 and δC 0.00) as an internal standard. 1H,13C, and two-dimensional homonuclear (1H and 1H) COSY, TOCSY, and ROESY, as well as (1H and 13C) HSQC-DEPT, coupled HSQC, and HMBC experiments were recorded at 27 °C with a Bruker DRX Avance 700-MHz spectrometer (operating frequencies 700.75 MHz for 1H NMR and 176.2 MHz for 13C NMR), equipped with a 5-mm CPQCI multinuclear inverse cryo-probe head with a z gradient, and applying standard Bruker software. COSY, TOCSY, and ROESY experiments were recorded using data sets (t1 by t2) of 4096 by 512 points, COSY with 1 and TOCSY and ROESY with 8 scans. The TOCSY experiment was carried out in the phase-sensitive mode with mixing times of 60 ms and ROESY of 300 ms. Additionally, NMR spectra of U2 were carried out on a Bruker Avance AVIII 500 spectrometer equipped with a TCI cryoprobe. All spectra were measured in MeOD/CDCl3 (16.66/83.37%) at 27 °C. Double quantum-filtered COSY, TOCSY, and ROESY experiments were recorded using data sets (t1 by t2) of 2048 by 512 points, with 16 scans. The TOCSY experiment was carried out in the phase-sensitive mode with mixing times of 120 ms and ROESY of 300 ms.
RESULTS
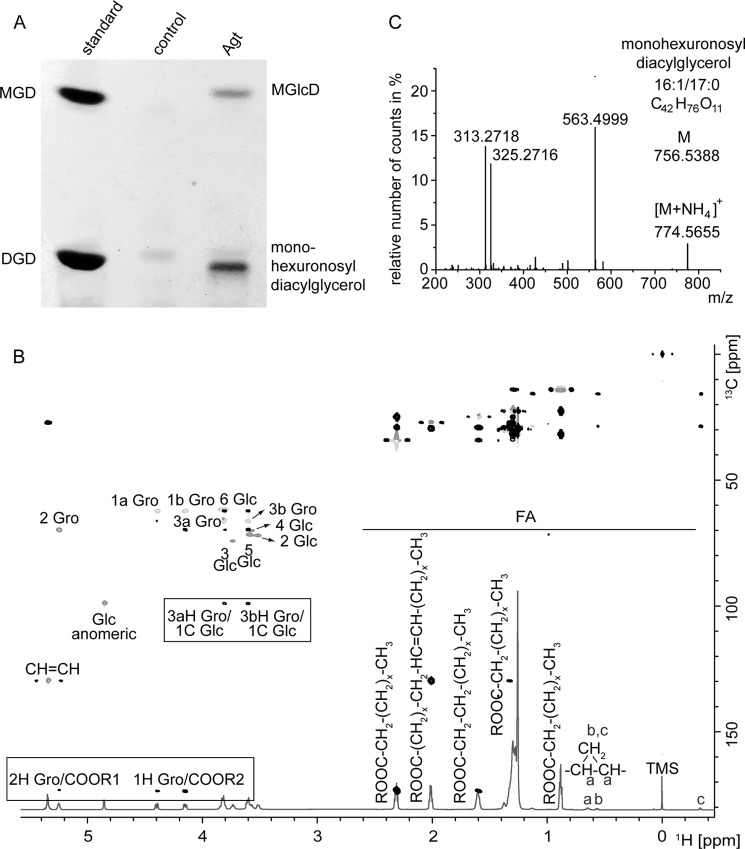
Expression of an Agrobacterial Glycosyltransferase (atu2297) Led to the Accumulation of MGlcD and Monohexuronosyl Diacylglycerol in E. coli
The presence of the unknown glycolipid U2 both in A. tumefaciens and M. loti (3, 4) suggests the existence of glycosyltransferases with homologous sequences in the two organisms. For the identification, we first searched for annotated glycosyltransferase sequences for A. tumefaciens in the CAZy database (8). To restrict the number of genes, we selected only candidates with homologous sequences in Agrobacterium and Mesorhizobium. This search revealed 14 sequences, which were all cloned from Agrobacterium and expressed in E. coli ElectroSHOX or BL21 (DE3). Only one of the open reading frames (atu2297; glycosyltransferase family 4, containing α-glycosyltransferases) led to the accumulation of two new glycolipids in E. coli BL21 (DE3) migrating slightly above MGD or below DGD, respectively (Fig. 1A). Expression in E. coli ElectroSHOX led to the accumulation of only one glycolipid migrating slightly above MGD (data not shown). One explanation for the absence of the second glycolipid may be a reduced availability of the respective sugar donor in this E. coli strain. Compositional and structural analysis with NMR spectroscopy confirmed one of the lipids as MGlcD with α-d-configuration of the glucose (Fig. 1B). The α-configuration was confirmed by the 3J(1,2) value of 3.6 Hz measured from the 1H spectrum. Therefore, the ORF atu2297 codes for an α-glycosyltransferase (Agt) synthesizing MGlcD. The second glycolipid expressed in E. coli migrates similar to U2 from Agrobacterium and Mesorhizobium on TLC plates (3, 4). It was further analyzed with Q-TOF MS/MS where it was detected as an ammonium adduct (in the positive ion mode), with one out of several main molecular species (Fig. 1C). The calculated m/z of the parental ion was 774.5727. The neutral loss of 211.0660 of the fragmented lipid indicates the loss of hexuronic acid as ammonia adduct representing the lipid headgroup. The different fragment ions in the spectrum are derived from DAG-16:0/17:0c (m/z 563.4999) and from monoacyl glyceryl-16:0 (m/z 313.2718) and −17:0c (m/z 325.2716). Therefore, this second glycolipid isolated from E. coli represents monohexuronosyl diacylglycerol.
FIGURE 1.
Accumulation of two new glycolipids in E. coli expressing Agt from Agrobacterium. A, TLC of lipid extracts from E. coli BL21 (DE3) expressing Agt or the empty vector as control. The new glycolipids were identified as MGlcD and monohexuronosyl diacylglycerol. B, overlay of 1H,HSQCdept and HMBC spectra of MGlcD. Spectra were recorded in CDCl3 at 27 °C utilizing the Bruker DRX Avance 700 MHz spectrometer. Important intra-residual scalar correlations are marked in the box. R1 and R2 indicate the following: 14:0; 16:1; 16:0; 17:0c (ω9,10); 18:1; 19:0c (ω9,10). TMS, tetramethylsilane; FA, fatty acids. C, Q-TOF MS/MS spectrum of monohexuronosyl diacylglycerol. Parental ions were selected as ammonium adducts and fragmented. The figure shows the spectrum of one main species with m/z 774.5727. The fragment with m/z 563.4999 represents DAG-16:0/17:0c (as protonated form with loss of H2O). The neutral loss of 211.0660 (m/z 774.5655 minus 563.4999) is derived from hexuronic acid (as ammonium adduct). The ions m/z 313.2718 and 325.2716 represent monoacylglycerol-16:0 and monoacylglycerol-17:0c, respectively, each as protonated form with loss of H2O.
U1 and U2 from Agrobacterium Contain Glucose or Glucuronic Acid in Their Headgroups, Respectively
To reveal the structural details of the agrobacterial glycolipids U1 and U2, we separated them via two-dimensional TLC and analyzed the isolated lipids with Q-TOF MS/MS. This analysis does not allow distinguishing between different epimeric and anomeric configurations of the sugars. The fragmentation spectra of a main molecular species of each agrobacterial lipid are shown in Fig. 2. The neutral losses of 197.0905 and 211.0748 are derived from hexose and hexuronic acid, respectively, as ammonia adducts (Fig. 2). The fragment ion m/z 617.5498 (or 617.5448) was derived from DAG-18:1/19:0c, whereas the fragment ions 339.2873 (or 339.2854) and 353.3052 (or 353.3032) were derived from monoacylglycerol-18:1 and −19:0c, respectively. Therefore, U1 and U2 represent two glycerolipids containing hexose or hexuronic acid in their headgroups, respectively (Fig. 2). These results were confirmed further by GC analyses together with NMR spectroscopy that allowed determining the nature as well as absolute and anomeric configuration of the headgroups of the two lipids. U1 could be identified as MGlcD (Table 1) with α-d-Glcp, and U2 as GlcAD with α-d-GlcpA linked to DAG (Table 2). The 1JH1,C1 value of 172.26 Hz proved the α-configuration of U2.
FIGURE 2.
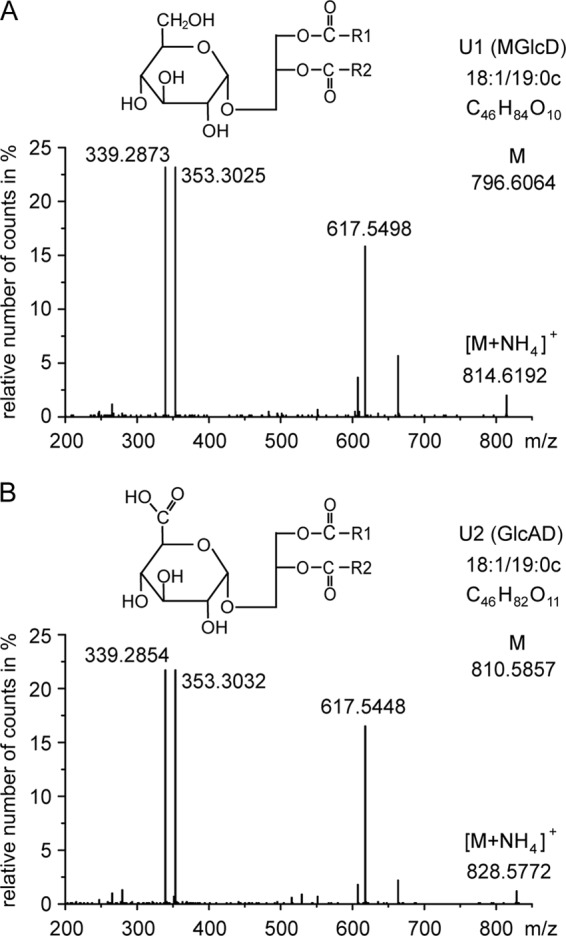
Q-TOF MS/MS spectra of monohexosyl diacylglycerol (U1) and monohexuronosyl diacylglycerol (U2). Parental ions (as ammonium adducts) of one main molecular species of U1 (calculated m/z 814.6403) and U2 (calculated m/z 828.6196) were selected in the positive mode (with a detection window of m/z 1.2) and fragmented. The fragments with m/z 617.5498 (or 617.5448) represent DAG-18:1/19:0c (as protonated form with loss of H2O). The neutral loss of 197.0905 (m/z 814.6403 minus 617.5498) is derived from hexose (as ammonia adduct); the neutral loss of 211.0748 (m/z 828.6196 minus 617.5448) is derived from hexuronic acid (as ammonia adduct). The ions m/z 339.28 and 353.30 represent monoacylglycerol-18:1 and monoacylglycerol-19:0c, respectively (each as protonated form with loss of H2O). COO-R1 and COO-R2 represent two different fatty acyl residues (18:1 or 19:0c) bound to the glycerol back bone. The headgroup structures of U1 (MGlcD) and U2 (GlcAD) were further determined by NMR containing α-glucose or α-glucuronic acid, respectively, as illustrated in the figure.
TABLE 1.
1H and 13C chemical shifts of U1 identified as 1,2-diacyl-3-α-d-Glcp-sn-Gro reported (internal standard, tetramethylsilane)
Spectra were recorded in CDCl3 at 27 °C utilizing a Bruker DRX Avance 700 spectrometer. Gro stands for glycerol.
| 1a | 1b | 2 | 3a | 3b | 4 | 5 | 6a | 6b | |
|---|---|---|---|---|---|---|---|---|---|
| α-d-Glcp | |||||||||
| H (δ) | 4.85 (3J1,2 2.6 Hz) | 3.50 | 3.72 | 3.58 | 3.59 | 3.81 | 3.83 | ||
| C (δ) | 99.13 | 72.16 | 74.30 | 70.05 | 71.85 | 61.84 | |||
| Gro | |||||||||
| H (δ) | 4.39 | 4.15 | 5.24 | 3.81 | 3.60 | ||||
| C (δ) | 62.41 | 69.85 | 66.35 | ||||||
TABLE 2.
1H and 13C chemical shifts of 1,2-diacyl-3-α-d-GlcpA-sn-Gro (U2) reported (internal standard, tetramethylsilane, δH 0.00, δC 0.00)
Spectra were recorded in MeOD/CDCl3 (16.66/83.37%) at 27 °C utilizing Bruker Avance AVIII 500 and DRX Avance 700 spectrometers.
| 1a | 1b | 2 | 3a | 3b | 4 | 5 | 6a | 6b | |
|---|---|---|---|---|---|---|---|---|---|
| α-d-GlcpA | |||||||||
| H (δ) | 4.90 | 3.51 | 3.69 | 3.59 | 4.10 | ||||
| C (δ) | 99.50 (1JH1, C1 173 Hz) | 71.61 | 73.31 | 71.89 | 70.95 | 172.26 | |||
| Gro | |||||||||
| H (δ) | 4.43 | 4.18 | 5.25 | 3.88 | 3.68 | ||||
| C (δ) | 62.47 | 70.03 | 66.78 | ||||||
Deletion of atu2297 Resulted in the Loss of the Two Glycolipids MGlcD and GlcAD
The accumulation of MGlcD and of monohexosyl diacylglycerol in E. coli transformed with the ORF atu2297 suggests that the native function of Agt encoded by this ORF is the synthesis of MGlcD and/or GlcAD in Agrobacterium. Furthermore, the gene coding for the glycosyltransferase involved in the formation of these two glycolipids is expected to be regulated by phosphate limitation. Vences-Guzmán et al. (14) have described the presence of four glycosyltransferase genes (inclusive atu2297) probably under the control of a pho box in A. tumefaciens. Interestingly, homologs to atu2297 present in other α-Proteobacteria are forming an operon with genes encoding a phospholipase C involved in lipid remodeling under phosphate starvation. Genes forming operons are in many cases encoding proteins acting in the same pathway. The phospholipase C would be responsible for DAG formation that could be used by a lipid glycosyltransferase for glycolipid formation. We deleted the glycosyltransferase gene in Agrobacterium by insertion of a gentamicin resistance cassette into the atu2297 locus via homologous recombination. The gene deletion was confirmed by PCR (data not shown). Wild type and deletion mutant Δagt were grown under phosphate starvation and the lipids separated via two-dimensional TLC. The disruption of atu2297 led to the loss of the two lipids MGlcD and GlcAD in the mutant (Fig. 3). Therefore, Agt is involved in the synthesis of both MGlcD and GlcAD in Agrobacterium.
FIGURE 3.
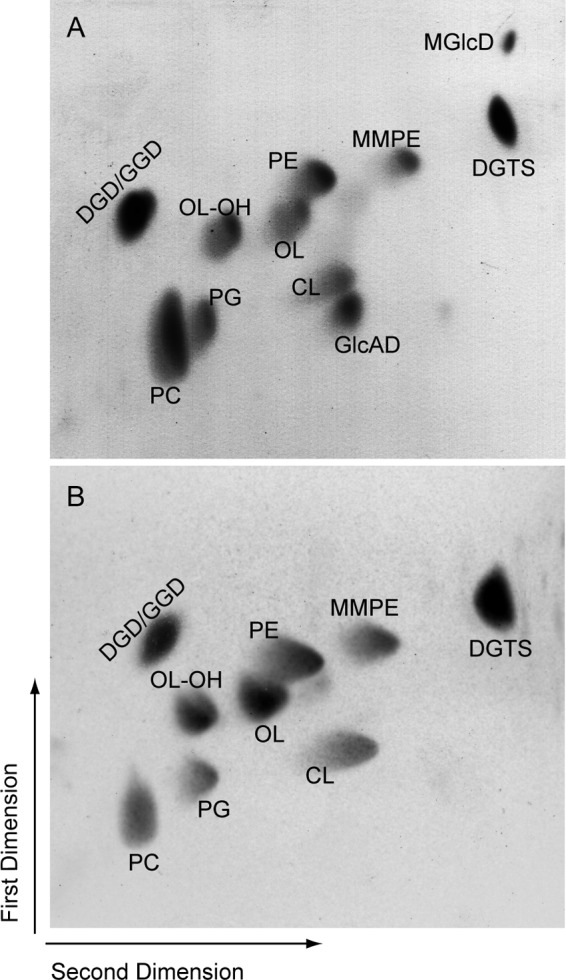
Two-dimensional TLC of lipid extracts from Agrobacterium wild type (A) or Δagt (B) grown under phosphate deprivation, with MGlcD and GlcAD being absent from the mutant. PE, phosphatidylethanolamine; MMPE, monomethyl-PE; PG, phosphatidylglycerol; CL, cardiolipin; OL-OH, hydroxylated form of OL (4).
Expression Experiments Show the Synthesis of the Two Lipids MGlcD and GlcAD by Agt
The accumulation of MGlcD and monohexuronosyl diacylglycerol in E. coli after expression of Agt, together with the absence of MGlcD and GlcAD in the Agrobacterium Δagt mutant, suggests that Agt is able to transfer both glucose and glucuronic acid onto diacylglycerol using the respective UDP-sugars. However, it may also be conceivable that GlcAD is formed from MGlcD in a second step by an unknown enzyme with glucose-6-dehydrogenase activity. To reveal the mechanism for the formation of MGlcD and GlcAD and to exclude the possibility that a further enzyme is involved in the formation of GlcAD by modification of MGlcD, further experiments were performed. To this end, we included in addition to Agt from Agrobacterium the glucosyltransferase alMGS from Acholeplasma (7) in our studies, which is known to synthesize only MGlcD but not GlcAD. To express the two enzymes in a glycolipid-free background, the Agrobacterium double knock-out mutant Δagt Δpgt was produced. This mutant was created by insertion of a kanamycin resistance cassette into the atu2297 locus of cells with a Δpgt background (4). The Δpgt mutants are characterized by the disruption of the processive glycosyltransferase Pgt and by the lack of GGD and DGD. The purpose of creation of this double knock-out was to abolish Agt activity and to prevent any activities resulting from Pgt, such as the synthesis of β-MGD or β-MGlcD (11), which cannot be distinguished in mass spectrometry (by their m/z and fragmentation pattern) from α-MGlcD synthesized by Agt or alMGS from Acholeplasma. Furthermore, the lack of DGD/GGD facilitates the detection and isolation of GlcAD, which migrates similar to DGD during TLC.
Agt or alMGS was then introduced into the Agrobacterium Δagt Δpgt double mutant. The two transformed strains are expected to produce MGlcD, and the strain expressing Agt is expected to contain in addition GlcAD. If the MGlcD lipid was the precursor for the synthesis of GlcAD by a second enzyme, then the alMGS-expressing strain should also contain GlcAD. However, if Agt itself is responsible for the synthesis of GlcAD, then GlcAD should be absent from cells expressing alMGS.
The cell lines were grown under phosphate starvation, because the glycolipid accumulation of expression cultures is higher compared with growth under phosphate-replete conditions (4); the lipids were analyzed via TLC and Q-TOF MS/MS. In the cells expressing Agt, we could detect both MGlcD and GlcAD as shown in Fig. 4. The empty vector control is free of any glycolipids. The line expressing alMGS from Acholeplasma accumulates MGlcD but lacks GlcAD (Fig. 4). Therefore, these results indicate that Agt synthesizes both MGlcD and GlcAD; the involvement of further MGlcD-modifying enzymes involved in GlcAD synthesis in Agrobacterium can be excluded.
FIGURE 4.
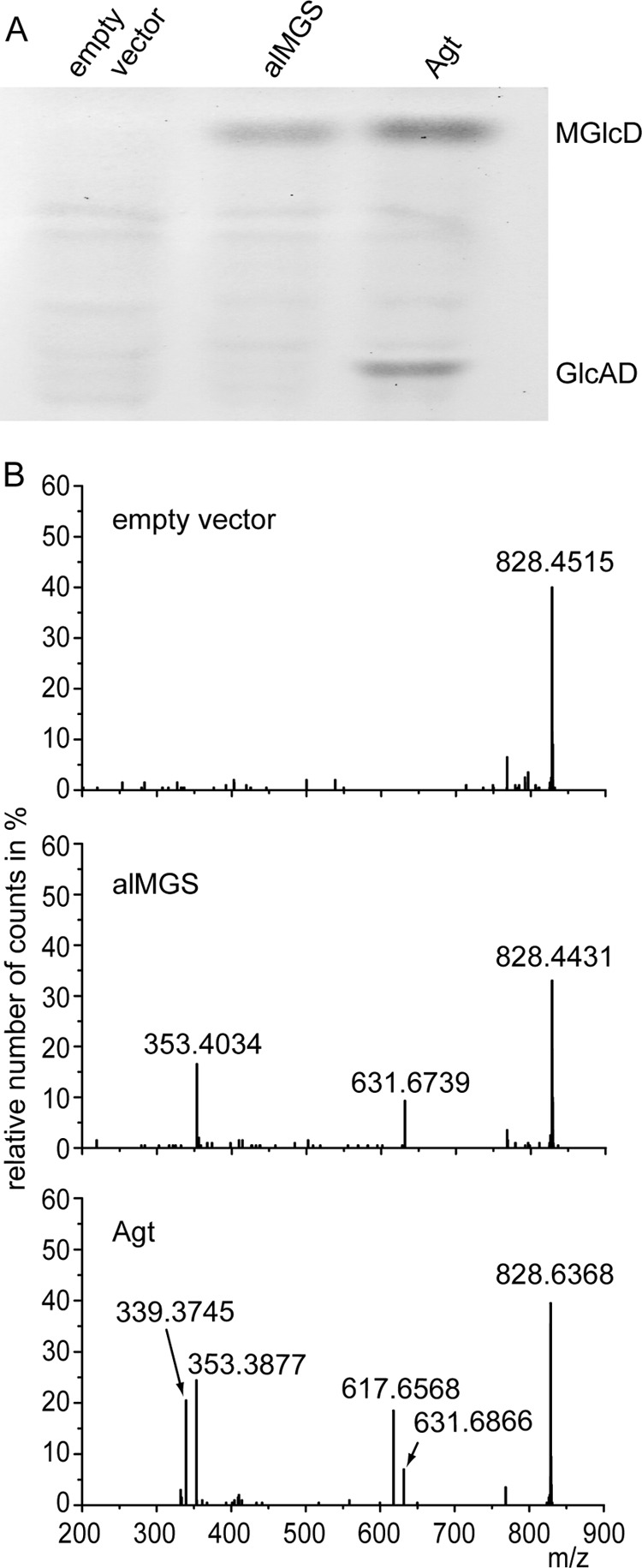
Accumulation of MGlcD and GlcAD in Agrobacterium double knock-out mutant Δagt Δpgt transformed with the empty vector, alMGS from Acholeplasma or Agt from Agrobacterium. A, TLC of lipid extracts from the three lines with expression of alMGS leading to the synthesis of MGlcD and Agt to the synthesis of both MGlcD and GlcAD. B, Q-TOF MS/MS spectra of MGlcD and GlcAD. Parental ions with a calculated m/z 828.6196 (with a detection window m/z 1.2) were selected in the positive mode. Because of the similarity of their m/z values, ions representing ammonium adducts of both GlcAD-18:1/19:0c (calculated m/z 828.6196) and MGlcD-19:0c/19:0c (calculated m/z 828.6560) were selected together for fragmentation in this experiment. The fragments with m/z 617.6 and 631.6 represent DAG-18:1/19:0c and DAG-19:0c/19:0c (as protonated form with loss of OH), respectively. The neutral loss of 197.0 (m/z 828.6 minus 631.6) is derived from glucose (as ammonium adduct); the neutral loss of 211.0 (m/z 828.6 minus 617.6) is derived from glucuronic acid (as ammonium adduct). Therefore, DAG-18:1/19:0c is derived from GlcAD, whereas DAG-19:0c/19:0c is derived from MGlcD. The peaks at m/z 353.3877/353.4034 or 339.3745 represent protonated monoacylglycerol (MAG) species containing a 19:0c or 18:1 fatty acid, respectively, with loss of H2O. The molecular species of MGlcD and GlcAD shown in the spectra represent one example each of different molecular species of the two glycolipids detected in Agrobacterium with all showing the same result that GlcAD is only present in cells expressing Agt and absent in cells expressing alMGS from Acholeplasma.
Enzyme Assays with Recombinant Agt Protein Confirm the Synthesis of MGlcD and GlcAD by Agt
For the confirmation of these results and further analysis of the sugar specificities of Agt, we performed enzyme assays with protein extracts prepared from E. coli cells expressing Agt or the empty vector as control. UDP-Glc or UDP-GlcUA were used as sugar donors, and DAG-14:0/14:0 as sugar acceptor. The formation of the respective glycolipids was analyzed via Q-TOF MS/MS. We detected MGlcD (14:0/14:0) or GlcAD (14:0/14:0) in the lipid extracts of enzyme assays of Agt, using UDP-Glc or UDP-GlcUA as substrates, respectively (Fig. 5). Protein extracts from E. coli harboring an empty vector were free of these two activities. These results demonstrate that GlcAD is not formed by modification of MGlcD but that the two lipids MGlcD and GlcAD are synthesized by Agt, exhibiting two different substrate specificities for UDP-Glc and UDP-GlcUA.
FIGURE 5.
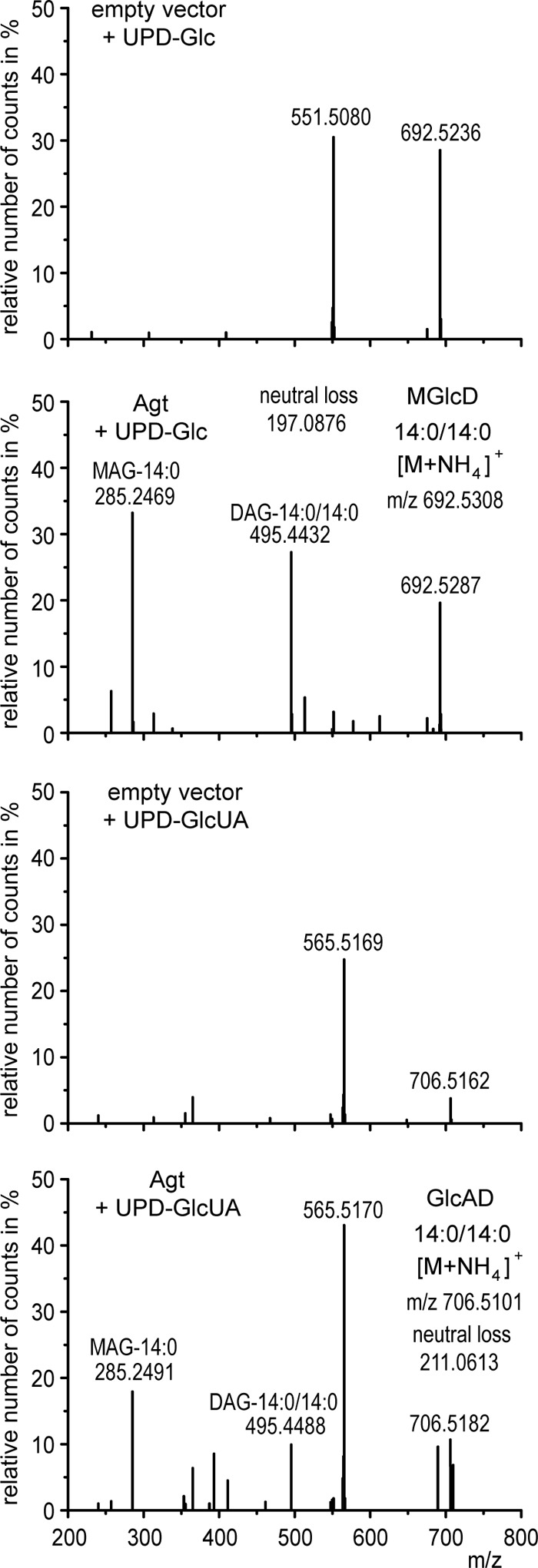
Q-TOF MS/MS spectra of MGlcD and GlcAD synthesized in enzyme assays with protein extracts from E. coli cells expressing Agt from Agrobacterium or harboring the empty vector as control. The assays were supplemented with DAG-14:0/14:0 as sugar acceptor and UDP-Glc or UDP-GlcUA as sugar donors. Parental ions with calculated m/z 692.5308 or 706.5101 representing ammonium adducts of MGlcD or GlcAD, respectively, were selected in the positive mode (with a detection window of m/z 1.2). The fragment ions with m/z 495.4432 (or 495.4488) represent the protonated form of DAG-14:0/14:0 (with loss of H2O), and m/z 285.2469 (or 285.4491) the protonated form of monoacylglycerol-14:0 (MAG-14:0) (with loss of H2O). The respective fragments were absent in the spectra of the empty vector control assays. The neutral losses of 197.0876 (692.5308 minus 495.4432) or 211.0613 (706.5101 minus 495.4488) represent the ammonia adducts of glucose or glucuronic acid, respectively. These spectra prove the formation of both MGlcD and GlcAD (by supplementation of UDP-Glc or UDP-GlcUA, respectively) by Agt. The detection of unspecific fragments (m/z 551.5 and 565.5) may result from the fragmentation of ions with similar m/z to the selected ions with m/z 692.5308 or 706.5101, which were also observed in the control assays.
MGlcD and GlcAD Are Replaced by DGD/GGD in the Δagt Single Mutant, although All Glycolipids Are Replaced by DGTS in the Δagt Δpgt Double Mutant
For investigation of the role of glycolipids in Agrobacterium, we analyzed the lipid composition of wild type, Δagt, and the glycolipid-free double knock-out mutant Δagt Δpgt. For this purpose, we grew the three lines under phosphate-replete and depleted conditions and quantified the lipids by measuring their fatty acid methyl esters via GC-MS. The different cell lines grown under phosphate-replete conditions were not distinguishable in their lipid compositions (Fig. 6). Differences could be observed under phosphate starvation in the different mutant lines compared with the wild type (Fig. 6). The single knock-out mutant Δagt compensates for the loss of MGlcD (1.5% in wild type) and GlcAD (5% in wild type) by a strong accumulation of the Pgt-dependent glycolipids GGD and DGD (from 10 in wild type to more than 20% in Δagt). This change in glycolipid composition is accompanied by a strong reduction of the phospholipid phosphatidylcholine (PC). All the other lipids are hardly affected. The double knock-out mutant compensates for the loss of all glycolipids by a strong accumulation of DGTS. Again, PC is strongly reduced.
FIGURE 6.
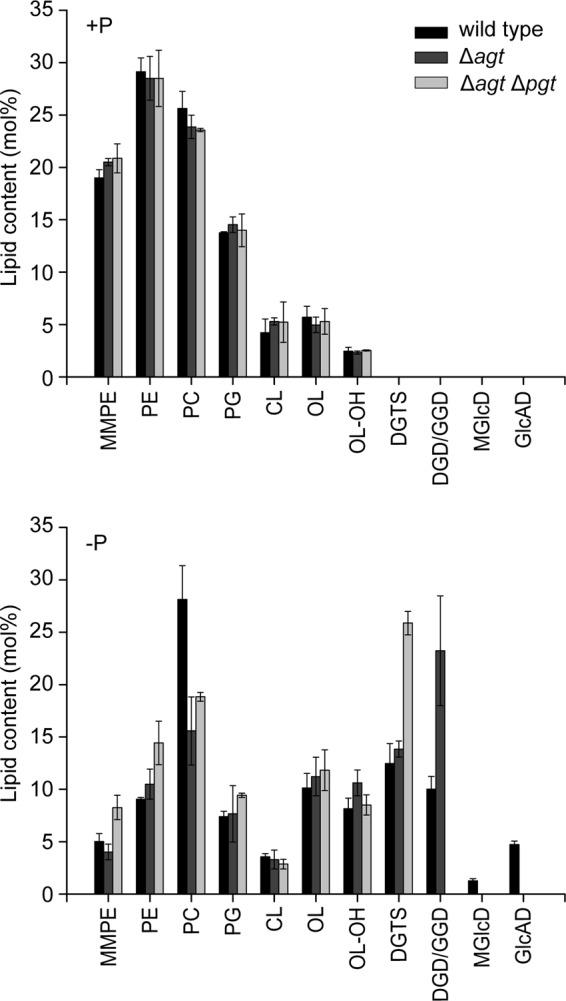
Lipid composition of Agrobacterium wild type, Δagt, and Δagt Δpgt grown under phosphate replete (+P) or depleted (−P) conditions. The bars represent mean values in mol % ± S.D. of three measurements. PE, phosphatidylethanolamine; MMPE, monomethyl-PE; PG, phosphatidylglycerol; CL, cardiolipin; OL-OH, hydroxylated form of OL (4) Lipids were quantified after isolation from the TLC plate, transmethylation, and measurements of fatty acid methyl esters by GC-MS.
Glycolipids Are Not Required for Growth or Virulence of Agrobacterium under Phosphate Deprivation
The accumulation the different glycolipids DGD, GGD, MGlcD, and GlcAD suggest that they are important for growth under phosphate deprivation. As shown in a previous study, the loss of DGD and GGD in the Δpgt mutant does not affect growth or virulence under phosphate starvation presumably because these two glycolipids are compensated for by DGTS and the remaining glycolipids MGlcD and GlcAD. In this study, we analyzed the growth and virulence of Δagt and Δagt Δpgt to reveal any effect of the lack of MGlcD/GlcAD or the complete loss of glycolipids. For growth curve experiments, wild type and the two mutant lines were grown in minimal medium with high (25 mm) or low (20 μm) phosphate. Growth of all lines is reduced under phosphate deprivation, but there is no difference between wild type, Δagt, and Δagt Δpgt (Fig. 7). The growth experiments were repeated four times. The virulence tests were performed three times with tobacco leaf discs inoculated separately with wild type, Δagt, or Δagt Δpgt under conditions with high or low phosphate. The number of calli formed on the leaf discs should give an indication for the virulence of the respective strains. Again, no differences could be observed between wild type and the two mutant lines (data not shown). These data suggest that glycolipids are not required for growth or virulence of Agrobacterium under normal and phosphate-deficient conditions.
FIGURE 7.
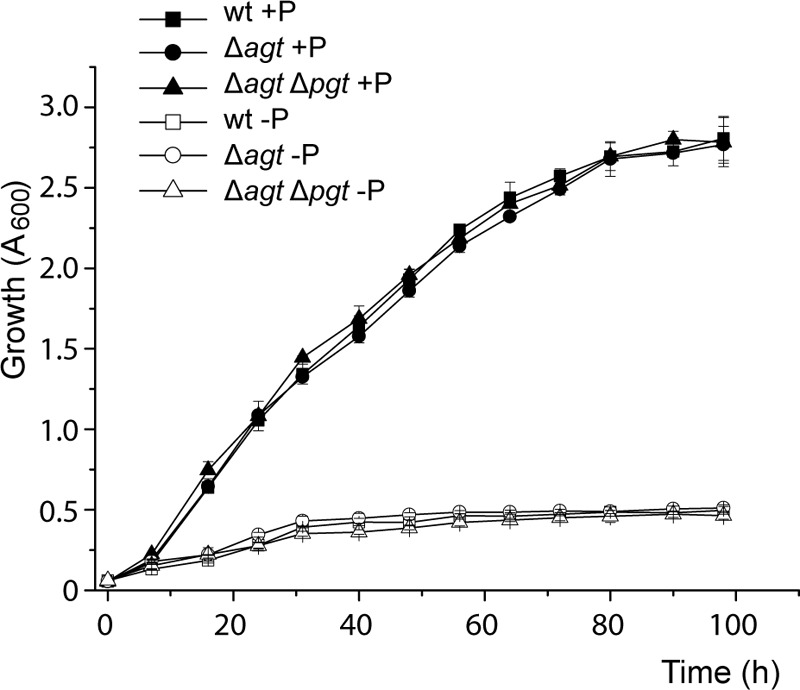
Growth curves of Agrobacterium cells grown with 25 mm (+P) or 20 μm (−P) phosphate show no differences between wild type (WT) and deletion mutants (Δagt and Δagt Δpgt) under the respective conditions. Mean values ± S.D. were calculated from three independent measurements.
DISCUSSION
A. tumefaciens accumulates four different glycolipids under phosphate starvation, which are GGD, DGD, and two unknown glycolipids U1 and U2 (4). In this study, we determined the structures of U1 (MGlcD) and U2 (GlcAD) and identified and characterized the responsible glycosyltransferase encoded by the ORF atu2297. This study is the first report on a lipid glycosyltransferase acting as a bifunctional enzyme by synthesizing neutral and acidic glycolipids. Furthermore, Agt is the first GlcAD synthase isolated.
In general, glycosyltransferases exhibit a high sugar donor specificity. The best studied glycosyltransferases include the MGD/MGlcD and DGD synthases from plants and cyanobacteria (15–18), the MGlcD and DGlcD synthases from Acholeplasma (7, 19), and the processive glycosyltransferases from the Gram-positive bacteria Staphylococcus or Bacillus (20, 21). There are only a few glycosyltransferases characterized as promiscuous enzymes, such as Pgt from Agrobacterium or Mesorhizobium, or the processive glycosyltransferases from the human pathogens Mycoplasma pneumoniae and Mycoplasma genitalum using UDP-Glc and UDP-galactose (UDP-Gal) as sugar donors (11, 22, 23). All these promiscuous glycosyltransferases transfer sugars with similar chemistry, although there are no reports on glycosyltransferases transferring sugars with different chemistry as shown for Agt.
The presence of MGlcD in representatives of the Rhizobiales has never been described before. Therefore, Agrobacterium is the first member of the Rhizobiales synthesizing MGlcD. With respect to other Gram-negative bacteria, MGlcD is mainly restricted to the cauliform bacteria and relatives (α-Proteobacteria), and to the Spirochaetes (1). It represents a bulk lipid in these organisms. MGlcD (with α- or β-anomeric configuration) is also found in many Gram-positive bacteria (1), often building the precursor for the synthesis of DGlcD (2).
There are only a few bacteria synthesizing the acidic glycolipid GlcAD. With respect to Rhizobiales, this lipid was described for the first time for the anoxygenic phototrophic bacterium Blastochloris viridis (24), but it has never been found before in nonphototrophic Rhizobiales. Therefore, Agrobacterium is the first representative of nonphototrophic Rhizobiales synthesizing GlcAD. It is further found in the cauliform bacteria and relatives (25–28). In most of these bacteria GlcAD represents a bulk lipid, and it is synthesized independently from phosphate supply (25, 29). There are only a few reports on GlcAD accumulating under phosphate deprivation as a surrogate for phospholipids (30, 31). Outside of Gram-negative bacteria, GlcAD or higher glycosylated forms exist only in a few other bacterial species (1, 32–34). The responsible GlcAD synthase in all these organisms have never been isolated. Enzyme assays revealed UDP-GlcUA as a sugar donor (29). GlcAD was recently also detected in plants, where it is probably synthesized by the SQD synthase SQD2 (35).
SQD, another acidic glycolipid, is widespread in phototrophic and nonphototrophic organisms. It is generally accepted that SQD especially replaces the acidic phospholipid phosphatidylglycerol under phosphate-limiting conditions (1, 6, 36, 37). Different species of Rhizobiales contain either SQD (Sinorhizobium and Rhizobium) or GlcAD (Blastochloris), but there are no reports on the presence of the two lipids in one species of Rhizobiales (1, 3). Thus, GlcAD and SQD might be counterpart lipids with similar functions in the different organisms.
The deletion of atu2297 with loss of MGlcD/GlcAD in Δagt or the complete loss of all glycolipids in Δagt Δpgt had no influence on growth or virulence of Agrobacterium. This may be explained by a mutual replacement of glycolipids and DGTS. A triple mutant of Sinorhizobium lacking SQD, OL, and DGTS was not impaired in its ability to form nodules on its host alfalfa (38). However, the loss of OL and DGTS or of all non-phosphorus lipids (OL, DGTS, and SQD) resulted in a decreased growth under phosphate starvation, suggesting that these lipids serve as bulk lipids. This may also be true for the glycolipids and DGTS in Agrobacterium. Therefore, a triple knock-out mutant of Agrobacterium lacking glycolipids and DGTS may be impaired in growth under phosphate-limited conditions. OL or hydroxy-OL may not be a surrogate for glycolipids in Agrobacterium (4) (Fig. 7).
In contrast to growth and virulence, the deletion of atu2297 had a strong effect on lipid composition under phosphate deprivation. Surprisingly, PC was strongly reduced in cells lacking MGlcD/GlcAD, although the amount of DGTS in Δagt was not changed. In contrast to Agrobacterium, the content of PC is negatively correlated to that of DGTS in Sinorhizobium (38, 39). The reduction of PC in Δagt or Δagt Δpgt may have different reasons as follows. (i) PC reduction may result from its degradation by a phospholipase C active under phosphate deprivation to provide DAG for the increased synthesis of GGD/DGD in Δagt or DGTS in Δagt Δpgt (4, 40). (ii) The reduction of PC may be necessary to sustain special membrane functions in a changed lipid environment caused by the lack of the acidic glycolipid GlcAD. Therefore, the differences in the lipid compositions in the different cell lines may reflect the ability and adaptability of Agrobacterium to compensate for the loss of the different glycolipids. With respect to this extended lipid redistribution caused by the loss of relatively small amounts of MGlcD/GlcAD, a special but unknown function especially of GlcAD may not be excluded.
A common but important function of membrane lipids is the maintenance of membrane structure and fluidity. A certain ratio of bilayer stabilizing and destabilizing lipids is crucial for optimal membrane functions. The bilayer-forming properties of membrane lipids can be regulated on different levels (41, 42). In Acholeplasma, the ratio of nonbilayer-forming to bilayer-forming lipids is regulated by glucosylation of the nonbilayer lipid MGlcD to form the bilayer-forming DGlcD with the DGlcD synthase as key enzyme (43–45). A similar mechanism was described for Mycoplasma. In this case, the regulative enzyme is a processive glycosyltransferase adjusting the ratio of MGlcD and DGlcD (22). A novel and alternative regulation mechanism would be the introduction of different headgroups into the glycolipid by a bifunctional glycosyltransferase as shown for Agt (this study), with GlcAD probably as a bilayer-forming glycolipid (46). Therefore, Agt may be an interesting candidate for a new type of glycosyltransferase playing a role for the membrane bilayer formation and function under phosphate deprivation.
Gene expression regulated by phosphate availability is often mediated via the two-component PhoB-PhoR regulatory system, with the PhoB protein binding in its phosphorylated form to a highly conserved sequence motif in the promoter region of the regulated gene. Such a PhoB regulon may activate Pgt and Agt in Agrobacterium (4, 47). Accumulation of glycolipids in Agrobacterium was suggested to depend further on the availability of DAG as the sugar acceptor (4). The regulatory role of the sugar donors UDP-Glc, UDP-Gal, and UDP-GlcUA in glycolipid accumulation has never been tested. Sugar nucleotides play a central role in the carbohydrate metabolism with participation in many different pathways in Agrobacterium, such as the formation of lipopolysaccharides in the outer membrane (48). The bulk of UDP-Glc is required for the synthesis of exo-polysaccharides (cellulose, succinoglycan, and cyclic β-glucans) found in biofilms (49–52). Biofilm production is increased under phosphate deprivation and regulated via the PhoB regulon (53). A key enzyme in the carbohydrate metabolism is UDP-Glc pyrophosphorylase, which produces UDP-Glc. UDP-Glc-4-epimerase and UDP-Glc dehydrogenase convert UDP-Glc to UDP-Gal or UDP-GlcUA, respectively. Agrobacterium contains ORFs (atu3778, atu3315, and atu4149) with sequence similarity to the respective enzymes in Sinorhizobium (54–57). Earlier results indicate that sugar nucleotides may have a regulatory role in the glycolipid accumulation in Agrobacterium during phosphate deprivation (11). Although Pgt has a higher preference for UDP-Gal compared with UDP-Glc, this preference is not reflected in the ratio of glucose- to galactose-containing glycolipids produced in Agrobacterium. The composition of these different glycolipids rather depends on the host employed for heterologous expression of Pgt and on the growth conditions (4, 11), indicating that the ratio of GGD to DGD in Agrobacterium might be regulated via the changing availability of UDP-Glc and UDP-Gal. Similarly, expression of Agt in different E. coli strains results in the production of glycolipids with different ratios of MGlcD and GlcAD (Fig. 1A, data not shown). Therefore, the incorporation of glucose or glucuronic acid into glycolipids by Agt might depend on the availability of the two substrates, UDP-Glc and UDP-GlcUA. The total amount of glycolipids synthesized may be influenced by up-regulation of UDP-Glc pyrophosphorylase, UDP-Glc-4-epimerase, and UDP-Glc dehydrogenase under phosphate deprivation.
Acknowledgments
We thank Katharina Jakob (Research Center Borstel) for excellent technical assistance; Hermann Moll (Research Center Borstel) for help with GC/MS analysis; Thomas Peters and Thorsten Biet (Institute of Chemistry, University of Lübeck, Germany) for NMR analyses; Heiko Kässner (Research Center Borstel) for NMR recordings, and Otto Holst (Research Center Borstel) for discussion and critical reading of the manuscript.
This work was supported by Deutsche Forschungsgemeinschaft Grant Ho 3870/1-3 (to G. H.). Part of this work was presented at the FEBS Workshop on Microbial Lipids: Diversity in Structure and Formation, May 16–19, 2013, Bern, Switzerland (BALI-009).
- DGTS
- diacylglyceryl trimethylhomoserine
- OL
- ornithine lipid
- SQD
- sulfoquinovosyl diacylglycerol
- DGD
- digalactosyl diacylglycerol
- GGD
- glucosylgalactosyl diacylglycerol
- MGlcD
- monoglucosyl diacylglycerol
- DGlcD
- diglucosyl diacylglycerol
- alMGS
- MGlcD synthase from A. laidlawii
- MGD
- monogalactosyl diacylglycerol
- DAG
- diacylglycerol
- UDP-GlcUA
- UDP-glucuronic acid
- UDP-Glc
- UDP-glucose
- Q-TOF MS
- quadrupole-time-of-flight mass spectrometry
- GlcAD
- glucuronosyl diacylglycerol
- PC
- phosphatidylcholine
- UDP-Gal
- UDP-galactose
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
REFERENCES
- 1. Hölzl G., Dörmann P. (2007) Structure and function of glycoglycerolipids in plants and bacteria. Prog. Lipid Res. 46, 225–243 [DOI] [PubMed] [Google Scholar]
- 2. Reichmann N. T., Gründling A. (2011) Location, synthesis and function of glycolipids and polyglycerolphosphate lipoteichoic acid in Gram-positive bacteria of the phylum Firmicutes. FEMS Microbiol. Lett. 319, 97–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Devers E. A., Wewer V., Dombrink I., Dörmann P., Hölzl G. (2011) A processive glycosyltransferase involved in glycolipid synthesis during phosphate deprivation in Mesorhizobium loti. J. Bacteriol. 193, 1377–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Geske T., Vom Dorp K., Dörmann P., Hölzl G. (2013) Accumulation of glycolipids and other non-phosphorous lipids in Agrobacterium tumefaciens grown under phosphate deprivation. Glycobiology 23, 69–80 [DOI] [PubMed] [Google Scholar]
- 5. Geiger O., González-Silva N., López-Lara I. M., Sohlenkamp C. (2010) Amino acid-containing membrane lipids in bacteria. Prog. Lipid Res. 49, 46–60 [DOI] [PubMed] [Google Scholar]
- 6. Weissenmayer B., Geiger O., Benning C. (2000) Disruption of a gene essential for sulfoquinovosyldiacylglycerol biosynthesis in Sinorhizobium meliloti has no detectable effect on root nodule symbiosis. Mol. Plant Microbe Interact. 13, 666–672 [DOI] [PubMed] [Google Scholar]
- 7. Berg S., Edman M., Li L., Wikström M., Wieslander A. (2001) Sequence properties of the 1,2-diacylglycerol 3-glucosyltransferase from Acholeplasma laidlawii membranes. Recognition of a large group of lipid glycosyltransferases in eubacteria and archaea. J. Biol. Chem. 276, 22056–22063 [DOI] [PubMed] [Google Scholar]
- 8. Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009) The carbohydrate-active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37, D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ostberg Y., Berg S., Comstedt P., Wieslander A., Bergström S. (2007) Functional analysis of a lipid galactosyltransferase synthesizing the major envelope lipid in the Lyme disease spirochete Borrelia burgdorferi. FEMS Microbiol. Lett. 272, 22–29 [DOI] [PubMed] [Google Scholar]
- 10. Hölzl G., Zähringer U., Warnecke D., Heinz E. (2005) Glycoengineering of cyanobacterial thylakoid membranes for future studies on the role of glycolipids in photosynthesis. Plant Cell Physiol. 46, 1766–1778 [DOI] [PubMed] [Google Scholar]
- 11. Hölzl G., Leipelt M., Ott C., Zähringer U., Lindner B., Warnecke D., Heinz E. (2005) Processive lipid galactosyl/glucosyltransferases from Agrobacterium tumefaciens and Mesorhizobium loti display multiple specificities. Glycobiology 15, 874–886 [DOI] [PubMed] [Google Scholar]
- 12. Schmidt-Eisenlohr H., Domke N., Angerer C., Wanner G., Zambryski P. C., Baron C. (1999) Vir proteins stabilize VirB5 and mediate its association with the T pilus of Agrobacterium tumefaciens. J. Bacteriol. 181, 7485–7492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gerwig G. J., Kamerling J. P., Vliegenthart J. F. (1979) Determination of the absolute configuration of mono-saccharides in complex carbohydrates by capillary G.L.C. Carbohydr. Res. 77, 10–17 [DOI] [PubMed] [Google Scholar]
- 14. Vences-Guzmán M. Á., Guan Z., Bermúdez-Barrientos J. R., Geiger O., Sohlenkamp C. (2013) Agrobacteria lacking ornithine lipids induce more rapid tumour formation. Environ. Microbiol. 15, 895–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kelly A. A., Froehlich J. E., Dörmann P. (2003) Disruption of the two digalactosyldiacylglycerol synthase genes DGD1 and DGD2 in Arabidopsis reveals the existence of an additional enzyme of galactolipid synthesis. Plant Cell 15, 2694–2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dörmann P., Balbo I., Benning C. (1999) Arabidopsis galactolipid biosynthesis and lipid trafficking mediated by DGD1. Science 284, 2181–2184 [DOI] [PubMed] [Google Scholar]
- 17. Awai K., Kakimoto T., Awai C., Kaneko T., Nakamura Y., Takamiya K., Wada H., Ohta H. (2006) Comparative genomic analysis revealed a gene for monoglucosyldiacylglycerol synthase, an enzyme for photosynthetic membrane lipid synthesis in cyanobacteria. Plant Physiol. 141, 1120–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Awai K., Maréchal E., Block M. A., Brun D., Masuda T., Shimada H., Takamiya K., Ohta H., Joyard J. (2001) Two types of MGDG synthase genes, found widely in both 16:3 and 18:3 plants, differentially mediate galactolipid syntheses in photosynthetic and nonphotosynthetic tissues in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 98, 10960–10965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Edman M., Berg S., Storm P., Wikström M., Vikström S., Ohman A., Wieslander A. (2003) Structural features of glycosyltransferases synthesizing major bilayer and nonbilayer-prone membrane lipids in Acholeplasma laidlawii and Streptococcus pneumoniae. J. Biol. Chem. 278, 8420–8428 [DOI] [PubMed] [Google Scholar]
- 20. Jorasch P., Warnecke D. C., Lindner B., Zähringer U., Heinz E. (2000) Novel processive and nonprocessive glycosyltransferases from Staphylococcus aureus and Arabidopsis thaliana synthesize glycoglycerolipids, glycophospholipids, glycosphingolipids and glycosylsterols. Eur. J. Biochem. 267, 3770–3783 [DOI] [PubMed] [Google Scholar]
- 21. Jorasch P., Wolter F. P., Zähringer U., Heinz E. (1998) A UDP glucosyltransferase from Bacillus subtilis successively transfers up to four glucose residues to 1,2-diacylglycerol: Expression of ypfP in Escherichia coli and structural analysis of its reaction products. Mol. Microbiol. 29, 419–430 [DOI] [PubMed] [Google Scholar]
- 22. Andrés E., Martínez N., Planas A. (2011) Expression and characterization of a Mycoplasma genitalium glycosyltransferase in membrane glycolipid biosynthesis: Potential target against Mycoplasma infections. J. Biol. Chem. 286, 35367–35379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Klement M. L., Ojemyr L., Tagscherer K. E., Widmalm G., Wieslander A. (2007) A processive lipid glycosyltransferase in the small human pathogen Mycoplasma pneumoniae: involvement in host immune response. Mol. Microbiol. 65, 1444–1457 [DOI] [PubMed] [Google Scholar]
- 24. Linscheid M., Diehl B. W., Overmöhle M., Riedl I., Heinz E. (1997) Membrane lipids of Rhodopseudomonas viridis. Biochim. Biophys. Acta 1347, 151–163 [DOI] [PubMed] [Google Scholar]
- 25. Abraham W. R., Strömpl C., Meyer H., Lindholst S., Moore E. R., Christ R., Vancanneyt M., Tindall B. J., Bennasar A., Smit J., Tesar M. (1999) Phylogeny and polyphasic taxonomy of Caulobacter species. Proposal of Maricaulis gen. nov. with Maricaulis maris (Poindexter) comb. nov. as the type species, and emended description of the genera Brevundimonas and Caulobacter. Int. J. Syst. Bacteriol. 49, 1053–1073 [DOI] [PubMed] [Google Scholar]
- 26. Abraham W. R., Strömpl C., Vancanneyt M., Bennasar A., Swings J., Lünsdorf H., Smit J., Moore E. R. (2004) Woodsholea maritima gen. nov., sp. nov., a marine bacterium with a low diversity of polar lipids. Int. J. Syst. Evol. Microbiol. 54, 1227–1234 [DOI] [PubMed] [Google Scholar]
- 27. Batrakov S. G., Nikitin D. I., Sheichenko V. I., Ruzhitsky A. O. (1997) Unusual lipid composition of the Gram-negative, freshwater, stalked bacterium Caulobacter bacteroides NP-105. Biochim. Biophys. Acta 1347, 127–139 [DOI] [PubMed] [Google Scholar]
- 28. Batrakov S. G., Nikitin D. I., Pitryuk I. A. (1996) A novel glycolipid, 1,2-diacyl-3-α-d-glucuronopyranosyl-sn-glycerol taurineamide, from the budding seawater bacterium Hyphomonas jannaschiana. Biochim. Biophys. Acta 1302, 167–176 [DOI] [PubMed] [Google Scholar]
- 29. Stern N., Tietz A. (1971) Biosynthesis of glucuronosyl diglyceride by a cell-free system obtained from a moderately halophilic-halotolerant bacterium. FEBS Lett. 19, 217–220 [DOI] [PubMed] [Google Scholar]
- 30. Segers P., Vancanneyt M., Pot B., Torck U., Hoste B., Dewettinck D., Falsen E., Kersters K., De Vos P. (1994) Classification of Pseudomonas diminuta Leifson and Hugh 1954 and Pseudomonas vesicularis Busing, Doll, and Freytag 1953 in Brevundimonas gen. nov. as Brevundimonas diminuta comb. nov., and Brevundimonas vesicularis comb. nov., respectively. Int. J. Syst. Bacteriol. 44, 499–510 [DOI] [PubMed] [Google Scholar]
- 31. Minnikin D. E., Abdolrahimzadeh H., Baddiley J. (1974) Replacement of acidic phospholipids by acidic glycolipids in Pseudomonas diminuta. Nature 249, 268–269 [DOI] [PubMed] [Google Scholar]
- 32. Batrakov S. G., Bergelson L. D. (1978) Lipids of the Streptomycetes. Structural investigation and biological interrelation, a review. Chem. Phys. Lipids 21, 1–29 [DOI] [PubMed] [Google Scholar]
- 33. Wolucka B. A., McNeil M. R., Kalbe L., Cocito C., Brennan P. J. (1993) Isolation and characterization of a novel glucuronosyl diacylglycerol from Mycobacterium smegmatis. Biochim. Biophys. Acta 1170, 131–136 [DOI] [PubMed] [Google Scholar]
- 34. Minnikin D. E., Abdolrahimzadeh H., Baddiley J. (1971) The interrelation of polar lipids in bacterial membranes. Biochim. Biophys. Acta 249, 651–655 [DOI] [PubMed] [Google Scholar]
- 35. Okazaki Y., Otsuki H., Narisawa T., Kobayashi M., Sawai S., Kamide Y., Kusano M., Aoki T., Hirai M. Y., Saito K. (2013) A new class of plant lipid is essential for protection against phosphorus depletion. Nat. Commun. 4, 1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. López-Lara I. M., Sohlenkamp C., Geiger O. (2003) Membrane lipids in plant-associated bacteria: Their biosyntheses and possible functions. Mol. Plant Microbe Interact. 16, 567–579 [DOI] [PubMed] [Google Scholar]
- 37. Cedergren R. A., Hollingsworth R. I. (1994) Occurrence of sulfoquinovosyl diacylglycerol in some members of the family Rhizobiaceae. J. Lipid Res. 35, 1452–1461 [PubMed] [Google Scholar]
- 38. López-Lara I. M., Gao J. L., Soto M. J., Solares-Pérez A., Weissenmayer B., Sohlenkamp C., Verroios G. P., Thomas-Oates J., Geiger O. (2005) Phosphorus-free membrane lipids of Sinorhizobium meliloti are not required for the symbiosis with alfalfa but contribute to increased cell yields under phosphorus-limiting conditions of growth. Mol. Plant Microbe Interact. 18, 973–982 [DOI] [PubMed] [Google Scholar]
- 39. Geiger O., Röhrs V., Weissenmayer B., Finan T. M., Thomas-Oates J. E. (1999) The regulator gene phoB mediates phosphate stress-controlled synthesis of the membrane lipid diacylglyceryl-N,N,N-trimethylhomoserine in Rhizobium (Sinorhizobium) meliloti. Mol. Microbiol. 32, 63–73 [DOI] [PubMed] [Google Scholar]
- 40. Zavaleta-Pastor M., Sohlenkamp C., Gao J. L., Guan Z., Zaheer R., Finan T. M., Raetz C. R., López-Lara I. M., Geiger O. (2010) Sinorhizobium meliloti phospholipase C required for lipid remodeling during phosphorus limitation. Proc. Natl. Acad. Sci. U.S.A. 107, 302–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Morein S., Andersson A., Rilfors L., Lindblom G. (1996) Wild-type Escherichia coli cells regulate the membrane lipid composition in a “Window” between gel and non-lamellar structures. J. Biol. Chem. 271, 6801–6809 [DOI] [PubMed] [Google Scholar]
- 42. Wieslander A., Karlsson O. P. (1997) Regulation of lipid syntheses in Acholeplasrna laidlawii. Curr. Top. Membr. 44, 517–540 [Google Scholar]
- 43. Vikström S., Li L., Karlsson O. P., Wieslander A. (1999) Key role of the diglucosyldiacylglycerol synthase for the nonbilayer-bilayer lipid balance of Acholeplasma laidlawii membranes. Biochemistry 38, 5511–5520 [DOI] [PubMed] [Google Scholar]
- 44. Vikström S., Li L., Wieslander A. (2000) The nonbilayer/bilayer lipid balance in membranes. Regulatory enzyme in Acholeplasma laidlawii is stimulated by metabolic phosphates, activator phospholipids, and double-stranded DNA. J. Biol. Chem. 275, 9296–9302 [DOI] [PubMed] [Google Scholar]
- 45. Karlsson O. P., Rytömaa M., Dahlqvist A., Kinnunen P. K., Wieslander A. (1996) Correlation between bilayer lipid dynamics and activity of the diglucosyldiacylglycerol synthase from Acholeplasma laidlawii membranes. Biochemistry 35, 10094–10102 [DOI] [PubMed] [Google Scholar]
- 46. Koynova R. D., Tenchov B. G., Kuttenreich H., Hinz H. J. (1993) Structure and Phase Behavior of a Charged Glycolipid (1,2-O-dialkyl-3-O-β-d-glucuronosyl-sn-glycerol). Biochemistry 32, 12437–12445 [DOI] [PubMed] [Google Scholar]
- 47. Yuan Z. C., Zaheer R., Morton R., Finan T. M. (2006) Genome prediction of PhoB regulated promoters in Sinorhizobium meliloti and twelve proteobacteria. Nucleic Acids Res. 34, 2686–2697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. De Castro C., Molinaro A., Lanzetta R., Silipo A., Parrilli M. (2008) Lipopolysaccharide structures from Agrobacterium and Rhizobiaceae species. Carbohydr. Res. 343, 1924–1933 [DOI] [PubMed] [Google Scholar]
- 49. Breedveld M. W., Miller K. J. (1994) Cyclic β-glucans of members of the family Rhizobiaceae. Microbiol. Rev. 58, 145–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Matthysse A. G., Thomas D. L., White A. R. (1995) Mechanism of cellulose synthesis in Agrobacterium tumefaciens. J. Bacteriol. 177, 1076–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cangelosi G. A., Hung L., Puvanesarajah V., Stacey G., Ozga D. A., Leigh J. A., Nester E. W. (1987) Common loci for Agrobacterium tumefaciens and Rhizobium meliloti exopolysaccharide synthesis and their roles in plant interactions. J. Bacteriol. 169, 2086–2091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tomlinson A. D., Ramey-Hartung B., Day T. W., Merritt P. M., Fuqua C. (2010) Agrobacterium tumefaciens ExoR represses succinoglycan biosynthesis and is required for biofilm formation and motility. Microbiology 156, 2670–2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Danhorn T., Hentzer M., Givskov M., Parsek M. R., Fuqua C. (2004) Phosphorus limitation enhances biofilm formation of the plant pathogen Agrobacterium tumefaciens through the PhoR-PhoB regulatory system. J. Bacteriol. 186, 4492–4501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kereszt A., Kiss E., Reuhs B. L., Carlson R. W., Kondorosi A., Putnoky P. (1998) Novel rkp gene clusters of Sinorhizobium meliloti involved in capsular polysaccharide production and invasion of the symbiotic nodule: the rkpK gene encodes a UDP-glucose dehydrogenase. J. Bacteriol. 180, 5426–5431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Glucksmann M. A., Reuber T. L., Walker G. C. (1993) Genes needed for the modification, polymerization, export, and processing of succinoglycan by Rhizobium meliloti: a model for succinoglycan biosynthesis. J. Bacteriol. 175, 7045–7055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Canter Cremers H. C., Batley M., Redmond J. W., Eydems L., Breedveld M. W., Zevehuizen L. P., Pees E., Wijffelman C. A., Lugtenberg B. J. (1990) Rhizobium leguminosarum exoB mutants are deficient in the synthesis of UDP-glucose 4′-epimerase. J. Biol. Chem. 265, 21122–21127 [PubMed] [Google Scholar]
- 57. Buendia A. M., Enenkel B., Köplin R., Niehaus K., Arnold W., Pühler A. (1991) The Rhizobium meliloti exoZl exoB fragment of megaplasmid 2: ExoB functions as a UDP-glucose 4-epimerase and ExoZ shows homology to NodX of Rhizobium leguminosarum biovar viciae strain TOM. Mol. Microbiol. 5, 1519–1530 [DOI] [PubMed] [Google Scholar]