Abstract
The third domain of life, the Archaea (formerly Archaebacteria), is populated by a physiologically diverse set of microorganisms, many of which reside at the ecological extremes of our global environment. Although ostensibly prokaryotic in morphology, the Archaea share much closer evolutionary ties with the Eukarya than with the superficially more similar Bacteria. Initial genomic, proteomic, and biochemical analyses have revealed the presence of “eukaryotic” protein kinases and phosphatases and an intriguing set of serine-, threonine-, and tyrosine-phosphorylated proteins in the Archaea that may offer new insights into this important regulatory mechanism.
Keywords: Archaea, Archaebacteria, Evolution, Protein Kinases, Protein Phosphatase, Protein Phosphorylation
A Maturing Evolutionary Perspective on Protein (De)phosphorylation
For several decades, the phosphorylation of proteins on serine and threonine residues was generally regarded as exclusively eukaryotic in origin and distribution, an adaptation to the coordination and communication requirements of a more complex compartmentalized cell form (1). In this scenario, tyrosine phosphorylation emerged to meet the expanded signal transduction needs of “higher” eukaryotes composed of multiple differentiated cells. The persistence and pervasiveness of this viewpoint are manifested by the continued use of the designator “eukaryotic protein kinase” (ePK)2 when referring to homologs of the prototype for this superfamily, the catalytic subunit of the cAMP-dependent protein kinase.
As the 1990s dawned, occasional reports surfaced indicating the presence of eukaryotic protein kinases and protein phosphatases in bacteria and viruses (1, 2). However, their close association with pathogenic organisms, tendency to target proteins endogenous to their eukaryotic hosts, and frequent coding by mobile elements such as plasmids were consistent with acquisition from the Eukarya by lateral gene transfer. It was not until genomics fueled a quantum leap in our understanding of the macromolecular populations within living organisms that it became apparent that many ostensibly eukaryotic protein kinases and protein phosphatases were in fact indigenous to some prokaryotes as well (3, 4).
Early Studies on Protein Phosphorylation in the Archaea
In the late 1970s, the Archaea emerged as a new player in the exploitation of phylogenetic diversity for tracing the evolution of biological macromolecules (5). Although morphologically prokaryotic, the rRNA sequences of the Archaea were more closely related to those of the Eukarya than to the superficially similar Bacteria, indicating that the first divergence from the last universal common ancestor separated the bacterial line of descent from a conjoint eukaryal/archaeal one.
The first indication that archaeal proteins were subject to regulation via covalent phosphorylation was reported by Spudich and Stoeckenius (6), who detected multiple radiolabeled polypeptides in extracts from cultures of the extreme halophile Halobacterium halobium that were grown in media containing 32P-labeled inorganic phosphate. The radiolabel remained associated with the polypeptides following exposure to either acid or hydroxylamine, indicating that the phosphoryl moieties were bound via phosphoester linkages. The apparent degree of phosphorylation of several polypeptides visibly decreased upon exposure to light, a phenomenon dependent on the presence of retinal (6). Skórko (7) employed a similar in vivo labeling approach to elucidate the presence of serine- and threonine-phosphorylated polypeptides in membrane fractions from the extreme acidothermophile Sulfolobus acidocaldarius.
A new twist was added when it was reported that multiple polypeptides from three diverse archaea (Haloferax volcanii, Methanosarcina thermophila TM-1, and Sulfolobus solfataricus P1) tested positive on Western blots probed with antibodies against phosphotyrosine (8). Immunoreactivity could be eliminated by prior treatment with alkaline phosphatase. This wholly unexpected finding was buttressed by the subsequent isolation of three phosphotyrosine-containing proteins from Thermococcus kodakaraensis KOD1 using a substrate-trapping version of an endogenous protein-tyrosine phosphatase (TkPTP): phenylalanyl-tRNA synthetase, an RNA terminal phosphate cyclase, and phosphomannomutase (9). Although highly suggestive of the existence of archaeal protein-tyrosine kinases, these findings were insufficient to rule out potential alternative explanations such as the hydrolytic breakdown of nucleotidylated tyrosine residues (10), trapping of phosphoenzyme intermediates (11), or adventitious autophosphorylation (12).
Protein-serine/threonine/tyrosine Kinases and Phosphatases: Demise of the “Eukaryotic” Paradigm
The first example of apparent domain trespass by a eukaryotic protein kinase or phosphatase in the Archaea was PP1-arch1, a PPP family protein-serine/threonine phosphatase. First detected by incubating soluble extracts of S. solfataricus P1 with 32P-phosphoproteins (13), subsequent purification and cloning of its gene revealed a monomeric enzyme sharing ≈30% identity with the catalytic domains of PP1 and PP2A from the Eukarya (14). Subsequently, genes encoding two additional archaeal PPPs, PP1-arch2 from M. thermophila TM-1 (15) and PyPP1 from Pyrodictium abyssi TAG11 (16), were cloned, and their recombinant protein products were demonstrated to possess protein-serine/threonine phosphatase activity. All three archaeal PPPs preferred Mn2+ as cofactor; little activity was detected in the presence of Mg2+. A dendrogram constructed using established PPP family protein phosphatases from the Archaea, Bacteria, and Eukarya mirrors the Woesian tree (Fig. 1), indicating that archaeal PPPs were inherited from the last universal common ancestor (17), not acquired by lateral gene transfer. Intriguingly, PP1-arch2 was inhibited by micromolar concentrations of okadaic acid, a classic diagnostic agent for PP1 and PP2A in eukaryotes (15). The primordial origins of this important family of signal transduction enzymes offered the first hint that protein (de)phosphorylation emerged at an earlier point in evolution than previously suspected.
FIGURE 1.
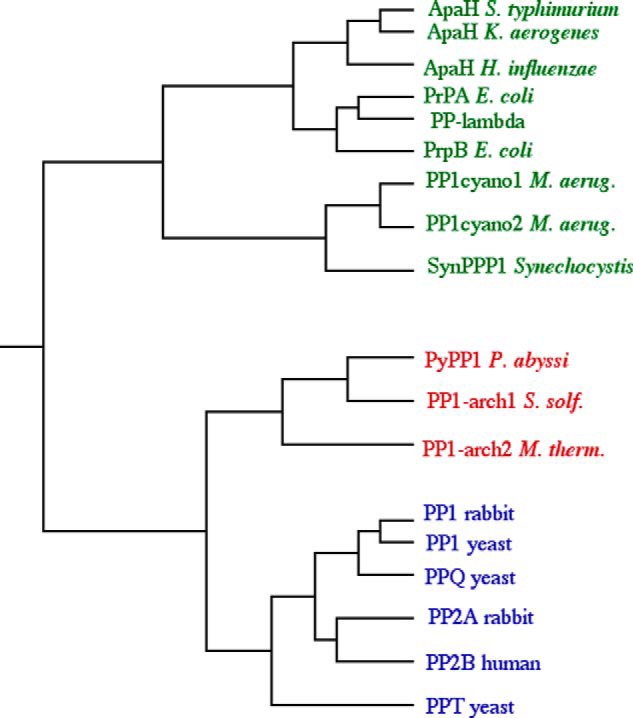
PPP tree. Shown is a dendrogram constructed using the sequences of assorted eukaryal (blue) and bacterial (green) PPP family protein phosphatases along with the three verified PPP family protein-serine/threonine phosphatases (red): PP1-arch1 from S. solfataricus (S. solf.) (14), PP1-arch2 from M. thermophila (M. therm.) TM-1 (15), and PyPP1 from P. abyssi TAG11 (16). PP, protein phosphatase; M. aerug., Microcystis aeruginosa.
Capitalizing on the initial wave of genome sequencing, Smith and King (18) reported the presence of ORFs encoding deduced ePK homologs in three methanogenic archaea: Methanococcus vannielii, Methanococcus voltae, and Methanobacterium thermoautotrophicum. As genome sequences continued to accumulate, it became increasingly apparent that virtually every member of the Archaea possessed an ORF encoding an ePK homolog (4, 19). However, although ubiquitous, the number of ePKs present in archaeal genomes was generally small, generally one to four (20). Perhaps more significantly, those ePKs previously stereotyped in the Eukarya as being “atypical” (21) dominated archaeal kinomes, often to the exclusion of typical ePKs (3, 22).
The distribution of protein-serine/threonine/tyrosine phosphatase homologs among the Archaea exhibits a strikingly mosaic pattern wherein no superfamily predominates (Table 1). Little correlation is evident between the number of deduced protein phosphatases in a given archaeal genome and the quantity of prospective ePKs. Even in those archaea wherein the number of deduced ePKs reached 10 or more, the number of deduced protein phosphatases rarely exceeded two (20). However, roughly three-quarters possess a conventional PTP (cPTP) and/or a low molecular weight (LMW) PTP (19, 23). Indeed, the deduced complement of protein-serine/threonine/tyrosine phosphatases for one-third of archaea listed in Table 1 consists exclusively of one or more PTPs. No representatives of the Cdc25 family of dual-specificity protein phosphatases have been detected in the Archaea, notwithstanding the presence of the Cdc25 evolutionary precursor rhodanese (24).
TABLE 1.
Distribution of eukaryotic protein-serine/threonine/tyrosine kinase/phosphatase superfamilies in the Archaea
Listed below are archaeal organisms whose genomes have been annotated by the Comprehensive Microbial Resource (J. Craig Venter Institute). “Yes” indicates that the genome in question contains one or more ORFs encoding a potential ePK, PPP family protein phosphatase, PPM family protein phosphatase, cPTP, or LMW PTP.
| Archaeon | ePK | PPP | PPM | cPTP | LMW PTP |
|---|---|---|---|---|---|
| Aeropyrum pernix K1 | Yes | Yes | |||
| Archaeoglobus fulgidus DSM4303 | Yes | Yes | Yes | ||
| Caldivirga maquilingensis IC-167 | Yes | Yes | Yes | ||
| Candidatus Korarchaeum cryptofilum OPF8 | Yes | Yes | Yes | Yes | |
| Candidatus Methanoregula boonei 6A8 | Yes | Yes | Yes | ||
| Haloacrula marismortui ATCC 43049 | Yes | Yes | Yes | ||
| Halobacterium NRC-1 | Yes | Yes | |||
| Halobacterium salinarium R1 | Yes | Yes | |||
| Halomicrobium mukohataei DSM12286 | Yes | Yes | Yes | Yes | |
| Haloquadratum walsbyi DSM16790 | Yes | Yes | |||
| Ignicoccus hospitalis KIN4/l | Yes | Yes | |||
| Metallosphaera sedula DSM5348 | Yes | Yes | Yes | ||
| Methanobacterium thermoautotrophicus Delta H | Yes | Yes | Yes | ||
| Methanococcoides burtonii DSM6242 | Yes | ||||
| Methanococcus aeolicus Nankai-3 | Yes | Yes | |||
| Methanococcus jannaschii DSM2661 | Yes | Yes | |||
| Methanococcus maripaludis C5 | Yes | Yes | |||
| Methanococcus maripaludis C6 | Yes | Yes | |||
| Methanococcus maripaludis C7 | Yes | Yes | |||
| Methanococcus maripaludis S2 | Yes | Yes | |||
| Methanocorpusculum labreanum Z | Yes | Yes | |||
| Methanoculleus marisnigri JR1 | Yes | Yes | |||
| Methanopyrus kandleri AV19 | Yes | ||||
| Methanosaeta thermophila PT | Yes | Yes | |||
| Methanosarcina acetivorans C2A | Yes | Yes | Yes | ||
| Methanosarcina barkeri Fusaro | Yes | Yes | |||
| Methanosarcina mazei Goe1 | Yes | Yes | Yes | ||
| Methanosphaera stadtmanae DSM3091 | Yes | ||||
| Methanospirillum hungatei JF-1 | Yes | Yes | Yes | ||
| Nanoarchaeum equitans KIN4-M | Yes | ||||
| Natronomonas pharaonis sp. | Yes | Yes | |||
| Picrophilus torridus DSM9790 | Yes | Yes | |||
| Pyrobaculum aerophilum IM2 | Yes | Yes | Yes | ||
| Pyrobaculum arsenaticum DSM13514 | Yes | Yes | Yes | ||
| Pyrobaculum islandicum DSM4184 | Yes | Yes | Yes | ||
| Pyrococcus abyssi GE5 | Yes | Yes | |||
| Pyrococcus furiosus DSM3638 | Yes | Yes | Yes | ||
| Pyrococcus horikoshii Shinkaj OT3 | Yes | Yes | |||
| Staphylothermus marinus F1 | Yes | Yes | |||
| Sulfolobus acidocaldarius DSM639 | Yes | Yes | Yes | ||
| Sulfolobus solfataricus P2 | Yes | Yes | Yes | ||
| Sulfolobus tokodaii strain 7 | Yes | Yes | Yes | ||
| Thermococcus kodakaraensis KOD1 | Yes | Yes | Yes | ||
| Thermofilum pendens Hrk5 | Yes | Yes | |||
| Thermoplasma acidophylum DSM1728 | Yes | ||||
| Thermoplasma volcanium GSS1 | Yes | Yes | Yes | ||
| Uncultured methanogenic archaeon RC-1 | Yes | Yes |
Two archaeal cPTPs have been characterized to date: TkPTP from T. kodakaraensis (9) and SsoPTP from S. solfataricus (25). Both proteins exhibited tyrosine phosphatase activity in vitro. In addition, TkPTP hydrolyzed free phosphoserine at rates comparable to free phosphotyrosine (9). X-ray crystallography of SsoPTP revealed considerable structural homology to VH1 family PTPs. The depth of its active site pocket suggests that SsoPTP is tyrosine-specific (25).
Of the two major protein-serine/threonine phosphatase families encountered in eukaryotic organisms (26), the PPP superfamily predominates over the PPM superfamily by a ratio of 3:1 (Table 1). The only archaeal PPM characterized to date, TvnPPM from Thermoplasma volcanium, exhibited both protein-tyrosine and protein-serine/threonine phosphatase activities in vitro, a property it shares with a handful of bacterial PPM homologs (27). The protein-tyrosine phosphatase activity of TvnPPM may be physiologically significant, as T. volcanium lacks deduced LMW PTPs or cPTPs.
Whence Eukaryotic Protein Kinases and Phosphatases?
What can the Archaea tell us about the origins and evolution of the protein kinases and protein phosphatases that lie at the core of the signal transduction networks in the Eukarya? The universal presence of specific families of atypical ePKs in both the Archaea and Eukarya, but not the Bacteria (3, 22, 28), indicates that the first recognizable member of the ePK superfamily appeared after the divergence of the combined archaeal/eukaryal line of descent from the bacterial but prior to the divergence of the Archaea from the Eukarya (3). The nearly universal presence of piD261/Bud32 and right open reading frame (RIO) protein kinases among the Archaea and Eukarya further suggests that these atypical ePKs most closely echo the archetypic ePK (22, 29, 30). Typical ePKs appear to have first emerged in the Eukarya (3), where they proliferated to an extent that far outstripped their older atypical siblings (21). From the Eukarya, typical ePKs spread to the Bacteria and, presumably, the Archaea via lateral gene transfer (4).
The two major families of eukaryotic protein-serine/threonine phosphatases offer a study in contrasts. The PPP family has a long evolutionary history dating to the last universal common ancestor (4, 31). PPPs appear to have spread throughout the three extant phylogenetic domains primarily via direct inheritance. In contrast, the PPM family emerged much later, in the Eukarya, and later spread to the Bacteria by lateral gene transfer (4, 32). It appears likely that the handful of archaeal PPMs were acquired “secondhand” from the Bacteria.
The widespread phylogenetic dispersal of the cPTPs and LMW PTPs is indicative of ancient origins that likely date back to the last universal common ancestor (33–35). Although rhodanese, the progenitor of the Cdc25 dual-specificity protein phosphatases, is pervasive among prokaryotic organisms, Cdc25 appears to be exclusively eukaryal in residence and, by extension, origin (24). The seemingly random distribution of these ancient protein phosphatases likely reflects the impact of reductive evolution (36, 37). Presumably the PPP, cPTP, and LMW PTP families were each represented in the original archaeal phosphatase suite. However, as they segregated to environmentally extreme but relatively monotonous habitats, many archaea downsized their sensor-response capabilities.
The Sulfolobales Kinome
Much of what we currently know concerning protein (de)phosphorylation in the Archaea has been generated from investigations of the members of the order Sulfolobales, in particular S. solfataricus. The S. solfataricus genome contains 10 ORFs encoding prospective ePKs, two of which exhibit recognizable transmembrane domains (Fig. 2). By eukaryal standards, ePKs account for only a minute portion (0.34%) of the S. solfataricus genome (38). However, relative to most other archaea, S. solfataricus is protein kinase-rich, with a kinome that even includes both typical and atypical ePKs. In contrast, like most archaea, its suite of recognizable protein phosphatases is relatively sparse, consisting of a single PPP family protein-serine/threonine phosphatase, PP1-arch1 (14), and a single cPTP (25). Of the 10 prospective ePKs from S. solfataricus, three have been studied in the laboratory: SsoPK2, SsoPK3, and SsoPK5 (Fig. 2). (The designator SsoPK1 was reserved for an ≈62-kDa membrane-associated glycoprotein with protein-serine/threonine kinase activity, possibly the product of ORF sso2291 (39, 40).3)
FIGURE 2.
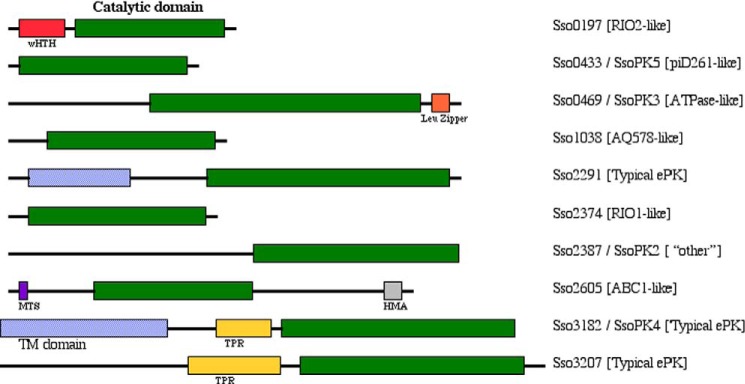
Known and deuced ePKs encoded by the genome of S. solfataricus P2. Shown are schematic diagrams of the known and deduced ePKs encoded by the genome of S. solfataricus (38). The identity of the ORF is given on the right along with the protein name, if any. The name of the most closely related protein kinase subfamily, as deduced from the sequence of the putative catalytic domains, is shown in parentheses using the classification scheme of Leonard et al. (3). The predicted catalytic, transmembrane, (TM), and tetratricopeptide repeat (TPR) domains are colored green, blue, and yellow, respectively. Other prospective features include winged helix-turn-helix (wHTH; red), leucine zipper (orange), methyltransferase-like (MTS; purple), and heavy metal-associated (HMA) domains.
SsoPK2 is the protein product of ORF sso2387. It is an atypical ePK that does not appear to group with any of the “major” atypical subfamilies. It also has been annotated as a secretory ATPase (41). The catalytic domain of SsoPK2 occupies the C-terminal half of a projected 583-residue polypeptide. Analysis of the sequence of the N-terminal region revealed no obvious ligand-binding sites, protein-protein interaction sites, or transmembrane domains, suggesting that anchoring occurs via association with a second, membrane-bound protein (41). Expression of the mRNA encoding SsoPK2 could be detected only in cells grown on rich media (41).
Recombinant SsoPK2 phosphorylated itself in vitro (41, 42), as well as exogenous proteins of grossly basic (histones and bovine serum albumin) and acidic (casein and reduced carboxyamidomethylated and maleylated lysozyme) character (42). Phosphorylation occurred exclusively on serine residues. No activity was detected using the classic tyrosine kinase substrates poly(Glu,Tyr) and poly(Glu4,Tyr) or using GTP as the phosphodonor substrate (42). SsoPK2 exhibited ATPase activity at high temperatures (41). Both the protein kinase and ATPase activities of SsoPK2 exhibited a preference for Mn2+ over Mg2+ as cofactor (41, 42). Although autophosphorylation takes place on multiple serine residues, substitution of one of these, Ser548, which resides in the predicted T loop located between subdomains VII and VIII of the catalytic domain, by alanine resulted in complete abolition of autophosphorylation in vitro (42). Phosphorylation of an exogenous substrate (casein) was not affected, however.
SsoPK3, the polypeptide product of ORF sso0469, was first identified as one of the phosphoproteins produced when membrane extracts of S. solfataricus were incubated with [γ-32P]ATP (43). SsoPK3 belongs to the ATPase-like subfamily of atypical ePKs. Like SsoPK2, its catalytic domain occupies the C-terminal portion of the polypeptide. Although membrane-associated, it exhibits no obvious transmembrane domain, suggesting that anchoring occurs via some other structural motif. Recombinant SsoPK3 autophosphorylated itself on threonine and exogenous substrates such as casein, bovine serum albumin, and histone H4 on serine and, to a lesser degree, threonine (43). No activity was detected toward poly(Glu,Tyr) or poly(Glu4,Tyr). Like SsoPK2, Mn2+ was preferred as cofactor (43).
SsoPK5, the product of ORF sso0433, is among the smallest ePKs yet encountered. It is a member of the most ancient extant family of ePKs, the piD261/Bud32 protein kinases, whose sole mammalian homolog is the p53-related protein kinase (30). A candidate for the direct lineal descendant of the primordial ePK, homologs of piD261/Bud32 are found throughout the Archaea and Eukarya but are absent from the Bacteria (34). Disruption of the gene for piD261/Bud32 in Saccharomyces cerevisiae confers a severe slow growth phenotype that can be partially rescued by the p53-related ePK (44). Despite its essential nature and significant evolutionary head start over virtually every other family of ePKs, the piD261/Bud32 family has undergone no detectable expansion over the eons (29).
In archaeal genomes, the sequences encoding piD261/Bud32 homologs are generally located adjacent to or fused with the gene encoding Kae1 (kinase-associated endopeptidase 1), which is universally present in the members of all three phylogenetic domains (45). Structural analyses (46, 47) have revealed that piD261/Bud32 and Kae1 are components of a larger multiprotein complex dubbed the EKC/KEOPS complex, which stands for endopeptidase-like kinase chromatin-associated/kinase, endopeptidase, and other proteins of small size (45). It has been suggested that, in yeast, the complex plays a dual role involving regulation of transcription and maintenance of telomere integrity (46–48). More recently, it has been discovered that the EKC/KEOPS complex catalyzes the formation of N6-threonylcarbamoyl adenosine, a universal modification of tRNAs recognizing codons that begin with an adenine nucleotide (49). Within the context of this complex, piD261/Bud32 has been observed to display switchable protein kinase and P-loop ATPase activity (50).
When SsoPK5, the homolog of piD261/Bud32 from S. solfataricus, was expressed in Escherichia coli, the recombinant protein exhibited detectable but relatively sluggish protein-serine/threonine kinase activity toward acidic proteins such as casein and reduced carboxyamidomethylated and maleylated lysozyme (51). No activity was detected toward two basic proteins: histones and myelin basic protein. SsoPK5 also phosphorylated itself on serine and threonine. Substitution of Thr159, a predicted site of autophosphorylation, by alanine resulted in a 50% decrease in self-phosphorylation and loss of activity toward exogenous protein substrates (51). Intriguingly, ADP-ribose, the breakdown product of the poly(ADP-ribose) chains that covalently modify many eukaryal proteins, particularly those found in chromatin, markedly stabilized SsoPK5, an effect that could be enhanced by millimolar levels of 5′-AMP. Enhancement was highly specific to 5′-AMP and independent of autophosphorylation (51).
Three ORFs in the S. solfataricus genome code for potential homologs of the typical ePKs, specifically the protein kinases that phosphorylate eIF2α in the Eukarya: sso2291, sso3182, and sso3207 (Fig. 2). Phosphorylation of eIF2α by eukaryal eIF2α protein kinases inhibits global polypeptide synthesis in response to a variety of cellular stresses (52). An eIF2α protein kinase homolog from the hyperthermophilic archaeon Pyrococcus horikoshii OT3 (Ph0512p) phosphorylates the archaeal homolog of eIF2α (aIF2α) in vitro (53). Phosphorylation occurred on Ser48, which aligns with the site of regulatory protein phosphorylation in eIF2α (Ser51). This serine residue is conserved in deduced aIF2α proteins in the archaea Methanococcus jannaschii, M. thermoautotrophicum, and Archaeoglobus fulgidus (53), as well as S. solfataricus (38).
Like S. solfataricus, Sulfolobus tokodaii is relatively rich in ORFs encoding deduced ePKs. Moreover, at least one of the eight potential ePKs identified, the protein product of st1565, resembles the typical ePKs that predominate in eukaryotes (54). Using two-hybrid screens and pulldown assays, it was determined that the ST1565 protein kinase complexes with the protein product of st0829, a forkhead-associated (FHA) domain-containing transcription factor that interacts with the promoter region of a gene cluster encoding flagella-like proteins (55). Moreover, phosphorylation of the FHA domain-containing protein by the ST1565 protein kinase abrogated the former's DNA-binding capacity in vitro (55).
ORFs sso0197 and sso2374 from S. solfataricus encode potential RIO ePKs. The RIOs constitute the primary rival of piD261/Bud32 for the title of most ancient extant ePK family (3, 28). The RIO ePKs were first encountered in yeast, where they have been implicated in ribosome biogenesis, cell cycle progression, and genome integrity (56). Although the S. solfataricus RIO ePKs remain uncharacterized, the crystal structure for Rio2 from A. fulgidus has been determined (57), and a second archaeal RIO ePK from H. volcanii has been shown to phosphorylate the α subunit of the 20 S proteasome on serine and threonine in vitro (58). For more on the RIO ePKs, see the accompanying minireview by LaRonde (71).
The Sulfolobales Phosphoproteome
Recent mass spectrometric analysis of the proteins from S. solfataricus resulted in the identification of 1318 different sites of covalent phosphorylation distributed among 540 different proteins (59). Modification by phosphorylation appeared to be both physically and functionally pervasive, as the phosphoproteins identified included members of 21 of the 26 archaeal clusters of orthologous groups (60) represented in the S. solfataricus genome (59). In addition to a broad spectrum of proteins that participate in core metabolic processes, the S. solfataricus phosphoproteome also includes a number of deduced transcription factors; many ribosomal proteins; a large number of aminoacyl-tRNA synthetases; a predicted adenylate cyclase; DNA helicases, gyrases, primases, topoisomerases, transposases, and polymerases; ATPases; translation initiation and elongation factors; assorted permeases and transporters; ePKs; and a large number of unidentified proteins.
Perhaps even more unexpected than the unanticipated surfeit of phosphoproteins was the abundance of phosphorylated tyrosine residues in the phosphoproteins from S. solfataricus (59). In fact, the number of sites of tyrosine phosphorylation marginally exceeded the number of combined sites of serine and threonine phosphorylation. By contrast, the only other reported archaeal phosphoproteome, that of the extreme halophile Halobacterium salinarium, encompassed only 81 sites of phosphorylation on 69 proteins, only one of which was identified, albeit tentatively, as tyrosine (61).
It has yet to be determined what proportion of these archaeal phosphoproteins are the product of regulatory phosphorylation events. The fact the S. solfataricus phosphoproteome exhibited marked shifts when the organism's carbon source was switched from glucose to Tryptone is suggestive of the dynamism characteristic of regulatory events (59). However, data affirming that covalent phosphorylation plays a regulatory role in the Archaea have emerged for only a handful of proteins to date, such as d-gluconate dehydratase (62) and phosphohexomutase from S. solfataricus (63), an FHA domain-containing protein from S. tokodaii (55), and the proteasome from H. volcanii (58).
The phosphorylation of d-gluconate dehydratase was detected by growing S. solfataricus in the presence of [32P]phosphate (62). Subsequent incubation of the phosphoprotein with broad-spectrum acid phosphatases resulted in a loss of catalytic activity, implicating phosphorylation as an essential prerequisite for function. The S. solfataricus phosphohexomutase was identified via mass peptide profiling of cellular proteins. Although formation of a covalent phosphoenzyme intermediate constitutes an integral step in the enzyme's catalytic mechanism, the catalytic serine (Ser97) was distinct from the site of phosphorylation identified by mass spectrometry (Ser309). Modeling the S. solfataricus protein using the x-ray structure of a homolog predicted that Ser309 resided near the mouth of the active site, suggesting that phosphorylation of this residue would inhibit binding of phosphohexose substrates by an electrosteric mechanism similar to that by which phosphorylation inhibits the isocitrate dehydrogenases of enteric bacteria (64). Substitution of Ser309 by aspartate reduced Vmax by 20-fold, indicating that phosphorylation at that site is inhibitory (63).
The FHA domain-containing protein was initially implicated by its capacity to bind to and to physically interact with a typical ePK, the product of ORF st1565 from S. tokodaii (54). Subsequent analysis quickly revealed that the FHA domain-containing protein was phosphorylated by the ST1565 ePK in vitro, although the identity and location of the amino acid residue modified were not determined. Subsequent studies revealed that phosphorylation of the FHA domain-containing protein adversely impacted its ability to bind DNA (55). Analysis of the 20 S proteasome from H. volcanii by mass spectrometry revealed numerous covalent modifications, including phosphorylation of the α1, α2, and β subunits on serine and/or threonine (58). Blocking phosphorylation of serine and threonine α1 subunits by substituting alanine produced variants that were unable to restore pigment production or salt tolerance to α1 knock-out strains (65).
Open Questions
Studies to date on the protein phosphorylation networks resident in the members of the domain Archaea have raised some intriguing questions regarding the origins and development of this highly versatile molecular regulatory mechanism (66). For example, typical ePKs appear to be absent from the Euryarchaea, one of the two major subgroups within the domain Archaea. Moreover, certain of the typical ePKs within the other subgroup, the Crenarchaea, are homologs of a specific subset of typical ePKs, the eIF2α protein kinases (4), suggesting that these crenarchaea acquired them by horizontal gene transfer from the Eukarya. Dissecting the circumstances under which such a transfer may have occurred may reveal much about the elusively complex evolutionary events that generated the mosaic of unique, Bacteria-like, and Eukarya-like features that characterize archaeal genomes (67).
Another looming question is how to reconcile the apparent existence of protein tyrosine phosphorylation in the Archaea with its skewed distribution within the Eukarya (68). For years, bona fide rather than dual-specificity tyrosine phosphorylation has been described as a late-emerging process that evolved specifically to meet the demands of multicellular specialization in higher eukaryotes (35, 69). If archaeal protein tyrosine phosphorylation is ancient in origin, why was it not passed on to all members of the Eukarya? Did fungi and other protists somehow lose this capability along the way, and if so, what evolutionary pressures promoted its retention among many of the Archaea? Did phosphotyrosine's immunity from β-elimination render it better suited for hyperthermophiles? Does it necessarily follow, however, that loss of thermophily is sufficient to explain its putative disappearance? If the pervasiveness of PTPs among the Archaea indicates that the cPTPs and LMW PTPs predate protein tyrosine phosphorylation, what purpose did they serve in primordial organisms? Might they still be serving these roles today? Ample evidence exists for the catalytic adaptability of both PTP scaffolds in the form of homologs that hydrolyze phosphoinositides (70), dephosphorylate serine and threonine residues (34), and reduce arsenate to arsenite (19). Does the seeming imbalance between the handful of ePKs and protein phosphatases in an archaeon like S. solfataricus and its massive phosphoproteome indicate that the Archaea harbor some as yet unrecognized types of protein-serine/threonine/tyrosine kinases and/or phosphatases?
This is the third article in the Thematic Minireview Series “Protein Serine/Threonine and Tyrosine Phosphorylation in Prokaryotes.”
B. H. Lower and P. J. Kennelly, unpublished data.
- ePK
- eukaryotic protein kinase
- PTP
- protein-tyrosine phosphatase
- PPP
- family of protein-serine/threonine phosphatases that include eukaryotic PP1, PP2A, and PP2B
- cPTP
- conventional PTP
- LMW
- low molecular weight
- PPM
- family of eukaryotic divalent metal ion-dependent protein-serine/threonine phosphatases whose exemplar is protein phosphatase 2C
- RIO
- right open reading frame
- FHA
- forkhead-associated.
REFERENCES
- 1. Kennelly P. J., Potts M. (1996) Fancy meeting you here! A fresh look at “prokaryotic” protein phosphorylation. J. Bacteriol. 178, 4759–4764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Guan K.-L., Dixon J. E. (1993) Bacterial and viral protein tyrosine phosphatases. Semin. Cell Biol. 4, 389–396 [DOI] [PubMed] [Google Scholar]
- 3. Leonard C. J., Aravind L., Koonin E. V. (1998) Novel families of putative protein kinases in bacteria and archaea: evolution of the “eukaryotic” protein kinase superfamily. Genome Res. 8, 1038–1047 [DOI] [PubMed] [Google Scholar]
- 4. Ponting C. P., Aravind L., Schultz J., Bork P., Koonin E. V. (1999) Eukaryotic signaling domain homologues in archaea and bacteria. Ancient ancestry and horizontal gene transfer. J. Mol. Biol. 289, 729–745 [DOI] [PubMed] [Google Scholar]
- 5. Woese C. R., Magrum L. J., Fox G. E. (1978) Archaebacteria. J. Mol. Evol. 11, 245–251 [DOI] [PubMed] [Google Scholar]
- 6. Spudich J. L., Stoeckenius W. (1980) Light-mediated retinal-dependent reversible phosphorylation of Halobacterium proteins. J. Biol. Chem. 255, 5501–5503 [PubMed] [Google Scholar]
- 7. Skórko R. (1984) Protein phosphorylation in the archaebacterium Sulfolobus acidocaldarius. Eur. J. Biochem. 145, 617–622 [DOI] [PubMed] [Google Scholar]
- 8. Smith S. C., Kennelly P. J., Potts M. (1997) Protein-tyrosine phosphorylation in the Archaea. J. Bacteriol. 179, 2418–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jeon S.-J., Fujiwara S., Takagi M., Tanaka T., Imanaka T. (2002) Tk-PTP, a protein-tyrosine phosphatase from the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1: enzymatic characteristics and identification of its substrate proteins. Biochem. Biophys. Res. Commun. 295, 508–514 [DOI] [PubMed] [Google Scholar]
- 10. Foster R., Thorner J., Martin G. S. (1989) Nucleotidylation, not phosphorylation, is the major source of the phosphotyrosine detected in enteric bacteria. J. Bacteriol. 171, 272–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Solow B., Bischoff K. M., Zylka M. J., Kennelly P. J. (1998) Archaeal phosphoproteins. Identification of a hexosephosphate mutase and the α-subunit of succinyl-CoA synthetase in the extreme acidothermophile Sulfolobus solfataricus. Protein Sci. 7, 105–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Almaula N., Lu Q., Delgado J., Belkin S., Inouye M. (1995) Nucleoside diphosphate kinase from Escherichia coli. J. Bacteriol. 177, 2524–2529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kennelly P. J., Oxenrider K. A., Leng J., Cantwell J. S., Zhao N. (1993) Identification of a serine/threonine-specific protein phosphatase from the archaebacterium Sulfolobus solfataricus. J. Biol. Chem. 268, 6505–6510 [PubMed] [Google Scholar]
- 14. Leng J., Cameron A. J., Buckel S., Kennelly P. J. (1995) Isolation and cloning of a protein-serine/threonine phosphatase from an archaeon. J. Bacteriol. 177, 6510–6517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Solow B., Young J. C., Kennelly P. J. (1997) Gene cloning and expression and characterization of a toxin-sensitive protein phosphatase from the methanogenic archaeon Methanosarcina thermophila TM-1. J. Bacteriol. 179, 5072–5075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mai B., Frey G., Swanson R. V., Mathur E. J., Stetter K. O. (1998) Molecular cloning and functional expression of a protein-serine/threonine phosphatase from the hyperthermophilic archaeon Pyrodictium abyssi TAG11. J. Bacteriol. 180, 4030–4035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bhaduri A., Sowdhamini R. (2005) A genome-wide survey of prokaryotic O-protein phosphatases. J. Mol. Biol. 352, 736–752 [DOI] [PubMed] [Google Scholar]
- 18. Smith R. F., King K. Y. (1995) Identification of a eukaryote-like protein kinase gene in Archaebacteria. Protein Sci. 4, 126–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shi L., Potts M., Kennelly P. J. (1998) The serine-, threonine-, and/or tyrosine-specific protein kinases and protein phosphatases of prokaryotic organisms: a family portrait. FEMS Microbiol. Rev. 22, 229–253 [DOI] [PubMed] [Google Scholar]
- 20. Kennelly P. J. (2003) Archaeal protein kinases and protein phosphatases–insights from genomics and biochemistry. Biochem. J. 370, 373–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Manning G., Whyte D. B., Martinez R., Hunter T., Sudarsanam S. (2002) The protein kinase complement of the human genome. Science 298, 1912–1934 [DOI] [PubMed] [Google Scholar]
- 22. Kannan N., Taylor S. S., Zhai Y., Venter J. C., Manning G. (2007) Structural and functional diversity of the microbial kinome. PLoS Biol. 5, e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Stravopodis D. J., Kyrpides N. C. (1999) Identification of protein-tyrosine phosphatases in Archaea. J. Mol. Evol. 48, 625–627 [DOI] [PubMed] [Google Scholar]
- 24. Bordo D., Bork P. (2002) The rhodanese/Cdc25 phosphatase superfamily. Sequence-structure-function relations. EMBO Rep. 3, 741–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chu H.-M., Wang A. H.-J. (2007) Enzyme-substrate interactions revealed by crystal structures of the archaeal Sulfolobus PTP-fold phosphatase and its phosphopeptide complexes. Proteins 66, 996–1003 [DOI] [PubMed] [Google Scholar]
- 26. Barford D. (1996) Molecular mechanisms of the protein-serine/threonine phosphatases. Trends Biochem. Sci. 21, 407–412 [DOI] [PubMed] [Google Scholar]
- 27. Dahche H., Abdullah A., Potters M. B., Kennelly P. J. (2009) A PPM-family protein phosphatase from Thermoplasma volcanium hydrolyzes protein-bound phosphotyrosine. Extremophiles 13, 371–377 [DOI] [PubMed] [Google Scholar]
- 28. Esser D., Siebers B. (2013) Atypical protein kinases of the RIO family in archaea. Biochem. Soc. Trans. 41, 399–404 [DOI] [PubMed] [Google Scholar]
- 29. Facchin S., Sarno S., Marin O., Lopreiato R., Sartori G., Pinna L. A. (2002) Acidophilic nature of yeast PID261/BUD32, a putative ancestor of eukaryotic protein kinases. Biochem. Biophys. Res. Commun. 296, 1366–1371 [DOI] [PubMed] [Google Scholar]
- 30. Facchin S., Lopreiato R., Ruzzene M., Marin O., Sartori G., Götz C., Montenarh M., Carignani G., Pinna L. A. (2003) Functional homology between yeast pID261/Bud32 and human PRPK: both phosphorylate p53 and PRPK partially complements piD261/Bud32 deficiency. FEBS Lett. 549, 63–66 [DOI] [PubMed] [Google Scholar]
- 31. Moorhead G. B. G., De Wever V., Templeton G., Kerk D. (2009) Evolution of protein phosphatases in plants and animals. Biochem. J. 417, 401–409 [DOI] [PubMed] [Google Scholar]
- 32. Zhang W., Shi L. (2004) Evolution of the PPM-family protein phosphatases in Streptomyces: duplication of catalytic domain and lateral recruitment of additional sensory domains. Microbiology 150, 4189–4197 [DOI] [PubMed] [Google Scholar]
- 33. Ramponi G., Stefani M. (1997) Structural, catalytic, and functional properties of low Mr phosphotyrosine protein phosphatases. Evidence of a long evolutionary history. Int. J. Biochem. Cell Biol. 29, 279–292 [DOI] [PubMed] [Google Scholar]
- 34. Alonso A., Burkhalter S., Sasin J., Tautz L., Bogetz J., Huynh H., Bremer M. C. D., Holsinger L. J., Godzik A., Mustelin T. (2004) The minimal essential core of a cysteine-based protein-tyrosine phosphatase revealed by a novel 16-kDa VH1-like phosphatase, VHZ. J. Biol. Chem. 279, 35768–35774 [DOI] [PubMed] [Google Scholar]
- 35. Alonso A., Sasin J., Bottini N., Freidberg I., Friedberg I., Osterman A., Godzik A., Hunter T., Dixon J., Mustelin T. (2004) Protein tyrosine phosphatases in the human genome. Cell 117, 699–711 [DOI] [PubMed] [Google Scholar]
- 36. Waters E., Hohn M. J., Ahel I., Graham D. E., Adams M. D., Barnstead M., Beeson K. Y., Bibbs L., Bolanos R., Keller M., Kretz K., Lin X., Mathur E., Ni J., Podar M., Richardson T., Sutton G. G., Simon M., Soll D., Stetter K. O., Short J. M., Noordewier M. (2003) The genome of Nanoarchaeum equitans: insights into early archaeal evolution and derived parasitism. Proc. Natl. Acad. Sci. U.S.A. 100, 12984–12988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wolf Y. I., Koonin E. V. (2013) Genome reduction as the dominant mode of evolution. BioEssays 35, 829–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. She Q., Singh R. K., Confalonieri F., Zivanovic Y., Allard G., Awayez M. J., Chan-Weiher C. C., Clausen I. G., Curtis B. A., De Moors A., Erauso G., Fletcher C., Gordon P. M., Heikamp-de Jong I., Jeffries A. C., Kozera C. J., Medina N., Peng X., Thi-Ngoc H. P., Redder P., Schenk M. E., Theriault C., Tolstrup N., Charlebois R. L., Doolittle W. F., Duguet M., Gaasterland T., Garrett R. A., Ragan M. A., Sensen C. W., Van der Oost J. (2001) The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl. Acad. Sci. U.S.A. 98, 7835–7840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lower B. H., Bischoff K. M., Kennelly P. J. (2000) The archaeon Sulfolobus solfataricus contains a membrane-associated protein kinase that preferentially phosphorylates threonine residues. J. Bacteriol. 182, 3452–3459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lower B. H., Kennelly P. J. (2002) The membrane-associated protein-serine/threonine kinase from Sulfolobus solfataricus is a glycoprotein. J. Bacteriol. 184, 2614–2619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Albers S.-V., Driessen A. J. M. (2005) Analysis of ATPases of putative secretion operons in the thermoacidophilic archaeon Sulfolobus solfataricus. Microbiology 151, 763–773 [DOI] [PubMed] [Google Scholar]
- 42. Lower B. H., Kennelly P. J. (2003) Open reading frame sso2387 from the archaeon Sulfolobus solfataricus encodes a polypeptide with protein-serine kinase activity. J. Bacteriol. 185, 3436–3445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lower B. H., Potters M. B., Kennelly P. J. (2004) A phosphoprotein from the archaeon Sulfolobus solfataricus with protein-serine kinase activity. J. Bacteriol. 186, 463–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sartori G., Mazzotta G., Stocchetto S., Pavanello A., Carignani G. (2000) Inactivation of six genes from chromosomes VII and XIV from Saccharomyces cerevisiae and basic phenotypic analysis of the mutants. Yeast 16, 255–265 [DOI] [PubMed] [Google Scholar]
- 45. Hecker A., Lopreiato R., Graille M., Collinet B., Forterre P., Libri D., van Tilbeurgh H. (2008) Structure of the archaeal Kae1/Bud32 fusion protein MJ1130: a model for the eukaryotic EKC/KEOPS complex. EMBO J. 27, 2340–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kisseleva-Romanova E., Lopreiato R., Baudin-Baillieu A., Rouselle J.-C., Ilan L., Hofmann K., Namane A., Mann C., Libri D. (2006) Yeast homolog of a cancer-testis antigen defines a new transcription complex. EMBO J. 25, 3576–3585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mao D. Y. L., Neculai D., Downey M., Orlicky S., Haffani Y. Z., Ceccarelli D. F., Ho J. S. L., Szilard R. K., Zhang W., Ho C. S., Wan L., Fares C., Rumpel S., Kurinov I., Arrowsmith C. H., Durocher D., Sicheri F. (2008) Atomic structure of the KEOPS complex: an ancient protein kinase-containing molecular machine. Mol. Cell 32, 259–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hecker A., Graille M., Madec E., Gadelle D., Le Cam E., van Tilbergh H., Forterre P. (2009) The universal Kae1 protein and the associated Bud32 kinase (PRPK), a mysterious protein couple probably essential for genome maintenance in Archaea and Eukarya. Biochem. Soc. Trans. 37, 29–35 [DOI] [PubMed] [Google Scholar]
- 49. Srinivasan M., Mehta P., Yu Y., Prugar E., Koonin E. V., Karzai A. W., Sternglanz R. (2011) The highly conserved KEOPS/EKC complex is essential for a universal tRNA modification, t6A. EMBO J. 30, 873–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Perrochia L., Guetta D., Hecker A., Forterre P., Basta T. (2013) Functional assignment of KEOPS/EKC complex subunits in biosynthesis of the universal t6A tRNA modification. Nucleic Acids Res. 41, 9484–9499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Haile J. D., Kennelly P. J. (2011) The activity of an ancient atypical protein kinase is stimulated by ADP-ribose in vitro. Arch. Biochem. Biophys. 511, 56–63 [DOI] [PubMed] [Google Scholar]
- 52. Donnelly N., Gorman A. M., Gupta S., Samali A. (2013) The eIF2α kinases: their structure and functions. Cell. Mol. Life Sci. 70, 3493–3511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tahara M., Ohsawa A., Saito S., Kimura M. (2004) In vitro phosphorylation of initiation factor 2α (aIF2α) from hyperthermophilic archaeon Pyrococcus horikoshii OT3. J. Biochem. 135, 479–485 [DOI] [PubMed] [Google Scholar]
- 54. Wang B., Yang S., Zhang L., He Z.-G. (2010) Archaeal eukaryote-like serine/threonine protein kinase interacts with and phosphorylates a forkhead-associated-domain-containing protein. J. Bacteriol. 192, 1956–1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Duan X., He Z.-G. (2011) Characterization of the specific interaction between archaeal FHA domain-containing protein and the promoter of a flagellar-like gene-cluster and its regulation by phosphorylation. Biochem. Biophys. Res. Commun. 407, 242–247 [DOI] [PubMed] [Google Scholar]
- 56. Angermayr M., Roidl A., Bandlow W. (2002) Yeast Rio1p is the founding member of a novel subfamily of protein serine kinases involved in control of cell cycle progression. Mol. Microbiol. 44, 309–324 [DOI] [PubMed] [Google Scholar]
- 57. LaRonde-LeBlanc N., Guszczynski T., Copeland T., Wlodawer A. (2005) Autophosphorylation of Archaeoglobus fulgidus Rio2 and crystal structures of its nucleotide-metal ion complexes. FEBS J. 272, 2800–2810 [DOI] [PubMed] [Google Scholar]
- 58. Humbard M. A., Reuter C. J., Zuobi-Hasona K., Zhou G., Maupin-Furlow J. A. (2010) Phosphorylation and methylation of proteasomal proteins of the haloarchaeon Haloferax volcanii. Archaea 2010, 481725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Esser D., Pham T. K., Reimann J., Albers S.-V., Siebers B., Wright P. C. (2012) Change of carbon source causes dramatic effects in the phospho-proteome of the archaeon Sulfolobus solfataricus. J. Proteome Res. 11, 4823–4833 [DOI] [PubMed] [Google Scholar]
- 60. Makarova K. S., Sorokin A. V., Novichkov P. S., Wolf Y. I., Koonin E. V. (2007) Clusters of orthologous genes for 41 archaeal genomes and implications for evolutionary genomics. Biol. Direct 2, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Aivaliotis M., Macek B., Gnad F., Reichelt P., Mann M., Oesterhelt D. (2009) Ser/Thr/Tyr phosphorylation in the archaeon Halobacterium salinarium–a representative of the third domain of life. PLoS ONE 4, e4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kim S., Lee S. B. (2005) Identification and characterization of Sulfolobus solfataricus d-gluconate dehydratase: a key enzyme in the non-phosphorylated Entner-Doudoroff pathway. Biochem. J. 387, 271–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ray W. K., Keith S. M., DeSantis A. M., Hunt J. P., Larson T. J., Helm R. F., Kennelly P. J. (2005) A phosphohexomutase from the archaeon Sulfolobus solfataricus is covalently modified by phosphorylation on serine. J. Bacteriol. 187, 4270–4275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hurley J. H., Dean A. M., Thorsness P. E., Koshland D. E., Jr., Stroud R. M. (1990) Regulation of isocitrate dehydrogenase by phosphorylation involves no long-range conformational change in the enzyme. J. Biol. Chem. 265, 3599–3602 [DOI] [PubMed] [Google Scholar]
- 65. Humbard M. A., Stevens S. M., Jr., Maupin-Furlow J. A. (2006) Posttranslational modification of the 20 S proteasomal proteins of the archaeon Haloferax volcanii. J. Bacteriol. 188, 7521–7530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hunter T. (2012) Why nature chose phosphate to modify proteins. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367, 2513–2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zhaxybayeva O., Lapierre P., Gogarten J. P. (2005) Ancient gene duplications and the root(s) of the tree of life. Protoplasma 227, 53–64 [DOI] [PubMed] [Google Scholar]
- 68. Gnad F., Forner F., Zielinska D. F., Birney E., Gunawardena J., Mann M. (2010) Evolutionary constraints of phosphorylation in eukaryotes, prokaryotes, and mitochondria. Mol. Cell. Proteomics 9, 2642–2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Manning G., Plowman G. D., Hunter T., Sudarsanam S. (2002) Evolution of protein kinase signaling from yeast to man. Trends Biochem. Sci. 27, 514–520 [DOI] [PubMed] [Google Scholar]
- 70. Maehama T., Taylor G. S., Dixon J. E. (2001) PTEN and myotubularin: novel phosphoinositide phosphatases. Annu. Rev. Biochem. 70, 247–279 [DOI] [PubMed] [Google Scholar]
- 71. LaRonde N. A. (2014) The ancient microbial RIO kinases. J. Biol. Chem. 289, 10.1074/jbc.R113.538090 [DOI] [PMC free article] [PubMed] [Google Scholar]


