Background: Prevention of adult articular cartilage and treatment of its lesions are essential.
Results: Microarray analyses identified Fgf18, which inhibits aggrecan release from cartilage and enhances proliferation of chondrocytes. The intra-articular injection of rhFGF18 prevented cartilage degeneration in a rat osteoarthritis model.
Conclusion: Fgf18 protects adult articular cartilage through Timp1 expression.
Significance: Fgf18 may represent a therapeutic agent for osteoarthritis.
Keywords: Cartilage, Chondrocytes, Fibroblast Growth Factor (FGF), Microarray, Osteoarthritis, Regeneration
Abstract
To identify genes that maintain the homeostasis of adult articular cartilage and regenerate its lesions, we initially compared four types of chondrocytes: articular (AA) versus growth plate (AG) cartilage chondrocytes in adult rats, and superficial layer (IS) versus deep layer (ID) chondrocytes of epiphyseal cartilage in infant rats. Microarray analyses revealed that 40 and 186 genes had ≥10-fold higher expression ratios of AA/AG and IS/ID, respectively, and 16 genes showed ≥10-fold of both AA/AG and IS/ID ratios. The results were validated by real-time RT-PCR analysis. Among them, Hoxd1, Fgf18, and Esm1 were expressed more strongly in AA than in IS. Fgf18 was the extracellular and secreted factor that decreased glycosaminoglycan release and depletion from the cartilage, and enhanced proliferation of articular chondrocytes. Fgf18 was strongly expressed in the articular cartilage chondrocytes of adult rats. In a surgical rat osteoarthritis model, a once-weekly injection of recombinant human FGF18 (rhFGF18) given 3 weeks after surgery prevented cartilage degeneration in a dose-dependent manner at 6 and 9 weeks after surgery, with significant effect at 10 μg/week of rhFGF18. As the underlying mechanism, rhFGF18 strongly up-regulated Timp1 expression in the cell and organ cultures, and inhibition of aggrecan release by rhFGF18 was restored by addition of an antibody to Timp1. In conclusion, we have identified Fgf18 as a molecule that protects articular cartilage by gene expression profiling, and the anticatabolic effects may at least partially be mediated by the Timp1 expression.
Introduction
Because adult articular cartilage has poor protective and regenerative capacity by itself (1, 2), its lesions do not heal spontaneously and often culminate in degenerative joint disorders like osteoarthritis. For their treatment, surgical interventions such as arthroscopic abrasion, transplantation of osteochondral grafts, autologous chondrocyte implantation, and finally, total joint replacement have been required (3). Hence, a strategy for prevention and treatment of the articular lesions using a less invasive approach like intra-articular injection of a protective or regenerative agent may greatly improve the treatment in the field of cartilage injury as well as osteoarthritis.
To identify such an agent, we performed whole genome gene expression microarray analyses and sought to identify a set of genes that maintain the homeostasis of adult articular cartilage and regenerate its lesions. Adult articular cartilage is a permanent cartilage, whereas growth plate cartilage is destined to be replaced by bone through the endochondral ossification process. Based on the hypothesis that distinct genes are expressed in the two cartilages and regulate specific functions of their chondrocytes, we compared a gene expression profile of adult articular cartilage chondrocytes with that of growth plate cartilage chondrocytes in rats to identify genes that maintain the homeostasis of the articular cartilage. Furthermore, to identify genes that have a potency to regenerate the articular cartilage, we compared another gene expression profile of superficial layer chondrocytes of infant epiphyseal cartilage which form articular cartilage with that of the deep layer chondrocytes which form growth plate cartilage. Because the identified genes may be of functional relevance for the regulation of articular chondrocyte activities, we believe that the obtained results will help to optimize the therapeutic concepts for the treatment of articular cartilage lesions.
EXPERIMENTAL PROCEDURES
Isolation of Four Types of Cartilage
All animal experiments were undertaken according to the guidelines of the Animal Care and Use Committee of the University of Tokyo and that of the Institute of Laboratory Animal Resources' Guide for the Care and Use of Laboratory Animals of the USA.
Sprague-Dawley rats (Sankyo Laboratories, Tokyo, Japan) were sacrificed by cervical dislocation under diethyl ether anesthesia. For histological analysis, knees of the proximal tibia were harvested from 6-day-old and 10-week-old rats, fixed in 10% formalin, decalcified in KCX solution (Falma, Tokyo), embedded in paraffin, cut into 10-μm sections, and stained with Alcian blue and nuclear fast red.
Adult articular cartilage (AA)2 and adult growth plate cartilage (AG) were harvested from four (two male and two female) 10-week-old rats, and the cartilages were separated from surrounding tissues under a dissecting microscope. Furthermore, after the epiphyseal cartilage of the proximal tibias were isolated under a dissecting microscope from one litter of 6-day-old rats, the surface one-sixth zone and the bottom one-fourth zone of the cartilages were isolated with scalpels as the infant superficial layer (IS) and the infant deep layer (ID), respectively.
The cartilages were digested in DMEM/Ham's F12 (Wako, Osaka, Japan) with 0.3% collagenase (Wako) for 1 h, minced with two scissors, and digested again in DMEM/Ham's F12 with 0.3% collagenase, 0.1% Pronase (Calbiochem), and 1% FBS (Invitrogen) for 3 h. The digests were then filtered through a 70-μm cell strainer (BD Biosciences), pelleted by centrifugation at 1500 rpm, washed with PBS, resuspended with culture medium composed of DMEM/Ham's F12 supplemented with 10% (v/v) FBS, 1% (v/v) penicillin-streptomycin (Sigma-Aldrich), and plated on a 6-cm culture dish. To remove the floating hematopoietic cells and purify chondrocytes, cells were then cultured for 5 days at 37 °C under 5% CO2 with change of the culture medium every day. Then, total RNAs were extracted with TRIZol Reagent (Invitrogen) followed by purification with an RNeasy Mini Kit (Qiagen) according to the manufacturers' instructions.
Real-time RT-PCR
Total RNAs of the four samples were reverse-transcribed using a QuantiTect Reverse Transcription Kit (Qiagen) according to the manufacturer's instructions. Partial length cDNA of Sox9, Col2a1, Prg4, Barx1, Runx2, and Col10a1 including PCR amplicon sequences was amplified by PCR, cloned into pCR-TOPO Zero II vectors (Invitrogen), and used as standard templates after linearization. Real-time RT-PCR was performed on a Thermal Cycler Dice Real-Time System (Takara, Shiga, Japan) using QuantiTect SYBR Green PCR Master Mix (Qiagen) according to the manufacturers' instructions. Copy numbers of target gene mRNA in each total RNA were calculated by reference to standard curves and were adjusted to the murine standard total RNA (Applied Biosystems) with the rodent GAPDH as an internal control. Primer sequence information is available upon request.
For validation of microarray results by real-time RT-PCR analysis and examination of expression levels of matrix metalloproteinase (Mmp), a disintegrin and metalloproteinase with thrombospondin motifs (Adamts), and tissue inhibitor of metalloproteinases (Timp) family genes, relative quantification was performed according to the ΔΔCT method using GAPDH as an internal control.
Microarray Analysis
Whole genome gene expression profiles of AA, AD, IS, and ID were measured using a SurePrint G3 Rat GE 8 X 60K Microarray (Agilent Technologies, Palo Alto, CA). Prior to microarray analysis, the quality of total RNAs of AA, AD, IS, and ID was assessed using a Bioanalyzer 2100 (Agilent Technologies) according to the manufacturer's instruction. RNAs were then labeled using a Low Input Quick Amp Labeling Kit, One-Color (Agilent Technologies) with Cy3. The microarray was hybridized with the targets for 17 h at 65 °C using a Gene Expression Hybridization kit (Agilent Technologies), washed using Gene Expression Wash Buffers Pack (Agilent Technologies), and scanned by a G2565CA Microarray Scanner (Agilent Technologies). Obtained data were processed with Feature Extraction Software (Agilent Technologies). Validity of the signal intensity of each microarray spot was judged by flags generated by this software. If flags IsSaturated, IsFeatNonUnifOL, and IsBGNonUnifOL were all 0 and flags IsPosAndSignif and IsWellAboveBG were both 1, the signal of a spot was judged as valid. For each probe, if signal intensity of a sample was 10-fold or higher than that of another probe and signal of the former was evaluated as valid, we judged that signal of the probe was up-regulated in the former compared with the latter. Each gene loaded on the array had 1–20 probes. If the signal of more than half the probes of a gene were up-regulated in one sample compared with another, we judged that the gene was up-regulated in the former compared with the latter. By this method, we determined genes up-regulated in AA compared with AG and/or in IS compared with ID. All results of the present microarray analyses are provided at ArrayExpress (accession number E-MTAB-1218).
Cell Culture
We isolated primary articular chondrocytes from mouse femoral heads, femoral condyles, and tibial plateau as described previously (4) and cultured them in DMEM/10% FBS with 250 ng/ml rhFGF18 (Peprotech), 600 ng/ml rhESM1 (Peprotech), or the same volume of PBS as the vehicle. We assessed cell proliferation using a CCK-8 assay kit (Dojindo) according to the manufacturer's instruction.
Organ Culture
We isolated 3-week-old mouse femoral head cartilage as described previously (5) and cultured it in DMEM/10% FBS with 250 ng/ml rhFGF18, 2 mg/ml rhESM1 (Lunginnov), 1 μg/ml rhTIMP1 (Peprotech), or the same volume of PBS as the vehicle. We then assessed released glycosaminoglycan amounts in the medium by the dimethylmethylene blue assay during 4-day cultures of mouse femoral heads as described previously (5). To neutralize Timp1, we added an antibody to Timp1 (Abcam) at the final concentration of 5 μg/ml.
Immunofluorescence
The proximal tibia of a 14-week-old Sprague-Dawley rat and mouse femoral head cartilage after the organ culture were harvested, and 10-μm paraffin-embedded sections were prepared as described above. After deparaffinization and blocking with 3% BSA in PBS, the sections were incubated with antibodies to Fgf18, Timp1, Timp3 (1:100; Santa Cruz Biotechnology, Santa Cruz, CA), aggrecan neoepitope DIPEN, NITEGE (1:100; MD Bioproducts, St. Paul, MN), ARG (1:100; Abcam) or nonimmune rabbit IgG (1:100; Millipore). We had performed immunofluorescence with or without the blocking procedure and confirmed that each specific signal was not influenced, and background signals were appropriately masked by this blocking. We detected the signal with a TSA Plus Fluorescence System (PerkinElmer Life Sciences).
Immunoblotting
We collected the medium after 3-day culture of mouse femoral heads and 1-day culture of mouse articular chondrocytes with or without rhFGF18 (2 μg/ml). We concentrated the medium 10 times by acetone precipitation and then analyzed by SDS-PAGE and immunoblotting using an antibody to Timp1.
Rat Osteoarthritis Model Experiments
For the rat experimental osteoarthritis model, a complete medial meniscal tear was surgically created in the right knee joint of isoflurane-anesthetized male Lewis rats (body weight 282–340 g; Harlan Laboratories Inc., Indianapolis, IN) using a previously described technique (6). Three weeks after the surgery, rats were divided into the following groups. The treatment regimens were: (i) a single intra-articular injection of rhFGF18 (0.3, 1, 3, or 10 μg/75 μl of saline) or the vehicle alone (75 μl of saline); (ii) once-weekly intra-articular injection of rhFGF18 for 3 weeks (0.3, 1, 3, or 10 μg/75 μl of saline/week) or the vehicle alone (75 μl of saline) into the right knee. Each group was sacrificed 6 weeks (n = 10 per group) or 9 weeks (n = 5 per group) after the surgery.
After the sacrifice, the right knee was removed and decalcified in 5% formic acid for 4–6 days before cutting in half in the frontal plane and embedding in paraffin wax. Three sections were cut in 200-μm steps, stained with toluidine blue, and analyzed using ImagePro PlusTM software (Media Cybernetics). In scoring the three sections of the joint, the worst case scenario for the two halves on each slide was determined for the cartilage lesion as the cartilage degeneration width (in micrometers). This reflects the areas of tibial cartilage lesion in which both chondrocyte and proteoglycan loss extend ≥50% of the cartilage thickness. The measurement was taken over the area of greatest lesion severity in each of the three zones across the tibial surface. The cartilage degeneration score was determined as described previously (7).
Samples of growth plate, ear, sternum, and trachea were also histologically examined after fixing in formalin. Cartilage thickness was measured using an ocular micrometer through a nontangential section area most representative of the thickness.
Statistical Analyses
Statistical analyses were performed using one-way analysis of variance followed by a Dunnett analysis. Histopathological parameters were compared using the Student's two-tailed t test with significance set at p < 0.05.
RESULTS
Sampling and Characteristics of Chondrocytes from Four Types of Cartilage
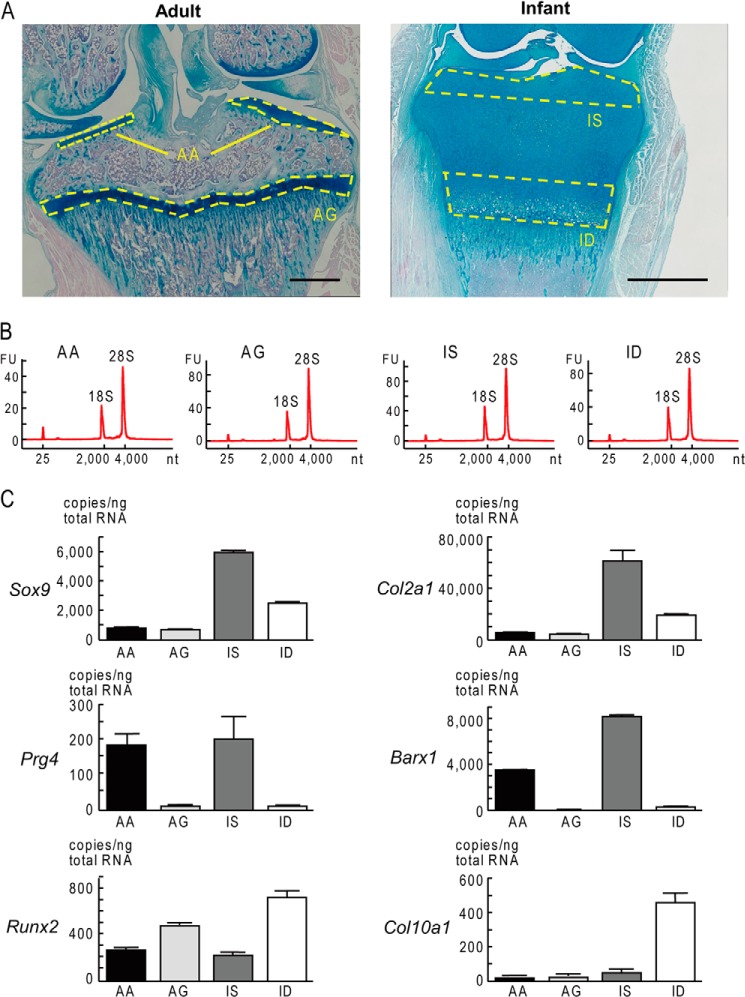
We initially harvested tissues from AA and AG in the proximal tibia of 10-week-old (adult) rats as well as from the IS and ID of epiphyseal cartilage in the same site of 6-day-old (infant) rats (Fig. 1A). After 5-day culture, the total RNA extracted from chondrocytes in the four types of cartilage was confirmed to be of good quality as judged by the wave shape including the ratio of 28S/18S ribosomal RNA (Fig. 1B) (8).
FIGURE 1.
Sampling of chondrocytes from four types of cartilages. A, Alcian blue-stained sections of the proximal tibia of 10-week-old (adult) and 6-day-old (infant) rats are shown. Cartilage samples were harvested from the indicated areas. Scale bars, 1 mm. B, total RNA extracted from chondrocytes in the four types of cartilages after 5-day culture was analyzed with the Agilent 2100 Bioanalyzer to examine their quality and quantity. The 28S and 18S ribosomal RNA peaks indicate good quality. C, mRNA levels of markers of cartilage formation (Sox9 and Col2a1, top row), articular cartilage (Prg4 and Barx1, middle row), and endochondral ossification (Runx2 and Col10a1, bottom row) were measured in chondrocytes from the four types of cartilage (AA, AG, IS, and ID) by real-time RT-PCR. Data are shown as means (bars) ± S.D. (error bars).
To know the characteristics of chondrocytes from the four cartilage types (AA, AG, IS, and ID), we measured expressions of markers of cartilage formation (Sox9 and Col2a1), articular cartilage (Prg4 and Barx1), and endochondral ossification (Runx2 and Col10a1) by real-time RT-PCR (Fig. 1C). The cartilage formation markers Sox9 and Col2a1 were more strongly expressed in infant cartilage chondrocytes (IS and ID) than adult chondrocytes (AA and AG), especially in IS, indicating that IS chondrocytes have the strongest potency of chondrogenic differentiation and cartilage formation. The articular cartilage markers Prg4 and Barx1 were confirmed to be more strongly expressed in AA and IS chondrocytes than in the other two cartilages. Contrarily, the endochondral ossification markers Runx2 and Col10a1 were higher in AG and ID chondrocytes, especially in ID, indicating the potency of terminal differentiation to form bone. These results demonstrate that the four types of chondrocyte maintain their specific characteristics and potencies depending on the site and age.
Comparison of Gene Expression Profiles
Initially, to identify genes that maintain the homeostasis of adult articular cartilage, we compared a gene expression profile of AA chondrocytes with that of AG chondrocytes by microarray analysis, and found that 40 genes showed ≥10-fold higher expression in AA than in AG (supplemental Table 1). Next, to identify genes that generate the articular cartilage, we compared a gene expression profile of IS chondrocytes with that of IP chondrocytes and found that 186 genes showed ≥10-fold higher expression in IS than in IP (supplemental Table 2). The number of genes that showed both potencies (AA/AG≥10 and IS/ID≥10) was 16.
Several Wnt signaling modulators like Wif1 (supplemental Table 1), Sfrp2, Gsc, Apc2, Ndp, Grem1, Frzb, Dkk3, Cpz, Aes (supplemental Table 2), and Barx1 (Table 1) were identified as predominant genes in the AA and IS cartilages (≥10-fold versus AG and IP, respectively). Of note, all of these genes but Ndp are negative regulators of the Wnt signaling. This is consistent with a recent report that the Wnt signaling pathway is suppressed in articular chondrocytes compared with growth plate chondrocytes through expression of the antagonists Frzb and Dkk1 (9). Also, insulin-like growth factor signal-related genes like Igf1, Igfbp5, Wisp2, Nov (supplemental Table 2) and Esm1 (Table 1) were highly expressed in the AA and IS cartilage. Furthermore, expressions of transforming growth factor-β (TGF-β) superfamily-related factors like Eng, Bmpr1b, Gdf15, Grem1 (supplemental Table 2) and Gdf10 (Table 1) were high in the AA and IS cartilage.
TABLE 1.
List of 16 genes of which both AA/AG and IS/ID were shown to be ≥10-fold by the microarray analysis (supplemental Tables 1 and 2)
The ratios of AA/AG, IS/ID, and AA/IS determined by the real-time RT-PCR analysis using the indicated primers are also shown. Three genes that were expressed more strongly in AA than in IS (AA/IS>1) are shown in red.
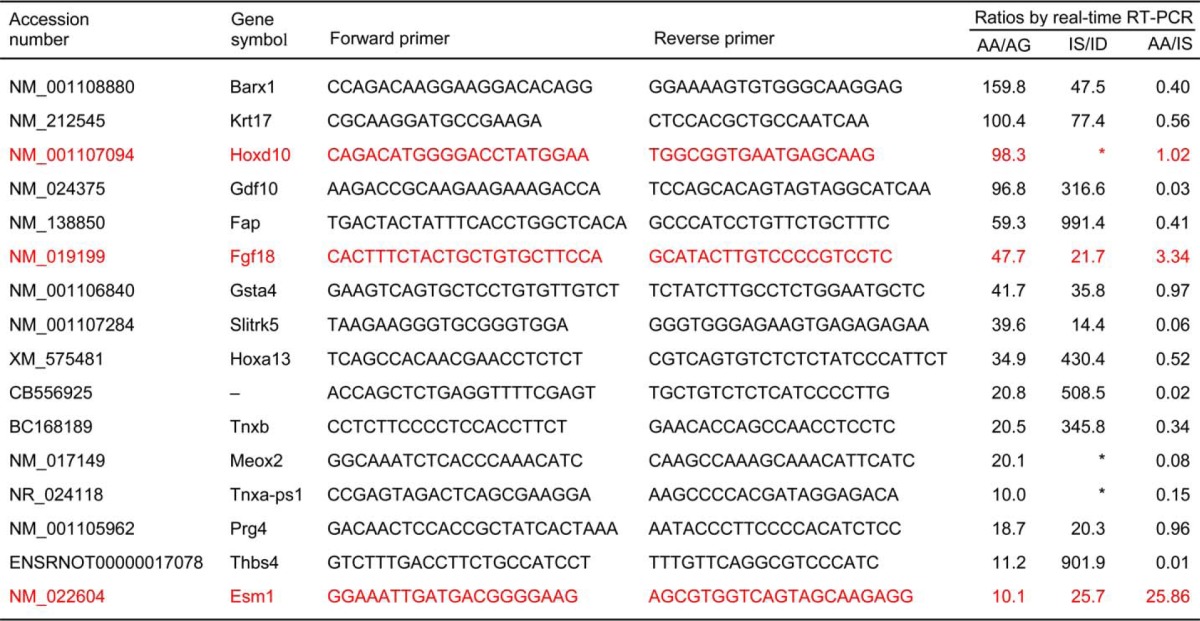
It is known that not all gene microarray data are reproducible because of sample processing errors or the technology per se (10, 11), so we examined the expression profiles of the 16 genes by a real-time RT-PCR analysis and confirmed that both AA/AG and IS/ID were ≥10-fold in all genes (Table 1). Furthermore, because our target genes are assumed to protect and regenerate adult articular cartilage rather than infant cartilage, three among the 16 genes we selected were expressed more strongly in AA than in IS (AA/IS>1): Hoxd10 (homeobox D10), Fgf18 (fibroblast growth factor-18), and Esm1 (endothelial cell-specific molecule-1) (Table 1). Of these three, Fgf18 and Esm1 are extracellular and secreted factors that might be more easily applied for the treatment of lesions of adult articular cartilage than intracellular Hoxd10. Furthermore, the AA/AG ratio of Fgf18 was the highest (47.9) by microarray analysis (supplemental Table 1) and the second highest (47.7) by real-time RT-PCR analysis (Table 1) in the three genes, with much greater levels than Esm1 (10.9 and 10.1, respectively).
rhFGF18 significantly decreased glycosaminoglycan release and depletion from the cartilage of mouse femoral heads whereas rhESM1 did not affect it (Fig. 2A). Furthermore, rhFGF18 more strongly enhanced proliferation of articular chondrocytes than rhESM1 (Fig. 2B). These indicate that Fgf18 may be more potent in protection of articular cartilage than Esm1. Immunofluorescence confirmed that Fgf18 was strongly expressed in the articular cartilage chondrocytes of adult rats but was hardly detected in the growth plate cartilage (Fig. 2C).
FIGURE 2.
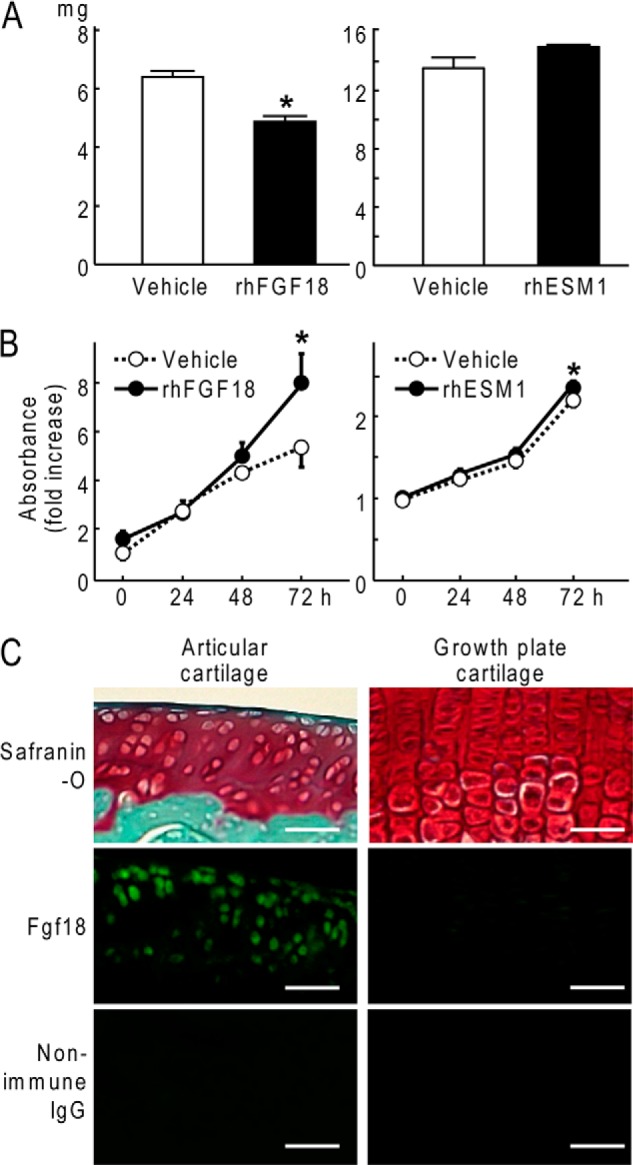
A, effects of rhFGF18 and rhESM1 on released glycosaminoglycan amounts in the medium determined by the dimethylmethylene blue assay during 4-day culture of mouse femoral heads. Data are shown as means (bars) ± S.D. (error bars) for 16 samples of rhFGF18 group and 3 samples of rhESM1 group. *, p < 0.05; significant difference from the vehicle group. B, time course of effects of rhFGF18 and rhESM1 on cell proliferation curves determined by the CCK-8 assay during 72-h cultures of mouse primary chondrocytes. Data are shown as means (symbols) ± S.D. (error bars) for three wells/group. *, p < 0.05; significant difference from the vehicle group. C, safranin O staining and immunofluorescence using an anti-Fgf18 antibody or the control nonimmune rabbit IgG in articular cartilage and growth plate cartilage of the proximal tibia of a 14-week-old rat. Scale bars, 50 μm.
Effect of rhFGF18 in the Rat Model of Osteoarthritis
A previous study has already shown that twice-weekly intra-articular injection of rhFGF18 for 3 weeks prevented cartilage degeneration in the surgically induced rat meniscal tear model (6). To confirm the therapeutic effect of rhFGF18 on cartilage lesions and to determine whether such frequent injections are necessary, we focused on the effect of a single and once-weekly intra-articular injection of rhFGF18 in the same model. Three weeks after the surgery of the meniscal tear to induce osteoarthritis in the right knee, we performed a single intra-articular injection of rhFGF18 (0.3, 1, 3, or 10 μg) or once-weekly intra-articular injection of rhFGF18 (0.3, 1, 3, or 10 μg/week) for 3 weeks into the knee and compared the cartilage lesion with those of an equivalent group of rats that received intra-articular injection of vehicle alone (75 μl of saline). Quantification was performed of the cartilage degeneration width (in micrometers) and the OARSI cartilage degeneration score of medial knees 6 and 9 weeks after the surgery (3 and 6 weeks after the start of injection). The cartilage degeneration progressed in the control saline-injected knees at 6 weeks after surgery and even more at 9 weeks (Fig. 3), and no group of any rhFGF18 concentration with a single injection significantly affected the degeneration compared with the saline group at either time point (Fig. 3A, left panel). However, once-weekly injection of rhFGF18 decreased the cartilage degeneration width in a dose-dependent manner both at 6 and 9 weeks, and the cartilage degeneration score at 9 weeks (Fig. 3A, right). A significant effect was seen with 10 μg/week of rhFGF18, which was confirmed by representative histological features of rat medial knees (Fig. 3B). We believe that rhFGF18 in this condition evokes very robust structural benefit because five of five studies showed reproducible results.
FIGURE 3.
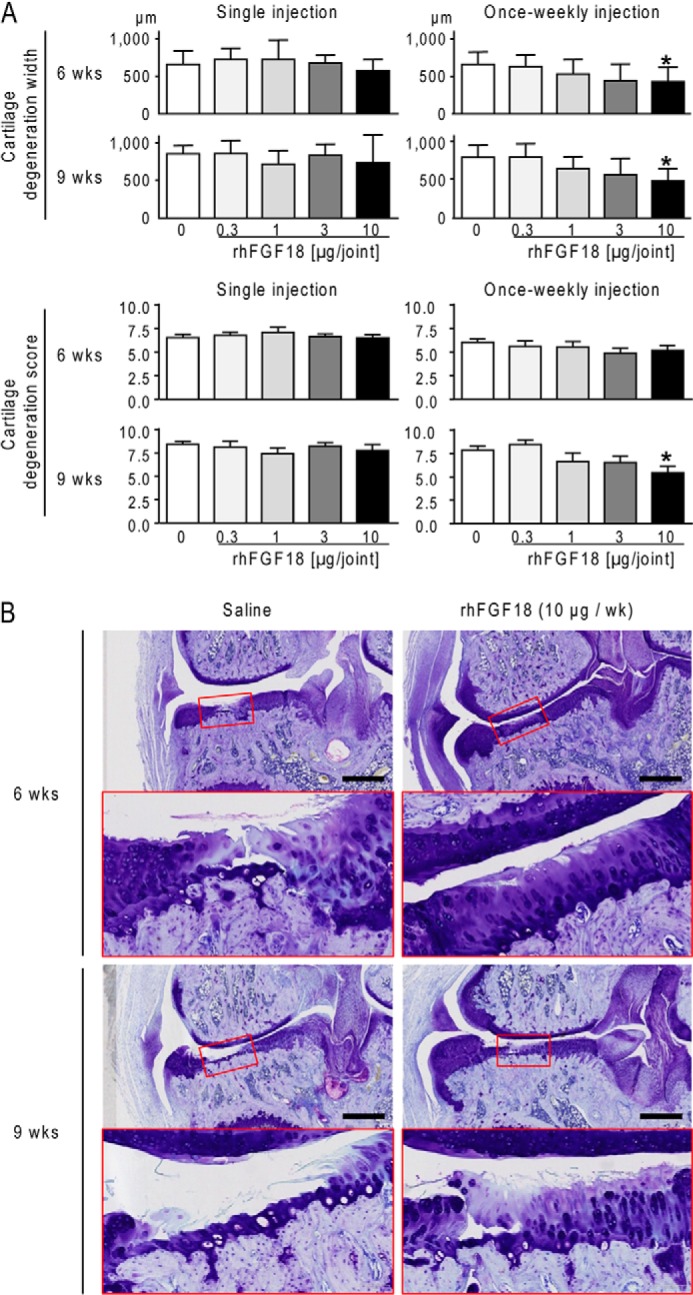
Therapeutic effect of rhFGF18 on cartilage lesions in an experimental rat osteoarthritis model. A, cartilage degeneration width (upper) and score (lower) of rat medial knees 6 weeks (n = 10/group) and 9 weeks (n = 5/group) after surgically induced osteoarthritis, following a single intra-articular injection of saline or rhFGF18 (0.3, 1, 3, and 10 μg) (left) or once-weekly intra-articular injection for 3 weeks (right) of saline or different dosage regimens of rhFGF18 (0.3, 1, 3, and 10 μg/week). Data are shown as means (bars) ± S.D. (error bars) for 10 (6 weeks) and 5 (9 weeks) mice/group. *, p < 0.05; significant difference from the saline group. B, representative toluidine blue stainings of rat medial knees 6 and 9 weeks after surgically induced osteoarthritis, following once-weekly intra-articular injection of saline or rhFGF18 (10 μg/week) for 3 weeks. Inset boxes indicate the regions shown in the enlarged images immediately below. Scale bars, 1 mm.
Our further histological examination in cartilaginous tissues other than articular cartilage revealed no effect by any of the single or once-weekly rhFGF18 regimens studied on growth plate, ear, sternum, or tracheal cartilage samples (data not shown). Furthermore, following intra-articular administration, levels of intact rhFGF18 in blood were minimal, suggesting that the potential systemic effects of rhFGF18 are likely to be slight and are unlikely to confound the therapeutic potential of rhFGF18 in cartilage repair (data not shown).
Mechanism Underlying the Anticatabolic Effect of Fgf18 on Articular Cartilage
To analyze the mechanism of the anticatabolic effect of Fgf18, mouse femoral head explants and mouse articular chondrocytes were cultured with and without rhFGF18, and mRNA levels of Mmp, Adamts, and Timp family genes were compared. Among them, Timp1 was significantly altered by rhFGF18 in both cultures (Fig. 4A). Immunoblotting confirmed that rhFGF18 induced Timp1 protein expression in the cultures of mouse femoral heads and mouse articular chondrocytes (Fig. 4B). Furthermore, a decrease of glycosaminoglycan release by rhFGF18 in the culture medium of mouse femoral heads was partially restored by addition of a neutralizing antibody to Timp1, indicating that the Fgf18 anticatabolic effect was at least partially mediated by Timp1 (Fig. 4C).
FIGURE 4.
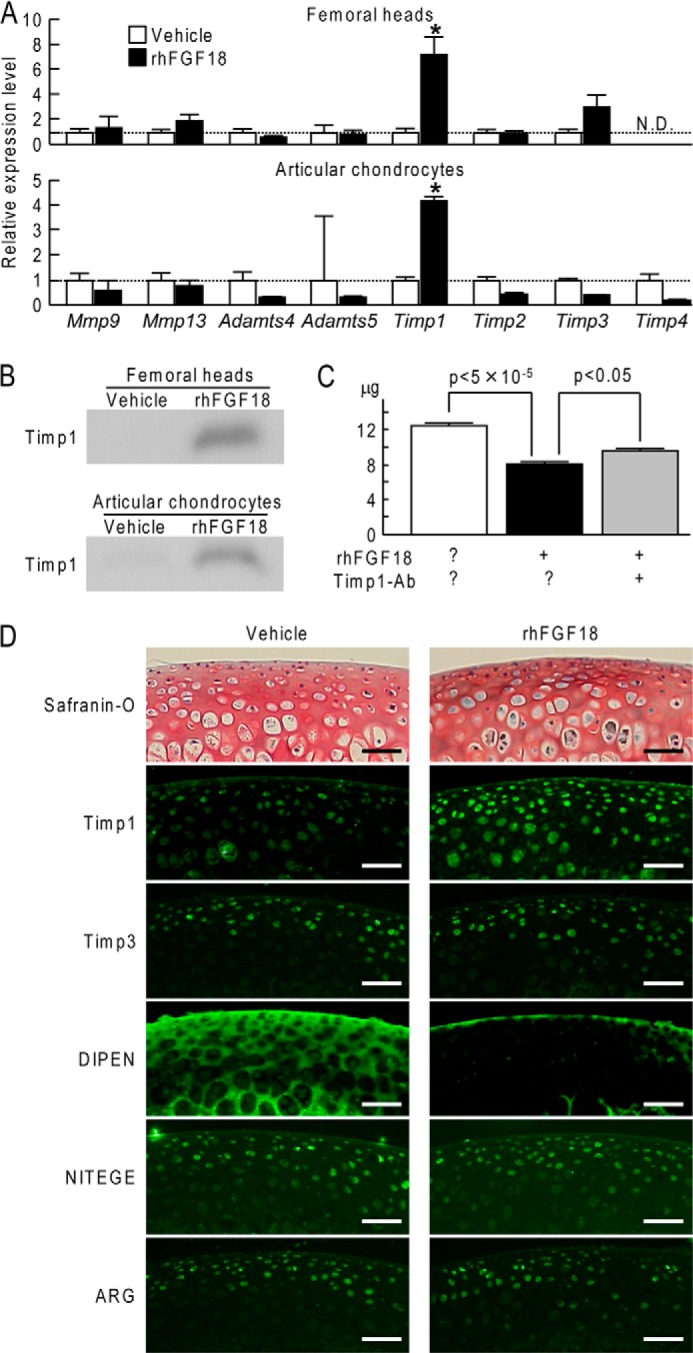
A, effects of rhFGF18 on mRNA levels of Mmp, Adamts, and Timp family genes after 3-day culture of mouse femoral heads (2 μg/ml rhFGF18) and 1-day culture of mouse articular chondrocytes (250 ng/ml rhFGF18). *, p < 0.01; significant difference from the vehicle group. N.D., not detected. B, immunoblotting of the medium after 3-day culture of mouse femoral heads (2 μg/ml rhFGF18) and 1-day culture of mouse articular chondrocytes (250 ng/ml rhFGF18) using an antibody to Timp1. The medium was concentrated 10 times by acetone precipitation. C, effects of rhFGF18 (250 ng/ml) and/or an antibody to Timp1 (Timp1-Ab) on released glycosaminoglycan amounts determined by the dimethylmethylene blue assay during a 4-day culture of mouse femoral heads. Data are shown as means (bars) ± S.D. (error bars) for seven samples/group. D, safranin O staining and immunofluorescence for Timp1, Timp3, DIPEN, NITEGE, and ARG in mouse femoral head cartilage after 4-day culture with or without rhFGF18. Scale bars, 50 μm.
In addition, we examined expressions of Timp1, Timp3, and several aggrecan neoepitopes in mouse femoral head cartilage after a 4-day culture with or without rhFGF18. Timp1 protein was confirmed to be increased by rhFGF18 treatment, whereas Timp3 protein was not altered (Fig. 4D). Notably, aggrecan neoepitope DIPEN, which is known to be generated by Mmp, was markedly suppressed by rhFGF18 treatment, whereas neither NITEGE nor ARG, which are generated by Adamts, was altered. These data indicate that Fgf18 suppressed aggrecan cleavage by Mmp through induction of Timp1.
Effect of Timp1 on Aggrecan Cleavage and Release
Because a functional role of Timp1 in articular cartilage remains unknown, we finally performed rhTIMP1 treatment in the culture of mouse femoral heads. Just like rhFGF18 treatment, rhTIMP1 significantly decreased glycosaminoglycan release and depletion from the cartilage (Fig. 5A). Furthermore, immunofluorescence confirmed suppression of DIPEN by rhTIMP1, similar to that by rhFGF18 (Fig. 5B).
FIGURE 5.
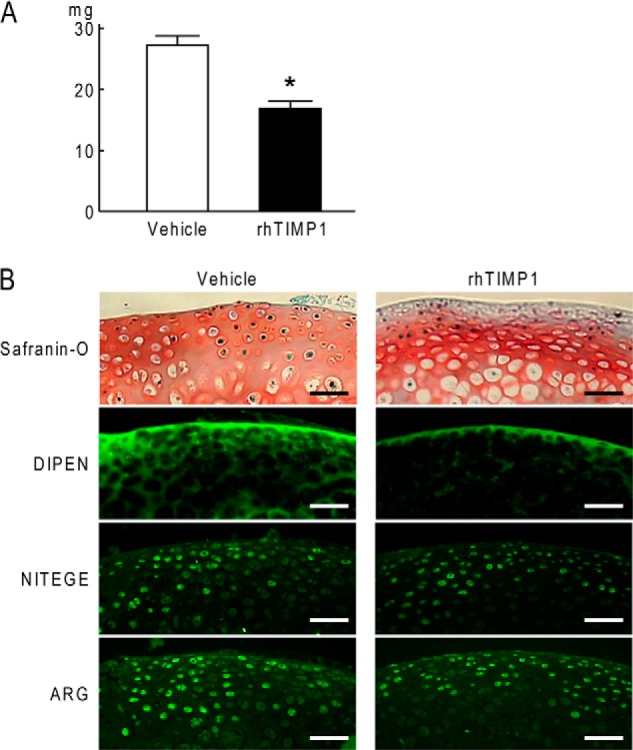
A, effects of rhTIMP1 on released glycosaminoglycan amounts in the medium determined by the dimethylmethylene blue assay during 4-day culture of mouse femoral heads. Data are shown as means (bars) ± S.D. (error bars) for three samples of each group. *, p < 0.05, significant difference from the vehicle group. B, safranin O staining and immunofluorescence for DIPEN, NITEGE, and ARG in mouse femoral head cartilage after 4-day culture with or without rhTIMP1. Scale bars, 50 μm.
DISCUSSION
In the present gene profiling study, both Prg4 and Barx1, well known articular cartilage markers, were included in the 16 genes of which both AA/AG and IS/ID were ≥10-fold by the microarray and real-time RT-PCR analyses (Table 1), indicating the accuracy of the present approach. Prg4, also called superficial zone protein or lubricin, is a secreted glycoprotein that provides lubrication and protects articular cartilage surfaces (12). The high expression of Prg4 in the articular chondrocytes is consistent with previous reports on gene profiling studies in cartilage (13, 14). Barx1, a homeobox-containing transcription factor, is known to be expressed in the developing joint interzone cells (15), which give rise to articular chondrocytes later (16, 17). High expression of Barx1 in the adult articular cartilage shown by the present study implies that it may also play an important role not only in the formation but also in the maintenance of articular cartilage. Meanwhile, we cultured the harvested cells for 5 days before the RNA extraction to exclude floating blood cells. We cannot deny the possibility that the in vitro condition during the culture might have influenced the gene expression patterns.
The present study identified several Wnt, Igf, and Tgf-β signal-related molecules as predominant genes in AA and IS cartilages. Previous gene profiling studies also showed higher expressions of these factors including Dkk3, Wif1, Sfrp5, Igfbp4/5, Nov, Gdf10, Grem1 and Frzb in articular chondrocytes than growth plate chondrocytes (9, 13, 18). We speculate that it may be more appropriate to use these signaling molecules in tissue engineering of articular cartilage because the articular chondrocytes under their physiological conditions in vivo are exposed to these factors and therefore may respond to them more optimally.
Like Fgf18, Esm1 was an extracellular and secreted factor of which both AA/AG and IS/ID were ≥10-fold and AA/IS was >1 in the 16 selected genes. Esm1 has been reported to be secreted mainly by endothelial cells of organs like lung or kidney and to function as a regulator of cell adhesion, inflammatory disorders, and tumor progression (19, 20). Similarly, Thbs4 is another extracellular matrix protein holding promise for the study of the role of articular cartilage integrity in the 16 selected genes because Thbs4 was also reported to show the highest specificity for articular cartilage expression in a previous RT-PCR screening (18). In fact, most of the other thrombospondins, especially Thbs1, Thbs2, and Thbs5 (also known as Comp), have been functionally studied extensively in cartilage (21, 22). Also, the identified expression profiles of other genes may provide new insights into understanding the physiological gene control of chondrocytic phenotype. Further studies of the individual genes of interest may generate clues for selecting physiologically relevant signals in tissue engineering of articular cartilage.
The present gene profiling analyses identified Fgf18 as a potent molecule that protects or regenerates adult articular cartilage. In fact, endogenous Fgf18 is known to play an important role in skeletal growth and development because mice lacking Fgf18 exhibit several malformations such as delayed closure of the calvarial sutures, enlargement of the growth plate, and impairment of osteogenic differentiation (23, 24). In contrast to its role in the growth plate where it inhibits chondrocyte proliferation and differentiation (23–25), Fgf18 has been reported to have anabolic effects on chondrocytes in other cartilaginous tissues like auricular cartilage, bronchial cartilage, and costal cartilage in animal models (25–27). These effects seem to be because of the direct action on mature chondrocytes rather than on the progenitor cells (25–27). In the present study, endogenous Fgf18 was shown to be abundantly expressed in mature chondrocytes of articular cartilage (Fig. 2C), suggesting the cell-autonomous or autocrine/paracrine effects. Although the receptors that mediate the Fgf18-induced regeneration of articular cartilage have not yet been established, Fgf18 is generally known to activate the IIIc splice variants of FgfR2 and FgfR3, and both of these receptors are known to be expressed in chondrocytes of human and murine articular cartilage (25, 26, 28–31).
The present study further identified Timp1 as a novel downstream molecule of Fgf18 in articular chondrocytes. Timps are endogenous protein regulators of the Mmp, a disintegrin and metalloproteinase (Adam), and Adamts family (32). In mammals, four Timps (Timp1–4) have been identified (32), and all of them are abundantly expressed in human normal and osteoarthritic cartilage (33). All four Timps inhibit proteinase family members, but with affinities that vary for different inhibitor-proteinase pairs. For example, Timp1 inhibits Mt1-mmp, Mt3-mmp, Mt5-mmp, Mmp19, and Adam10 (32). However, Timp1 was not identified by the present microarray analysis, and its expression levels in the four types of samples were similar. We speculate that the Timp1 induction by Fgf18 is not essential in 6-day-old and 10-week-old rat cartilages, but that Timp1 is one of the mediators of the rhFGF18 action. Similar suppressions of aggrecan neoepitope DIPEN and no obvious change of NITEGE and ARG between rhFGF18 and rhTIMP1 in the present study suggest that Fgf18 exerts its anticatabolic effect through inhibition of Mmp activity by Timp1 induction. In addition to their proteinase inhibitory activity, Timps are known to have various biological functions such as promoting cell proliferation, inhibiting angiogenesis, and modulating apoptosis (34). Hence, Timp1 might mediate not only the anticatabolic effect of Fgf18, but also its proanabolic effect, although the precise mechanism remains unclarified.
Several growth factors other than Fgf18 have been reported in animal models to have anabolic effects on cartilage including insulin-like growth factor-I, osteogenic protein-1, TGF-β, hepatocyte growth factor, and Fgf2 (35–42). Although the apparent efficacy of these factors has been proved in osteochondral defect models and partial thickness chondral defect models, Fgf18 was the first reported to show the protective or regenerative effect in the surgically induced rat meniscal tear model by Moore et al. (6). They showed that twice-weekly intra-articular injection of rhFGF18 (0, 0.1, 1.0, and 5.0 μg) for 3 weeks prevented cartilage degeneration in a dose-dependent manner (6). In the present study, the effect seemed not to be achieved by a single injection; however, once-weekly injection of rhFGF18 (10 μg/week) could maintain the protective effect at least for 6 weeks after the injection without showing side or adverse effects on other cartilages. These findings support the further evaluation of rhFGF18 as an innovative approach to the treatment of osteoarthritis by disease modification rather than by symptom management.
Acknowledgments
We thank R. Yamaguchi, H. Kawahara, and D. Mori for technical assistance and the contribution and expertise of BolderBioPath, Inc., especially A. Bendele, for performing the rat mensical tear study.
The gene expression profiling study was supported by Grants-in-aid for Scientific Research 19109007 and 23689065 from the Japanese Ministry of Education, Culture, Sports, Science and Technology. The rat osteoarthritis study was sponsored by Merck Serono S.A., Geneva, Switzerland (a branch of Merck Serono S.A., Coinsins, Switzerland, an affiliate of Merck KGaA, Darmstadt, Germany). rhFGF18 (INN: sprifermin) is in clinical development and is not currently approved by any regulatory authority, including the European Medicines Agency (EMA) or the US Food and Drug Administration (FDA). C. H. L. and H. G. are employees of Merck KGaA.

This article contains supplemental Tables 1 and 2.
- AA
- adult articular cartilage
- Adamts
- a disintegrin and metalloprotease with thrombospondin motifs
- AG
- adult growth plate cartilage
- ID
- infant deep layer
- IS
- infant superficial layer
- Mmp
- matrix metalloprotainase
- rhFGF18
- recombinant human fibroblast growth factor-18
- Timp
- tissue inhibitor of metalloproteinases.
REFERENCES
- 1. Darling E. M., Athanasiou K. A. (2003) Articular cartilage bioreactors and bioprocesses. Tissue Eng. 9, 9–26 [DOI] [PubMed] [Google Scholar]
- 2. Hunziker E. B. (1999) Articular cartilage repair: are the intrinsic biological constraints undermining this process insuperable? Osteoarthritis Cartilage 7, 15–28 [DOI] [PubMed] [Google Scholar]
- 3. Farr J., Cole B., Dhawan A., Kercher J., Sherman S. (2011) Clinical cartilage restoration: evolution and overview. Clin. Orthop. Relat. Res. 469, 2696–2705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gosset M., Berenbaum F., Thirion S., Jacques C. (2008) Primary culture and phenotyping of murine chondrocytes. Nat. Protoc. 3, 1253–1260 [DOI] [PubMed] [Google Scholar]
- 5. Stanton H., Golub S. B., Rogerson F. M., Last K., Little C. B., Fosang A. J. (2011) Investigating ADAMTS-mediated aggrecanolysis in mouse cartilage. Nat. Protoc. 6, 388–404 [DOI] [PubMed] [Google Scholar]
- 6. Moore E. E., Bendele A. M., Thompson D. L., Littau A., Waggie K. S., Reardon B., Ellsworth J. L. (2005) Fibroblast growth factor-18 stimulates chondrogenesis and cartilage repair in a rat model of injury-induced osteoarthritis. Osteoarthritis Cartilage 13, 623–631 [DOI] [PubMed] [Google Scholar]
- 7. Gerwin N., Bendele A. M., Glasson S., Carlson C. S. (2010) The OARSI histopathology initiative: recommendations for histological assessments of osteoarthritis in the rat. Osteoarthritis Cartilage 18, S24–S34 [DOI] [PubMed] [Google Scholar]
- 8. Schroeder A., Mueller O., Stocker S., Salowsky R., Leiber M., Gassmann M., Lightfoot S., Menzel W., Granzow M., Ragg T. (2006) The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol. Biol. 7, 3–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Leijten J. C., Emons J., Sticht C., van Gool S., Decker E., Uitterlinden A., Rappold G., Hofman A., Rivadeneira F., Scherjon S., Wit J. M., van Meurs J., van Blitterswijk C. A., Karperien M. (2012) Gremlin 1, frizzled-related protein, and Dkk-1 are key regulators of human articular cartilage homeostasis. Arthritis. Rheum. 64, 3302–3312 [DOI] [PubMed] [Google Scholar]
- 10. Chuaqui R. F., Bonner R. F., Best C. J., Gillespie J. W., Flaig M. J., Hewitt S. M., Phillips J. L., Krizman D. B., Tangrea M. A., Ahram M., Linehan W. M., Knezevic V., Emmert-Buck M. R. (2002) Post-analysis follow-up and validation of microarray experiments. Nat. Genet. 32, 509–514 [DOI] [PubMed] [Google Scholar]
- 11. Rajeevan M. S., Vernon S. D., Taysavang N., Unger E. R. (2001) Validation of array-based gene expression profiles by real-time (kinetic) RT-PCR. J. Mol. Diagn. 3, 26–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rhee D. K., Marcelino J., Baker M., Gong Y., Smits P., Lefebvre V., Jay G. D., Stewart M., Wang H., Warman M. L., Carpten J. D. (2005) The secreted glycoprotein lubricin protects cartilage surfaces and inhibits synovial cell overgrowth. J. Clin. Invest. 115, 622–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yamane S., Cheng E., You Z., Reddi A. H. (2007) Gene expression profiling of mouse articular and growth plate cartilage. Tissue Eng. 13, 2163–2173 [DOI] [PubMed] [Google Scholar]
- 14. Schumacher B. L., Block J. A., Schmid T. M., Aydelotte M. B., Kuettner K. E. (1994) A novel proteoglycan synthesized and secreted by chondrocytes of the superficial zone of articular cartilage. Arch. Biochem. Biophys. 311, 144–152 [DOI] [PubMed] [Google Scholar]
- 15. Church V., Yamaguchi K., Tsang P., Akita K., Logan C., Francis-West P. (2005) Expression and function of Bapx1 during chick limb development. Anat. Embryol. 209, 461–469 [DOI] [PubMed] [Google Scholar]
- 16. Rountree R. B., Schoor M., Chen H., Marks M. E., Harley V., Mishina Y., Kingsley D. M. (2004) BMP receptor signaling is required for postnatal maintenance of articular cartilage. PLoS Biol. 2, e355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Koyama E., Shibukawa Y., Nagayama M., Sugito H., Young B., Yuasa T., Okabe T., Ochiai T., Kamiya N., Rountree R. B., Kingsley D. M., Iwamoto M., Enomoto-Iwamoto M., Pacifici M. (2008) A distinct cohort of progenitor cells participates in synovial joint and articular cartilage formation during mouse limb skeletogenesis. Dev. Biol. 316, 62–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hissnauer T. N., Baranowsky A., Pestka J. M., Streichert T., Wiegandt K., Goepfert C., Beil F. T., Albers J., Schulze J., Ueblacker P., Petersen J. P., Schinke T., Meenen N. M., Pörtner R., Amling M. (2010) Identification of molecular markers for articular cartilage. Osteoarthritis Cartilage 18, 1630–1638 [DOI] [PubMed] [Google Scholar]
- 19. Bechard D., Meignin V., Scherpereel A., Oudin S., Kervoaze G., Bertheau P., Janin A., Tonnel A., Lassalle P. (2000) Characterization of the secreted form of endothelial-cell-specific molecule 1 by specific monoclonal antibodies. J. Vasc. Res. 37, 417–425 [DOI] [PubMed] [Google Scholar]
- 20. Sarrazin S., Adam E., Lyon M., Depontieu F., Motte V., Landolfi C., Lortat-Jacob H., Bechard D., Lassalle P., Delehedde M. (2006) Endocan or endothelial cell specific molecule-1 (ESM-1): a potential novel endothelial cell marker and a new target for cancer therapy. Biochim. Biophys. Acta 1765, 25–37 [DOI] [PubMed] [Google Scholar]
- 21. Agah A., Kyriakides T. R., Lawler J., Bornstein P. (2002) The lack of thrombospondin-1 (TSP1) dictates the course of wound healing in double-TSP1/TSP2-null mice. Am. J. Pathol. 161, 831–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bornstein P., Agah A., Kyriakides T. R. (2004) The role of thrombospondins 1 and 2 in the regulation of cell-matrix interactions, collagen fibril formation, and the response to injury. Int. J. Biochem. Cell Biol. 36, 1115–1125 [DOI] [PubMed] [Google Scholar]
- 23. Ohbayashi N., Shibayama M., Kurotaki Y., Imanishi M., Fujimori T., Itoh N., Takada S. (2002) FGF18 is required for normal cell proliferation and differentiation during osteogenesis and chondrogenesis. Genes Dev. 16, 870–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu Z., Xu J., Colvin J. S., Ornitz D. M. (2002) Coordination of chondrogenesis and osteogenesis by fibroblast growth factor 18. Genes Dev. 16, 859–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ellsworth J. L., Berry J., Bukowski T., Claus J., Feldhaus A., Holderman S., Holdren M. S., Lum K. D., Moore E. E., Raymond F., Ren H., Shea P., Sprecher C., Storey H., Thompson D. L., Waggie K., Yao L., Fernandes R. J., Eyre D. R., Hughes S. D. (2002) Fibroblast growth factor-18 is a trophic factor for mature chondrocytes and their progenitors. Osteoarthritis Cartilage 10, 308–320 [DOI] [PubMed] [Google Scholar]
- 26. Whitsett J. A., Clark J. C., Picard L., Tichelaar J. W., Wert S. E., Itoh N., Perl A. K., Stahlman M. T. (2002) Fibroblast growth factor 18 influences proximal programming during lung morphogenesis. J. Biol. Chem. 277, 22743–22749 [DOI] [PubMed] [Google Scholar]
- 27. Shimoaka T., Ogasawara T., Yonamine A., Chikazu D., Kawano H., Nakamura K., Itoh N., Kawaguchi H. (2002) Regulation of osteoblast, chondrocyte, and osteoclast functions by fibroblast growth factor (FGF)-18 in comparison with FGF-2 and FGF-10. J. Biol. Chem. 277, 7493–7500 [DOI] [PubMed] [Google Scholar]
- 28. Davidson D., Blanc A., Filion D., Wang H., Plut P., Pfeffer G., Buschmann M. D., Henderson J. E. (2005) Fibroblast growth factor (FGF) 18 signals through FGF receptor 3 to promote chondrogenesis. J. Biol. Chem. 280, 20509–20515 [DOI] [PubMed] [Google Scholar]
- 29. Im H. J., Muddasani P., Natarajan V., Schmid T. M., Block J. A., Davis F., van Wijnen A. J., Loeser R. F. (2007) Basic fibroblast growth factor stimulates matrix metalloproteinase-13 via the molecular cross-talk between the mitogen-activated protein kinases and protein kinase Cδ pathways in human adult articular chondrocytes. J. Biol. Chem. 282, 11110–11121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Muddasani P., Norman J. C., Ellman M., van Wijnen A. J., Im H. J. (2007) Basic fibroblast growth factor activates the MAPK and NFκB pathways that converge on Elk-1 to control production of matrix metalloproteinase-13 by human adult articular chondrocytes. J. Biol. Chem. 282, 31409–31421 [DOI] [PubMed] [Google Scholar]
- 31. Yan D., Chen D., Cool S. M., van Wijnen A. J., Mikecz K., Murphy G., Im H. J. (2011) Fibroblast growth factor receptor 1 is principally responsible for fibroblast growth factor 2-induced catabolic activities in human articular chondrocytes. Arthritis Res. Ther. 13, R130. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32. Murphy G. (2011) Tissue inhibitors of metalloproteinases. Genome Biol. 12, 233–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kevorkian L., Young D. A., Darrah C., Donell S. T., Shepstone L., Porter S., Brockbank S. M., Edwards D. R., Parker A. E., Clark I. M. (2004) Expression profiling of metalloproteinases and their inhibitors in cartilage. Arthritis Rheum. 50, 131–141 [DOI] [PubMed] [Google Scholar]
- 34. Brew K., Nagase H. (2010) The tissue inhibitors of metalloproteinases (TIMPs): an ancient family with structural and functional diversity. Biochim. Biophys. Acta 1803, 55–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hunziker E. B. (2001) Growth-factor-induced healing of partial-thickness defects in adult articular cartilage. Osteoarthritis Cartilage 9, 22–32 [DOI] [PubMed] [Google Scholar]
- 36. Nixon A. J., Fortier L. A., Williams J., Mohammed H. (1999) Enhanced repair of extensive articular defects by insulin-like growth factor-I-laden fibrin composites. J. Orthop. Res. 17, 475–487 [DOI] [PubMed] [Google Scholar]
- 37. Louwerse R. T., Heyligers I. C., Klein-Nulend J., Sugihara S., van Kampen G. P., Semeins C. M., Goei S. W., de Koning M. H., Wuisman P. I., Burger E. H. (2000) Use of recombinant human osteogenic protein-1 for the repair of subchondral defects in articular cartilage in goats. J. Biomed. Mater. Res. 49, 506–516 [DOI] [PubMed] [Google Scholar]
- 38. Jelic M., Pecina M., Haspl M., Kos J., Taylor K., Maticic D., McCartney J., Yin S., Rueger D., Vukicevic S. (2001) Regeneration of articular cartilage chondral defects by osteogenic protein-1 (bone morphogenetic protein-7) in sheep. Growth Factors 19, 101–113 [DOI] [PubMed] [Google Scholar]
- 39. Frenkel S. R., Saadeh P. B., Mehrara B. J., Chin G. S., Steinbrech D. S., Brent B., Gittes G. K., Longaker M. T. (2000) Transforming growth factor β superfamily members: role in cartilage modeling. Plast. Reconstr. Surg. 105, 980–990 [DOI] [PubMed] [Google Scholar]
- 40. Wakitani S., Imoto K., Kimura T., Ochi T., Matsumoto K., Nakamura T. (1997) Hepatocyte growth factor facilitates cartilage repair: full thickness articular cartilage defect studied in rabbit knees. Acta Orthop. Scand. 68, 474–480 [DOI] [PubMed] [Google Scholar]
- 41. Cuevas P., Burgos J., Baird A. (1988) Basic fibroblast growth factor (FGF) promotes cartilage repair in vivo. Biochem. Biophys. Res. Commun. 156, 611–618 [DOI] [PubMed] [Google Scholar]
- 42. Tanaka H., Mizokami H., Shiigi E., Murata H., Ogasa H., Mine T., Kawai S. (2004) Effects of basic fibroblast growth factor on the repair of large osteochondral defects of articular cartilage in rabbits: dose-response effects and long-term outcomes. Tissue Eng. 10, 633–641 [DOI] [PubMed] [Google Scholar]