Abstract
In bacterial pathogenesis, monitoring and adapting to the dynamically changing environment in the host and an ability to disrupt host immune responses are critical. The virulence determinants of pathogenic bacteria include the sensor/signaling proteins of the serine/threonine protein kinase (STPK) family that have a dual role of sensing the environment and subverting specific host defense processes. STPKs can sense a wide range of signals and coordinate multiple cellular processes to mount an appropriate response. Here, we review some of the well studied bacterial STPKs that are essential virulence factors and that modify global host responses during infection.
Keywords: Bacterial Signal Transduction, Host-Pathogen Interactions, Protein Secretion, Serine/Threonine Protein Kinase, Virulence Factors
Introduction
Successful adaptation to a changing environment requires efficient monitoring and a rapid response. The cascade of chemical reactions culminate in gene transcription and a fast metabolic adaptation. These rapid changes are important especially when responses have to be orchestrated from different cellular compartments. Thus, given the importance of signaling in the normal functioning of the host cell, it is not surprising that pathogens exploit host cell signaling networks to optimize their infectious cycles. Signaling systems are commonly involved in regulation of the expression of virulence factors of pathogenic bacteria during disease progression. Previously, the two-component systems were the only tools known for environmental sensing in bacteria (1, 2). In contrast, signaling in eukaryotes is accomplished primarily by a network of protein phosphorylation cascades that require the coordinated action of a number of serine/threonine and tyrosine kinases and their associated phosphatases. These protein kinases transfer a phosphate group from ATP or GTP onto specific serine, threonine, and/or tyrosine residues of a protein substrate. Typically, the phosphorylation functionally activates the substrate to perform either a specific activity or cellular localization and/or transfer the phosphate group to a downstream effector, initiating a cascade of signal-response reactions. The reverse reaction of dephosphorylation restores the activators and effectors to their initial functional state (not phosphorylated), preparing the system for the next signaling event. Thus, kinases and phosphatases function as on/off switches, modulating specific signal transduction pathways (3).
Until recently, it was assumed that signaling in prokaryotic and eukaryotic organisms was mediated by distinct mechanisms. However, recent advances in genetic strategies and genome sequencing have revealed the existence of “eukaryote-like” serine/threonine protein kinases (STPKs)2 and phosphatases in a number of prokaryotic organisms, including pathogens such as Streptococcus spp. (4–7), Mycobacterium (8–13), Yersinia spp. (14, 15), Listeria monocytogenes (16, 17), Pseudomonas aeruginosa (18), Enterococcus faecalis (19), and Staphylococcus aureus (20–22). In fact, the so-called eukaryote-like Ser/Thr kinases identified in prokaryotes share characteristic signature sequence motifs with the eukaryotic protein kinase superfamily based on sequence homology between their kinase domains (23). These domains are typically organized into 12 subdomains (Hanks domains) that fold in a characteristic two-lobed catalytic core structure, with the catalytic active site lying in a deep cleft formed between the two lobes (23, 24). The kinase catalytic domain can be defined by the presence of specific conserved motifs and nearly invariant residues that are directly or indirectly involved in positioning the phosphate donor ATP molecule and the protein substrate for catalysis. The structural conservation of the catalytic domain between different kinases is remarkable and is maintained across kingdoms.
The discovery of eukaryote-like signaling systems in bacterial pathogens has sparked an interest in understanding their function. This is due partly to the fact that eukaryotic protein kinases are currently the largest group of drug targets, second only to G-protein-coupled receptors (25, 26). A large number of STPK inhibitors have been approved by the Food and Drug Administration for use in humans (26), and ∼150 kinase inhibitors are also being tested in clinical trials (27, 28). In addition, STPKs are also being investigated as potential tools in therapeutic strategies (29, 30). Therefore, studies on prokaryotic STPKs in human pathogens have gained interest owing to the prospect of exploiting these signaling components in future anti-infective therapies.
The contribution of STPKs to bacterial growth and pathogenesis is multifaceted, as has been observed for other signaling systems. However, the mechanisms by which these kinases mediate diverse functions in a coordinated fashion remain to be completely understood, particularly their role(s) during host invasion/persistence, as we propose to detail in this minireview. The STPK-directed host-pathogen interactions known so far appear to be of different types: those in which the bacterial STPK phosphorylates a host substrate(s), those in which host defense is disrupted by STPK activity, and those in which the role of STPK is essential but the mechanism of interaction has not yet been clarified.
Bacterial Ser/Thr Kinases That Phosphorylate Eukaryotic Host Proteins
Yersinia YpkA
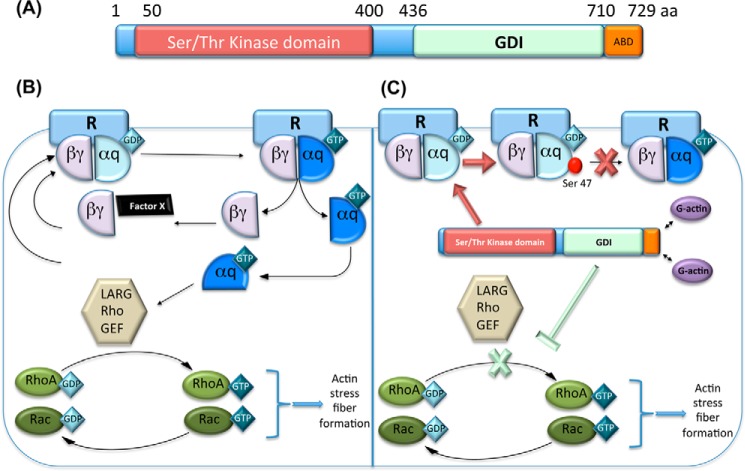
Bacteria from the genus Yersinia (Y. pestis, Y. pseudotuberculosis, and Y. enterocolitica) secrete the STPK Yersinia protein kinase A (YpkA, also named YopO) into host target cells via a type III secretion system (T3SS) (31). This kinase has been shown to disrupt the actin cytoskeleton and contribute to resisting phagocytosis by macrophages (32, 33). YpkA is a multidomain protein harboring an N-terminal Ser/Thr kinase domain and a C-terminal guanine nucleotide dissociation inhibitor (GDI) domain, followed by an actin-binding domain (Fig. 1A) (14, 15, 32, 34, 35). Once secreted into the host, YpkA localizes at the inner surface of the cytoplasmic membrane in eukaryotic cells (15, 34). Its kinase domain seems essential for virulence, as strains harboring an internal deletion in this domain show reduced lethality in infected mice (14, 36). In vitro kinase activity has been demonstrated for YpkA as being dependent on its interaction with globular actin (32, 37). Moreover, using J774 macrophages cell extracts, Juris et al. (38) have shown that YpkA phosphorylates actin and otubain, a cysteine protease involved in ubiquitin signaling and in the macrophage activation cascade, although the regulatory role of phosphorylation in these interactions was not clearly demonstrated. Although the relation between YpkA and actin depolymerization remains to be clarified, it seems that the kinase activity of YpkA is necessary for host cell shape and phagocytosis inhibition (36, 37).
FIGURE 1.
Y. pestis YpkA phosphorylates a host substrate and interferes with the host RhoA/Rac pathway. A, YpkA is a multidomain protein harboring an STPK domain, a GDI domain, and an actin-binding domain (ABD). aa, amino acids. B, in an inactive form, the heterotrimeric G-protein Gαq-βγ is associated with G-protein-coupled receptors (R), with Gαq bound to GDP. Upon activation of the receptor, GDP is exchanged for GTP on Gαq, which induces dissociation of the trimer from the receptor, and into Gαq and Gβγ subunits. Gαq-GTP activates the RhoA/Rac pathway through the LARG Rho guanine nucleotide exchange factor (leukemia-associated RhoGEF), which triggers formation of actin stress fibers. The GTPase activity of Gαq then hydrolyzes GTP to GDP, and the system reverts to the inactivate state. C, when YpkA is secreted into the host cell, its STPK domain is activated through interaction with host actin, and it phosphorylates Gαq. The latter can no longer bind GTP, which eventually leads to disruption of cytoskeletal integrity. Concurrently, the GDI domain of YpkA interacts with RhoA and Rac, leading to a switch off of the RhoA/Rac pathways.
The Gαq family of heterotrimeric G-proteins is known to activate RhoA-mediated pathways and plays a central regulatory role in a number of cellular activities requiring cytoskeletal rearrangements such as phagocytosis and motility (Fig. 1B) (39). Navarro et al. (40) demonstrated that YpkA phosphorylates Gαq on Ser-47, a key residue located in the binding loop of the G-protein, inhibiting GTP binding and thereby inhibiting Gαq signal transduction (Fig. 1C). Moreover, YpkA has also been shown to interact with other host proteins without phosphorylating them. The YpkA kinase carries a C-terminal Rho GTPase-binding domain that specifically binds and inactivates the small GTPases RhoA and Rac-1, two proteins of the Rho family involved in cytoskeleton integrity (34, 35, 41). YpkA thus mimics a host GDI (35) to “switch off” the RhoA/Rac pathways, causing cytoskeletal disruption and distortion of cell shape. Thus, the kinase- and GTPase-binding domains of YpkA act synergistically to impair specific host cellular functions. These studies highlight the role of YpkA in promoting the immune system failure at various levels.
Staphylococcus Stk1
S. aureus is considered mostly as an extracellular pathogen, but it can invade a variety of mammalian non-professional phagocytes such as epithelial cells (42) and keratinocytes (43) and survive phagocytosis by professional phagocytes such as neutrophils (44) and macrophages (45). S. aureus displays various protective and offensive responses that facilitate its persistence in the host (46, 47). Interestingly, Stk1 (also named PknB) has been shown to be important for infection in mice in an abscess model (48) as well as in a cutaneous model (49), and it is also required for antibiotic resistance (49). Stk1 was thought to be strictly membrane-associated until Miller et al. (50) demonstrated that the full-length protein could be detected in the extracellular medium, although the mechanism remains unknown. In this elegant study, the authors used a peptide microarray loaded with human peptides and identified 68 potential host-phosphorylated substrates (50). Interestingly, the substrates identified in this assay are involved in different cellular pathways such as signal transduction and stress or immune response. Among these is ATF-2, a DNA-binding protein known to regulate the expression of a broad set of genes coding for proteins involved in different processes such as cell cycle molecules (cyclin D1) (51), anti-apoptotic factors (52), and invasion-related molecules (53). Although it remains to be confirmed biochemically, other host proteins such as the pro-apoptotic Bcl-2-interacting protein Bim and the cytoskeleton-associated protein paxillin seem to be phosphorylated by Stk1.
Salmonella SteC
Salmonella enterica serovar Typhimurium (S. typhimurium) causes gastrointestinal disease in humans and a typhoid-like systemic infection in certain mouse strains. Its essential virulence factors include two T3SSs, encoded by genes from two gene clusters called SPI (Salmonella pathogenicity island)-1 and SPI-2, respectively. Effector proteins translocated by the SPI-1-encoded T3SS interfere with the actin cytoskeleton to induce bacterial invasion and contribute to the early maturation of the Salmonella-containing vacuole (SCV) (54). The SCV matures into an organelle that has some properties of late endosomes but lacks degradative enzymes of lysosomes. A few hours after bacterial entry, the SPI-2-encoded T3SS is activated and delivers a second set of effector proteins across the vacuolar membrane. These further modify the SCV and enable intravacuolar bacterial replication (55). In the course of these events, S. typhimurium causes major alterations to the microtubule, intermediate filament, and actin networks. The kinase activity of the STPK SteC, which is secreted by the SPI-2 T3SS, induces a meshwork of F-actin that is formed around SCVs and bacterial microcolonies (56). Recently, Odendall et al. (55) demonstrated that SteC promotes actin cytoskeleton reorganization by activating a signaling pathway involving the MAPKs MEK and ERK, myosin light chain kinase, and myosin IIB. Specifically, SteC phosphorylates MEK directly on Ser-200, a previously unknown phosphorylation site. Ser-200 phosphorylation is predicted to displace a negative regulatory helix causing autophosphorylation on the known MEK activatory residues, Ser-218 and Ser-222. Both steC-null and kinase-deficient mutant strains display enhanced replication in infected cells, suggesting that the effects of SteC on the actin cytoskeleton limit bacterial growth (55).
Bacterial STPKs That Disrupt Host NF-κB Pathways
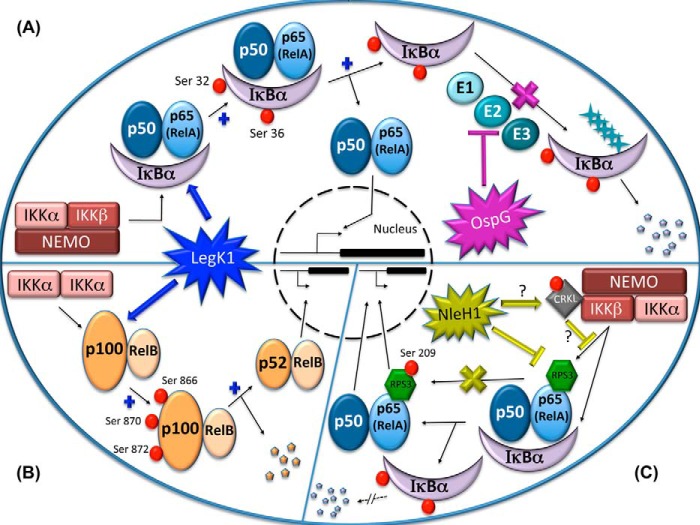
In a host cell, the transcription factor NF-κB protein complex is critically important in triggering an immune/inflammatory response. In the absence of cognate stimuli, NF-κB is prevented from translocating to the nucleus by inhibitors of the IκB family. In response to stimuli such as a bacterial infection, NF-κB is translocated to the nucleus, where it transcriptionally induces specific genes involved in a variety of processes aimed at eliminating the pathogen. In contrast, certain pathogenic bacteria present mechanisms to counter these attacks, some of which involve bacterial STPKs. Structurally, the different members of the NF-κB family are composed of combinations of five subunits, p50, p52, RelA (p65), RelB, and c-Rel, of which p50 and p52 are derived from p105 and p100, respectively. STPKs from different pathogenic bacteria seem to interact with host factors to disrupt the NF-κB signaling pathways and downstream processes as discussed below (Fig. 2).
FIGURE 2.
Interference with the host NF-κB signaling pathways by STPKs of pathogenic bacteria: LegK1, OspG, and NleH1. In the canonical NF-κB pathway, in response to stimulation, the IKK trimer (IKKα/IKKβ/NEMO) phosphorylates IκB inhibitor (here, IκBα), which normally sequesters the NF-κB (p50/p65) dimer in the cytoplasm. Once phosphorylated, IκBα dissociates from NF-κB dimer, which then translocates into the nucleus and activates genes implicated in the immune response. The dissociated phospho-IκBα is ubiquitinated by the ubiquitinylation system (E1, E2, and E3) and is addressed to and degraded in the proteasome. In the non-canonical pathway, the IKKα dimer phosphorylates p100, the precursor of p52 that is an inhibitor of the NF-κB dimer. Once phosphorylated, p100 is processed to p52, and the p52/RelB NF-κB dimer thus activated is translocated into the nucleus. The Legionella STPK LegK1 mimics IKKs in both canonical (A, left) and non-canonical (B) pathways and induces activation of the NF-κB pathways in the host. A (right), the Shigella sp. STPK OspG interacts with the ubiquitin-conjugating enzyme E2, blocking phospho-IκBα degradation and thus silencing the inflammatory response of the host. C, the RPS3 protein interacts with the p65 subunit of NF-κB, and when phosphorylated by IKKα, RPS3 is translocated into the nucleus and determines the regulatory specificity of NF-κB for target genes. The E. coli STPK NleH1 inhibits RPS3 phosphorylation and thus inhibits the host NF-κB response mechanism. CRKL could be involved in this inhibition, but the exact mechanism remains to be elucidated.
Legionella LegK1
Legionella pneumophila infects the lung macrophages and causes the so-called Legionnaires' disease. Once in the phagosome, this pathogen is able to redirect the classical bacterial phagolysosomal elimination to establish a replicative niche within an endoplasmic reticulum-derived compartment, named the Legionella-containing vacuole, and evade host cell defenses (57–59).
Different species of Legionella harbor three to five genes encoding putative STPKs (legK1–legK3 for L. pneumophila Philadelphia-1 (60) and legK1–legK5 for L. pneumophila Lens (61)). LegK1–LegK4 are known to be secreted by a type IV secretion system called Dot/Icm, which is essential for intracellular growth (58, 59). Only LegK1 has been shown to interfere with the NF-κB pathway, acting as an inflammatory activator. Using an NF-κB-specific luciferase reporter system, Ge et al. (62) demonstrated that ectopic expression of LegK1 in HEK-293T cells triggers activation of the NF-κB cascade (Fig. 2, A (left) and B). The same assay conducted with LegK2, LegK3, and a LegK1 kinase-dead mutant (carrying a mutation in the kinase domain) resulted in no activation of the NF-κB system. Therefore, LegK1 seems to be the only STPK able to interfere with the NF-κB pathway, and its kinase activity is required. Moreover, LegK1 activity is specific to NF-κB and does not affect other innate immune signaling pathways such as MAPK and IFN (62). Moreover, in the same study, the authors used a cell-free reconstitution system to show that LegK1 is able to phosphorylate IκBα, a central regulator of the NF-κB pathway (Fig. 2A), on Ser-32 and Ser-36 in an IκB kinase (IKK)-independent manner. In vitro assays with IκBα and a variety of LegK1 derivatives confirmed phosphorylation of IκBα by LegK1, indicating that the N-terminal “pre-kinase” part is critical for IκBα phosphorylation. The authors also demonstrated that p100, the precursor of the non-canonical NF-κB complex, is processed into p52 when phosphorylated by LegK1 (Fig. 2B). Taken together, these data highlight the function of the LegK1 kinase as a mimic of the host IKKs in the NF-κB response activation. Thus, L. pneumophila LegK1 is an STPK that phosphorylates host substrates (IκBα and p100) and disrupts the NF-κB pathway, thereby modulating host innate defenses and inflammatory responses during infection.
Shigella OspG
Shigella spp. are the agents of shigellosis in humans, a disease that is characterized by the destruction of the colonic epithelium and that is responsible for 1 million deaths per year (63). Shigella spp. use a T3SS to enter epithelial cells, thus triggering apoptosis (64). About 20 proteins have been identified as substrates of the T3SS (65). One of these, OspG, is an STPK known to manipulate the host innate immune system by down-regulating the canonical NF-κB pathway (Fig. 2A right). Using in vivo and in vitro approaches, Kim et al. (66) demonstrated that OspG is an STPK able to bind to ubiquitinylated E2 enzymes such as UbcH7 and UbcH5 without phosphorylating them. They showed that this sequestration leads to a decrease in IκBα degradation, thus blocking the activation pathway of NF-κB. The action of OspG seems to be dependent on its kinase activity, as the inactivated kinase mutant does not generate a similar attenuation of NF-κB signaling. In addition, infection assays carried out on the ligated ileal loop in rabbits confirmed that inactivation of ospG in Shigella strains induces a stronger inflammatory response in vivo (66). Moreover, in a recent study, OspG was found to be able to bind ubiquitin and polyubiquitin. This interaction seems to activate OspG kinase activity in vitro and to be required for attenuating the host NF-κB signaling pathway in vivo (67).
Escherichia NleH1 and NleH2
Enteropathogenic Escherichia coli (EPEC) and enterohemorrhagic E. coli (EHEC) are two diarrheagenic strains of E. coli that contribute to the health burden of food-borne disease. EHEC and EPEC are known to express and secrete effectors into intestinal epithelial cells through a T3SS (68). NleH1 and NleH2 have been biochemically characterized as STPKs and secreted in HeLa cells through a T3SS. These STPKs are able to interact with RPS3, a host protein known to bind the p65 subunit of NF-κB and regulate its affinity for its target genes (Fig. 2C) (69, 70). The interaction of RPS3 and NleH1, but not NleH2, reduces the nuclear abundance of RPS3, causing inhibition of the NF-κB-dependent transcriptional activity. The NF-κB activity seems also to be decreased by NleH2 when IKKβ is overexpressed (71, 72). In vivo tests showed that the EHEC ΔnleH1 strain, but not the ΔnleH2 strain, is hypervirulent in a gnotobiotic piglet infection model, indicating that NleH1 and NleH2 differentially regulate the inflammatory response of the host (70). In a mouse intestine model, EPEC NleH1 and NleH2 seemed to increase colonization and decrease inflammation (71). Phosphorylation of RPS3 on Ser-209 by IKKβ enhances its association with importin-α, thus mediating RPS3 entry into the karyopherin pathway for nuclear translocation (73). The interaction of RPS3 with NleH1 leads to the inhibition of its phosphorylation by IKKβ. Recently, a high-throughput screen to identify a host cell substrate of NleH1 yielded the CRKL (v-Crk sarcoma virus CT10 oncogene-like) protein (74). According to the proposed model, CRKL interacts dually with NleH1 and IKKβ. Interaction of a kinase-active form of NleH1 with CRKL is essential for the ability of NleH1 to inhibit RPS3 phosphorylation by IKKβ.
Bacterial STPKs with Unidentified Host Substrates
In some of the host-pathogen interactions known so far, the kinase activity of the bacterial STPKs is required, but the substrate has not been identified. In addition to E. coli NleH1 and NleH2 described above, this category also includes Legionella LegK2. Similarly, the substrates of most of the mycobacterial STPKs that participate in host-pathogen interactions have not yet been identified.
Legionella LegK2
LegK2 has been identified to be involved in the virulence of L. pneumophila by a combination of in vitro and in vivo approaches (61). A ΔlegK2 mutant showed no defects in in vitro growth but presented less cytotoxicity and delayed intracellular replication in amoebas compared with the wild-type strains. Moreover, vacuoles containing the mutant strain showed less efficient recruitment of endoplasmic reticulum markers. Complementation assays performed with wild-type and kinase-dead proteins provided evidence that LegK2 kinase activity is required for the normal infectious phenotype of L. pneumophila (61). Although no host targets have yet been identified, LegK2 seems to be a crucial virulence determinant involved in the establishment of the replicative niche in the macrophage.
Mycobacterium tuberculosis STPKs
M. tuberculosis is the causative agent of tuberculosis. It is capable of infection and long-term survival in host macrophages. The bacterium possesses several virulence factors that are expressed at different steps of infection all the way to establishing a latent infection and an eventual resuscitation from dormancy. Genome sequence analyses revealed 11 STPKs (8), four of which have been demonstrated to be involved in virulence in vivo: PknH, PknI, PknK, and PknG. Although these STPKs are important virulence factors, their host cell interactors have not yet been identified, except for that of PknG. Studies with genetic mutants of the above STPKs have revealed their roles in establishing an infection. In a mice model, the pknH mutant was found to survive and replicate to a higher bacillary load in mouse organs compared with its parental strain (75). Similarly, a pknI-null mutant showed increased intracellular growth inside THP-1 macrophage cells and hypervirulence in immunodeficient mice (76). More recently, Malhotra et al. (77) showed that a pknK deletion results in increased resistance of the mutant to acidic pH, hypoxia, and oxidative and stationary phase stress in vitro and increased survival during persistent infection in mice. Moreover, assays performed on host immune effectors suggested an immunomodulatory function of PknK during acute infection in mice (77).
PknG is a soluble STPK expressed by pathogenic or attenuated mycobacteria such as M. tuberculosis and Mycobacterium bovis bacillus Calmette-Guérin, but not by Mycobacterium smegmatis, a non-pathogenic species. PknG is known to play a role in persistence inside macrophages, presumably by inhibiting the critical step of phagosome-lysosome fusion, as shown for M. bovis and M. smegmatis in cultured macrophages (78, 79) and in a mouse model (80). Interestingly, the basis for the PknG-mediated enhanced survival in macrophages appears to be its interaction with, but not phosphorylation of, PKCα, an STPK from the host cell that is known to regulate phagolysosome formation (81). Other studies have also highlighted the role of PknG in interacting and disturbing host defense pathways (82–85). Recently, in Mycobacterium marinum, SecA2 was identified as the secretion system that likely introduces PknG into the host cell (86).
Conclusions
Thus, the emerging theme from the above examples is that not only are the STPKs in pathogenic bacteria essential for regulating important bacterial processes, but some are secreted such that they can interact with host substrates also, subverting essential host functions such as immune responses and cell shape and integrity. It is not yet clear whether these STPK interactions all involve phosphorylation of a host substrate. In some cases, the phosphorylation of a host substrate has been demonstrated, whereas in others, the STPK kinase activity is seen to be essential, but a phosphorylated substrate has not been identified. The biochemical mechanisms of these pathogen-directed targeted perturbations in the host cell signaling network are being actively investigated. Thus, bacterial STPKs are proving to be molecular switches that play key roles in host-pathogen interactions.
Acknowledgment
We thank Anuradha Alahari for helpful comments.
This work was supported by French National Research Agency Grant ANR-09-MIEN-004 and the ATIP-Avenir Program (to V. M. and M. J. C.). This is the second article in the Thematic Minireview Series “Protein Serine/Threonine and Tyrosine Phosphorylation in Prokaryotes.”
- STPK
- serine/threonine protein kinase
- T3SS
- type III secretion system
- GDI
- guanine nucleotide dissociation inhibitor
- SCV
- Salmonella-containing vacuole
- IKK
- IκB kinase
- EPEC
- enteropathogenic E. coli
- EHEC
- enterohemorrhagic E. coli.
REFERENCES
- 1. Stock A. M., Robinson V. L., Goudreau P. N. (2000) Two-component signal transduction. Annu. Rev. Biochem. 69, 183–215 [DOI] [PubMed] [Google Scholar]
- 2. Foussard M., Cabantous S., Pédelacq J., Guillet V., Tranier S., Mourey L., Birck C., Samama J. (2001) The molecular puzzle of two-component signaling cascades. Microbes Infect. 3, 417–424 [DOI] [PubMed] [Google Scholar]
- 3. Huse M., Kuriyan J. (2002) The conformational plasticity of protein kinases. Cell 109, 275–282 [DOI] [PubMed] [Google Scholar]
- 4. Rajagopal L., Clancy A., Rubens C. E. (2003) A eukaryotic type serine/threonine kinase and phosphatase in Streptococcus agalactiae reversibly phosphorylate an inorganic pyrophosphatase and affect growth, cell segregation, and virulence. J. Biol. Chem. 278, 14429–14441 [DOI] [PubMed] [Google Scholar]
- 5. Hussain H., Branny P., Allan E. (2006) A eukaryotic-type serine/threonine protein kinase is required for biofilm formation, genetic competence, and acid resistance in Streptococcus mutans. J. Bacteriol. 188, 1628–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Echenique J., Kadioglu A., Romao S., Andrew P. W., Trombe M. C. (2004) Protein serine/threonine kinase StkP positively controls virulence and competence in Streptococcus pneumoniae. Infect. Immun. 72, 2434–2437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jin H., Pancholi V. (2006) Identification and biochemical characterization of a eukaryotic-type serine/threonine kinase and its cognate phosphatase in Streptococcus pyogenes: their biological functions and substrate identification. J. Mol. Biol. 357, 1351–1372 [DOI] [PubMed] [Google Scholar]
- 8. Av-Gay Y., Everett M. (2000) The eukaryotic-like Ser/Thr protein kinases of Mycobacterium tuberculosis. Trends Microbiol. 8, 238–244 [DOI] [PubMed] [Google Scholar]
- 9. Chaba R., Raje M., Chakraborti P. K. (2002) Evidence that a eukaryotic-type serine/threonine protein kinase from Mycobacterium tuberculosis regulates morphological changes associated with cell division. Eur. J. Biochem. 269, 1078–1085 [DOI] [PubMed] [Google Scholar]
- 10. Wehenkel A., Bellinzoni M., Graña M., Duran R., Villarino A., Fernandez P., Andre-Leroux G., England P., Takiff H., Cerveñansky C., Cole S. T., Alzari P. M. (2008) Mycobacterial Ser/Thr protein kinases and phosphatases: physiological roles and therapeutic potential. Biochim. Biophys. Acta 1784, 193–202 [DOI] [PubMed] [Google Scholar]
- 11. Molle V., Kremer L. (2010) Division and cell envelope regulation by Ser/Thr phosphorylation: Mycobacterium shows the way. Mol. Microbiol. 75, 1064–1077 [DOI] [PubMed] [Google Scholar]
- 12. Alber T. (2009) Signaling mechanisms of the Mycobacterium tuberculosis receptor Ser/Thr protein kinases. Curr. Opin. Struct. Biol. 19, 650–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Greenstein A. E., Grundner C., Echols N., Gay L. M., Lombana T. N., Miecskowski C. A., Pullen K. E., Sung P. Y., Alber T. (2005) Structure/function studies of Ser/Thr and Tyr protein phosphorylation in Mycobacterium tuberculosis. J. Mol. Microbiol. Biotechnol. 9, 167–181 [DOI] [PubMed] [Google Scholar]
- 14. Galyov E. E., Håkansson S., Forsberg A., Wolf-Watz H. (1993) A secreted protein kinase of Yersinia pseudotuberculosis is an indispensable virulence determinant. Nature 361, 730–732 [DOI] [PubMed] [Google Scholar]
- 15. Håkansson S., Galyov E. E., Rosqvist R., Wolf-Watz H. (1996) The Yersinia YpkA Ser/Thr kinase is translocated and subsequently targeted to the inner surface of the HeLa cell plasma membrane. Mol. Microbiol. 20, 593–603 [DOI] [PubMed] [Google Scholar]
- 16. Archambaud C., Gouin E., Pizarro-Cerda J., Cossart P., Dussurget O. (2005) Translation elongation factor EF-Tu is a target for Stp, a serine-threonine phosphatase involved in virulence of Listeria monocytogenes. Mol. Microbiol. 56, 383–396 [DOI] [PubMed] [Google Scholar]
- 17. Lima A., Durán R., Schujman G. E., Marchissio M. J., Portela M. M., Obal G., Pritsch O., de Mendoza D., Cerveñansky C. (2011) Serine/threonine protein kinase PrkA of the human pathogen Listeria monocytogenes: biochemical characterization and identification of interacting partners through proteomic approaches. J. Proteomics 74, 1720–1734 [DOI] [PubMed] [Google Scholar]
- 18. Wang J., Li C., Yang H., Mushegian A., Jin S. (1998) A novel serine/threonine protein kinase homologue of Pseudomonas aeruginosa is specifically inducible within the host infection site and is required for full virulence in neutropenic mice. J. Bacteriol. 180, 6764–6768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kristich C. J., Wells C. L., Dunny G. M. (2007) A eukaryotic-type Ser/Thr kinase in Enterococcus faecalis mediates antimicrobial resistance and intestinal persistence. Proc. Natl. Acad. Sci. U.S.A. 104, 3508–3513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Beltramini A. M., Mukhopadhyay C. D., Pancholi V. (2009) Modulation of cell wall structure and antimicrobial susceptibility by a Staphylococcus aureus eukaryote-like serine/threonine kinase and phosphatase. Infect. Immun. 77, 1406–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Truong-Bolduc Q. C., Ding Y., Hooper D. C. (2008) Posttranslational modification influences the effects of MgrA on norA expression in Staphylococcus aureus. J. Bacteriol. 190, 7375–7381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Truong-Bolduc Q. C., Hooper D. C. (2010) Phosphorylation of MgrA and its effect on expression of the NorA and NorB efflux pumps of Staphylococcus aureus. J. Bacteriol. 192, 2525–2534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hanks S. K., Hunter T. (1995) Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 9, 576–596 [PubMed] [Google Scholar]
- 24. Kornev A. P., Taylor S. S. (2010) Defining the conserved internal architecture of a protein kinase. Biochim. Biophys. Acta 1804, 440–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bogoyevitch M. A., Barr R. K., Ketterman A. J. (2005) Peptide inhibitors of protein kinases–discovery, characterisation and use. Biochim. Biophys. Acta 1754, 79–99 [DOI] [PubMed] [Google Scholar]
- 26. Cheng H., Force T. (2010) Molecular mechanisms of cardiovascular toxicity of targeted cancer therapeutics. Circ. Res. 106, 21–34 [DOI] [PubMed] [Google Scholar]
- 27. Sachsenmaier C. (2001) Targeting protein kinases for tumor therapy. Onkologie 24, 346–355 [DOI] [PubMed] [Google Scholar]
- 28. Cohen P. (1991) Classification of protein-serine/threonine phosphatases: identification and quantitation in cell extracts. Methods Enzymol. 201, 389–398 [DOI] [PubMed] [Google Scholar]
- 29. McConnell J. L., Wadzinski B. E. (2009) Targeting protein serine/threonine phosphatases for drug development. Mol. Pharmacol. 75, 1249–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lu J., Kovach J. S., Johnson F., Chiang J., Hodes R., Lonser R., Zhuang Z. (2009) Inhibition of serine/threonine phosphatase PP2A enhances cancer chemotherapy by blocking DNA damage-induced defense mechanisms. Proc. Natl. Acad. Sci. U.S.A. 106, 11697–11702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cornelis G. R., Boland A., Boyd A. P., Geuijen C., Iriarte M., Neyt C., Sory M. P., Stainier I. (1998) The virulence plasmid of Yersinia, an antihost genome. Microbiol. Mol. Biol. Rev. 62, 1315–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Juris S. J., Rudolph A. E., Huddler D., Orth K., Dixon J. E. (2000) A distinctive role for the Yersinia protein kinase: actin binding, kinase activation, and cytoskeleton disruption. Proc. Natl. Acad. Sci. U.S.A. 97, 9431–9436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Grosdent N., Maridonneau-Parini I., Sory M. P., Cornelis G. R. (2002) Role of Yops and adhesins in resistance of Yersinia enterocolitica to phagocytosis. Infect. Immun. 70, 4165–4176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dukuzumuremyi J. M., Rosqvist R., Hallberg B., Akerström B., Wolf-Watz H., Schesser K. (2000) The Yersinia protein kinase A is a host factor inducible RhoA/-binding virulence factor. J. Biol. Chem. 275, 35281–35290 [DOI] [PubMed] [Google Scholar]
- 35. Prehna G., Ivanov M. I., Bliska J. B., Stebbins C. E. (2006) Yersinia virulence depends on mimicry of host Rho-family nucleotide dissociation inhibitors. Cell 126, 869–880 [DOI] [PubMed] [Google Scholar]
- 36. Wiley D. J., Nordfeldth R., Rosenzweig J., DaFonseca C. J., Gustin R., Wolf-Watz H., Schesser K. (2006) The Ser/Thr kinase activity of the Yersinia protein kinase A (YpkA) is necessary for full virulence in the mouse, mollifying phagocytes, and disrupting the eukaryotic cytoskeleton. Microb. Pathog. 40, 234–243 [DOI] [PubMed] [Google Scholar]
- 37. Trasak C., Zenner G., Vogel A., Yüksekdag G., Rost R., Haase I., Fischer M., Israel L., Imhof A., Linder S., Schleicher M., Aepfelbacher M. (2007) Yersinia protein kinase YopO is activated by a novel G-actin binding process. J. Biol. Chem. 282, 2268–2277 [DOI] [PubMed] [Google Scholar]
- 38. Juris S. J., Shah K., Shokat K., Dixon J. E., Vacratsis P. O. (2006) Identification of otubain 1 as a novel substrate for the Yersinia protein kinase using chemical genetics and mass spectrometry. FEBS Lett. 580, 179–183 [DOI] [PubMed] [Google Scholar]
- 39. Van Aelst L., D'Souza-Schorey C. (1997) Rho GTPases and signaling networks. Genes Dev. 11, 2295–2322 [DOI] [PubMed] [Google Scholar]
- 40. Navarro L., Koller A., Nordfelth R., Wolf-Watz H., Taylor S., Dixon J. E. (2007) Identification of a molecular target for the Yersinia protein kinase A. Mol. Cell 26, 465–477 [DOI] [PubMed] [Google Scholar]
- 41. Barz C., Abahji T. N., Trülzsch K., Heesemann J. (2000) The Yersinia Ser/Thr protein kinase YpkA/YopO directly interacts with the small GTPases RhoA and Rac-1. FEBS Lett. 482, 139–143 [DOI] [PubMed] [Google Scholar]
- 42. Dziewanowska K., Patti J. M., Deobald C. F., Bayles K. W., Trumble W. R., Bohach G. A. (1999) Fibronectin binding protein and host cell tyrosine kinase are required for internalization of Staphylococcus aureus by epithelial cells. Infect. Immun. 67, 4673–4678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kintarak S., Whawell S. A., Speight P. M., Packer S., Nair S. P. (2004) Internalization of Staphylococcus aureus by human keratinocytes. Infect. Immun. 72, 5668–5675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Voyich J. M., Braughton K. R., Sturdevant D. E., Whitney A. R., Saïd-Salim B., Porcella S. F., Long R. D., Dorward D. W., Gardner D. J., Kreiswirth B. N., Musser J. M., DeLeo F. R. (2005) Insights into mechanisms used by Staphylococcus aureus to avoid destruction by human neutrophils. J. Immunol. 175, 3907–3919 [DOI] [PubMed] [Google Scholar]
- 45. Kubica M., Guzik K., Koziel J., Zarebski M., Richter W., Gajkowska B., Golda A., Maciag-Gudowska A., Brix K., Shaw L., Foster T., Potempa J. (2008) A potential new pathway for Staphylococcus aureus dissemination: the silent survival of S. aureus phagocytosed by human monocyte-derived macrophages. PLoS ONE 3, e1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Foster T. J. (2005) Immune evasion by staphylococci. Nat. Rev. Microbiol. 3, 948–958 [DOI] [PubMed] [Google Scholar]
- 47. Sinha B., Fraunholz M. (2010) Staphylococcus aureus host cell invasion and post-invasion events. Int. J. Med. Microbiol. 300, 170–175 [DOI] [PubMed] [Google Scholar]
- 48. Débarbouillé M., Dramsi S., Dussurget O., Nahori M. A., Vaganay E., Jouvion G., Cozzone A., Msadek T., Duclos B. (2009) Characterization of a serine/threonine kinase involved in virulence of Staphylococcus aureus. J. Bacteriol. 191, 4070–4081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tamber S., Schwartzman J., Cheung A. L. (2010) The role of PknB kinase in antibiotic resistance and virulence in community acquired methicillin-resistant Staphylococcus aureus strain USA300. Infect. Immun. 78, 3637–3646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Miller M., Donat S., Rakette S., Stehle T., Kouwen T. R., Diks S. H., Dreisbach A., Reilman E., Gronau K., Becher D., Peppelenbosch M. P., van Dijl J. M., Ohlsen K. (2010) Staphylococcal PknB as the first prokaryotic representative of the proline-directed kinases. PLoS ONE 5, e9057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Beier F., Taylor A. C., LuValle P. (2000) Activating transcription factor 2 is necessary for maximal activity and serum induction of the cyclin A promoter in chondrocytes. J. Biol. Chem. 275, 12948–12953 [DOI] [PubMed] [Google Scholar]
- 52. Ma Q., Li X., Vale-Cruz D., Brown M. L., Beier F., LuValle P. (2007) Activating transcription factor 2 controls Bcl-2 promoter activity in growth plate chondrocytes. J. Cell. Biochem. 101, 477–487 [DOI] [PubMed] [Google Scholar]
- 53. Song H., Ki S. H., Kim S. G., Moon A. (2006) Activating transcription factor 2 mediates matrix metalloproteinase-2 transcriptional activation induced by p38 in breast epithelial cells. Cancer Res. 66, 10487–10496 [DOI] [PubMed] [Google Scholar]
- 54. Steele-Mortimer O., Brumell J. H., Knodler L. A., Méresse S., Lopez A., Finlay B. B. (2002) The invasion-associated type III secretion system of Salmonella enterica serovar Typhimurium is necessary for intracellular proliferation and vacuole biogenesis in epithelial cells. Cell. Microbiol. 4, 43–54 [DOI] [PubMed] [Google Scholar]
- 55. Odendall C., Rolhion N., Förster A., Poh J., Lamont D. J., Liu M., Freemont P. S., Catling A. D., Holden D. W. (2012) The Salmonella kinase SteC targets the MAP kinase MEK to regulate the host actin cytoskeleton. Cell Host Microbe 12, 657–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Poh J., Odendall C., Spanos A., Boyle C., Liu M., Freemont P., Holden D. W. (2008) SteC is a Salmonella kinase required for SPI-2-dependent F-actin remodelling. Cell. Microbiol. 10, 20–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. McDade J. E., Shepard C. C., Fraser D. W., Tsai T. R., Redus M. A., Dowdle W. R. (1977) Legionnaires' disease: isolation of a bacterium and demonstration of its role in other respiratory disease. N. Engl. J. Med. 297, 1197–1203 [DOI] [PubMed] [Google Scholar]
- 58. Kaufmann A. F., McDade J. E., Patton C. M., Bennett J. V., Skaliy P., Feeley J. C., Anderson D. C., Potter M. E., Newhouse V. F., Gregg M. B., Brachman P. S. (1981) Pontiac fever: isolation of the etiologic agent (Legionella pneumophila) and demonstration of its mode of transmission. Am. J. Epidemiol. 114, 337–347 [DOI] [PubMed] [Google Scholar]
- 59. Horwitz M. A. (1983) Cell-mediated immunity in Legionnaires' disease. J. Clin. Invest. 71, 1686–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. de Felipe K. S., Glover R. T., Charpentier X., Anderson O. R., Reyes M., Pericone C. D., Shuman H. A. (2008) Legionella eukaryotic-like type IV substrates interfere with organelle trafficking. PLoS Pathog. 4, e1000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hervet E., Charpentier X., Vianney A., Lazzaroni J. C., Gilbert C., Atlan D., Doublet P. (2011) Protein kinase LegK2 is a type IV secretion system effector involved in endoplasmic reticulum recruitment and intracellular replication of Legionella pneumophila. Infect. Immun. 79, 1936–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ge J., Xu H., Li T., Zhou Y., Zhang Z., Li S., Liu L., Shao F. (2009) A Legionella type IV effector activates the NF-κB pathway by phosphorylating the IκB family of inhibitors. Proc. Natl. Acad. Sci. U.S.A. 106, 13725–13730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kotloff K. L., Winickoff J. P., Ivanoff B., Clemens J. D., Swerdlow D. L., Sansonetti P. J., Adak G. K., Levine M. M. (1999) Global burden of Shigella infections: implications for vaccine development and implementation of control strategies. Bull. World Health Organ. 77, 651–666 [PMC free article] [PubMed] [Google Scholar]
- 64. Cossart P., Sansonetti P. J. (2004) Bacterial invasion: the paradigms of enteroinvasive pathogens. Science 304, 242–248 [DOI] [PubMed] [Google Scholar]
- 65. Buchrieser C., Glaser P., Rusniok C., Nedjari H., D'Hauteville H., Kunst F., Sansonetti P., Parsot C. (2000) The virulence plasmid pWR100 and the repertoire of proteins secreted by the type III secretion apparatus of Shigella flexneri. Mol. Microbiol. 38, 760–771 [DOI] [PubMed] [Google Scholar]
- 66. Kim D. W., Lenzen G., Page A. L., Legrain P., Sansonetti P. J., Parsot C. (2005) The Shigella flexneri effector OspG interferes with innate immune responses by targeting ubiquitin-conjugating enzymes. Proc. Natl. Acad. Sci. U.S.A. 102, 14046–14051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zhou Y., Dong N., Hu L., Shao F. (2013) The Shigella type three secretion system effector OspG directly and specifically binds to host ubiquitin for activation. PLoS ONE 8, e57558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hueck C. J. (1998) Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62, 379–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wan F., Anderson D. E., Barnitz R. A., Snow A., Bidere N., Zheng L., Hegde V., Lam L. T., Staudt L. M., Levens D., Deutsch W. A., Lenardo M. J. (2007) Ribosomal protein S3: a KH domain subunit in NF-κB complexes that mediates selective gene regulation. Cell 131, 927–939 [DOI] [PubMed] [Google Scholar]
- 70. Gao X., Wan F., Mateo K., Callegari E., Wang D., Deng W., Puente J., Li F., Chaussee M. S., Finlay B. B., Lenardo M. J., Hardwidge P. R. (2009) Bacterial effector binding to ribosomal protein S3 subverts NF-κB function. PLoS Pathog. 5, e1000708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Royan S. V., Jones R. M., Koutsouris A., Roxas J. L., Falzari K., Weflen A. W., Kim A., Bellmeyer A., Turner J. R., Neish A. S., Rhee K. J., Viswanathan V. K., Hecht G. A. (2010) Enteropathogenic E. coli non-LEE encoded effectors NleH1 and NleH2 attenuate NF-κB activation. Mol. Microbiol. 78, 1232–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Pham T. H., Gao X., Tsai K., Olsen R., Wan F., Hardwidge P. R. (2012) Functional differences and interactions between the Escherichia coli type III secretion system effectors NleH1 and NleH2. Infect. Immun. 80, 2133–2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wan F., Weaver A., Gao X., Bern M., Hardwidge P. R., Lenardo M. J. (2011) IKKβ phosphorylation regulates RPS3 nuclear translocation and NF-κB function during infection with Escherichia coli strain O157:H7. Nat. Immunol. 12, 335–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Pham T. H., Gao X., Singh G., Hardwidge P. R. (2013) Escherichia coli virulence protein NleH1 interaction with the v-Crk sarcoma virus CT10 oncogene-like protein (CRKL) governs NleH1 inhibition of the ribosomal protein S3 (RPS3)/NF-κB pathway. J. Biol. Chem. 288, 34567–34574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Papavinasasundaram K. G., Chan B., Chung J. H., Colston M. J., Davis E. O., Av-Gay Y. (2005) Deletion of the Mycobacterium tuberculosis pknH gene confers a higher bacillary load during the chronic phase of infection in BALB/c mice. J. Bacteriol. 187, 5751–5760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Gopalaswamy R., Narayanan S., Chen B., Jacobs W. R., Av-Gay Y. (2009) The serine/threonine protein kinase PknI controls the growth of Mycobacterium tuberculosis upon infection. FEMS Microbiol. Lett. 295, 23–29 [DOI] [PubMed] [Google Scholar]
- 77. Malhotra V., Arteaga-Cortés L. T., Clay G., Clark-Curtiss J. E. (2010) Mycobacterium tuberculosis protein kinase K confers survival advantage during early infection in mice and regulates growth in culture and during persistent infection: implications for immune modulation. Microbiology 156, 2829–2841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Walburger A., Koul A., Ferrari G., Nguyen L., Prescianotto-Baschong C., Huygen K., Klebl B., Thompson C., Bacher G., Pieters J. (2004) Protein kinase G from pathogenic mycobacteria promotes survival within macrophages. Science 304, 1800–1804 [DOI] [PubMed] [Google Scholar]
- 79. Scherr N., Müller P., Perisa D., Combaluzier B., Jenö P., Pieters J. (2009) Survival of pathogenic mycobacteria in macrophages is mediated through autophosphorylation of protein kinase G. J. Bacteriol. 191, 4546–4554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Tiwari D., Singh R. K., Goswami K., Verma S. K., Prakash B., Nandicoori V. K. (2009) Key residues in Mycobacterium tuberculosis protein kinase G play a role in regulating kinase activity and survival in the host. J. Biol. Chem. 284, 27467–27479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Chaurasiya S. K., Srivastava K. K. (2009) Downregulation of protein kinase C-α enhances intracellular survival of Mycobacteria: role of PknG. BMC Microbiol. 9, 271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. St-Denis A., Caouras V., Gervais F., Descoteaux A. (1999) Role of protein kinase C-α in the control of infection by intracellular pathogens in macrophages. J. Immunol. 163, 5505–5511 [PubMed] [Google Scholar]
- 83. Itoh S., Suzuki K., Nishihata J., Iwasa M., Oku T., Nakajin S., Nauseef W. M., Toyoshima S. (2002) The role of protein kinase C in the transient association of p57, a coronin family actin-binding protein, with phagosomes. Biol. Pharm. Bull. 25, 837–844 [DOI] [PubMed] [Google Scholar]
- 84. Holm A., Tejle K., Gunnarsson T., Magnusson K. E., Descoteaux A., Rasmusson B. (2003) Role of protein kinase Cα for uptake of unopsonized prey and phagosomal maturation in macrophages. Biochem. Biophys. Res. Commun. 302, 653–658 [DOI] [PubMed] [Google Scholar]
- 85. Ng Yan Hing J. D., Desjardins M., Descoteaux A. (2004) Proteomic analysis reveals a role for protein kinase C-α in phagosome maturation. Biochem. Biophys. Res. Commun. 319, 810–816 [DOI] [PubMed] [Google Scholar]
- 86. van der Woude A. D., Stoop E. J., Stiess M., Wang S., Ummels R., van Stempvoort G., Piersma S. R., Cascioferro A., Jimenez C. R., Houben E. N., Luirink J., Pieters J., van der Sar A. M., Bitter W. (2014) Analysis of SecA2-dependent substrates in Mycobacterium marinum identifies protein kinase G (PknG) as a virulence effector. Cell. Microbiol. 16, 280–295 [DOI] [PubMed] [Google Scholar]