Abstract
The RIO kinases existed before the split between Archaea and Eubacteria and are essential in eukaryotes. Although much has been elucidated in the past few years regarding the function of these proteins in eukaryotes, questions remain about their role in prokaryotes. Comparison of structure and sequence suggests that the ancient RIO kinases may have similar functional properties in prokaryotes as they do in eukaryotes. The conservation of charge distribution, functional residues, and overall structure supports a role for these proteins in ribosome interactions, as is their purpose in eukaryotes. However, a lack of study in this area has left little direct evidence in support of this function.
Keywords: ATPases, Bacterial Protein Kinases, Prokaryotic Protein Kinases, Ribosomal RNA Processing, Ribosome Assembly, Serine/Threonine Protein Kinase, RIO Kinase, Archaeal Protein Kinases
Introduction
The RIO kinases belong to an ancient group of proteins with an atypical protein kinase-like fold (1, 2). Of this group, which also contains the homologs of Bud32 and KdoK, RIO kinases are the most studied. The RIO kinase family clearly contains at least four subfamilies, named Rio1, Rio2, Rio3, and RioB (3, 4). Rio1 and Rio2 are found in most archaea and all eukaryotes, Rio3 is found in multicellular eukaryotes, and RioB is found in some archaea and some eubacteria (3, 4). The existence of these proteins in prokaryotes establishes that they are at least 1.5 billion years older that canonical eukaryotic protein kinases (ePKs).2 In fact, in a study on the evolution of the protein kinases, Leonard et al. (5) suggested that the RIO kinases could be the ancestors of the canonical ePKs. Although the role of the RIO kinases in prokaryotes is not known, in eukaryotes, it is clear that RIO kinases are essential and are required for the synthesis of new ribosomes (6–8). In particular, they are required for the last maturation step of the small subunit of the ribosome. This minireview will document what is known thus far about this family in microbes and provide support for the probability that archaeal RIO kinases perform similar biological roles as observed in eukaryotes. However, the role of RIO kinases in the eubacteria in which they exist remains a compelling unanswered question for which there are few clues.
Structures of the RIO Kinases of Archaea
Structural characterization of the RIO kinases was first performed using the homologs from Archaeoglobus fulgidus (Fig. 1) (9, 10). These structures firmly established that the core RIO kinase domain is similar in structure to canonical ePKs. However, the RIO kinase domain from these structures is a trimmed version of the conserved ePK core and, as such, lacks some key regions, including the activation loop (or subdomain VIII) and helices H and I (11). These regions are important in ePKs for interaction with substrate and converting the enzyme from an inactive to an active state. Like the ePK kinase domain for which a structure was first determined for cAMP-dependent protein kinase in 1991, the RIO kinase fold consists of two subdomains, called the N- and C-terminal lobes, connected by a hinge region (9, 12). In the cleft between these domains, the ATP and magnesium bind, and the reaction is catalyzed (10, 13). The invariant catalytic residues present in the C-terminal lobe of ePKs, including asparagine and aspartic acid residues on the catalytic loop (C-loop) and an aspartic acid residue on the metal-binding loop (M-loop), are all conserved in the RIO kinase domains, suggesting that they catalyze phosphoryl transfer as well (Fig. 2). RIO kinases also have a distinct phosphate-binding loop (P-loop) that is subfamily-specific (Fig. 2A) and a flexible loop (F-loop) region that is located in the N-terminal lobe. In addition, RIO kinases contain subfamily-specific domains outside the RIO domain required for function. All Rio2 proteins contain an N-terminal winged helix-turn-helix (wHTH) domain, and eukaryotic Rio1 and Rio2 proteins (but not archaeal) contain conserved, highly basic, C-terminal extensions, whereas Rio3 contains a conserved N-terminal domain of unknown structure that binds to ubiquitin in human cells (3, 14). The functions of these domains have not been clearly delineated, except for the role of the Rio2 wHTH domain in binding to the pre-40 S particle.
FIGURE 1.
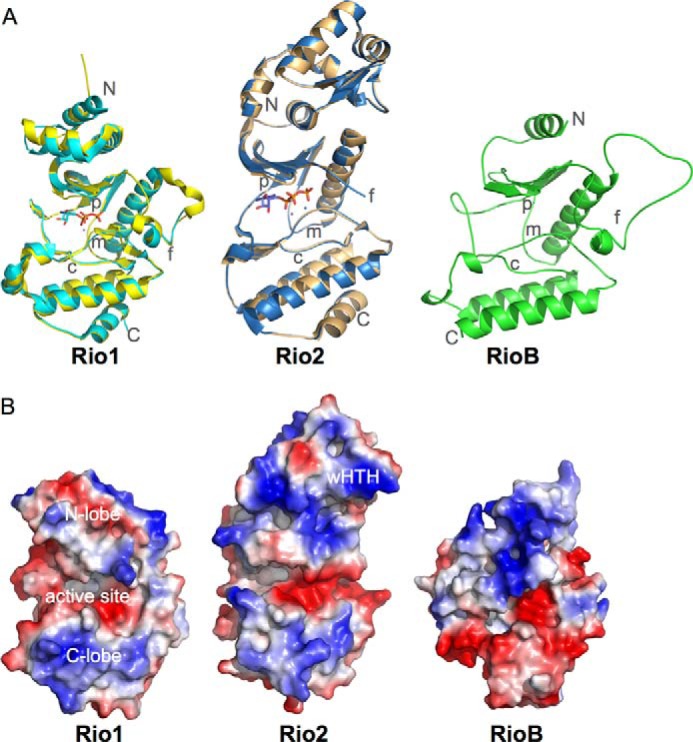
Structure and electrostatic surfaces for prokaryotic RIO kinases. A, the crystal structures of A. fulgidus Rio1 (cyan; Protein Data Bank ID 1ZP9) and Rio2 (blue; ID 1ZAO) bound to ATP superposed on their structures in the absence of nucleotide (yellow and orange, respectively; ID 1ZTF and 1TQI) and a homology model for Tetrasphaera elongata RioB (UniProt N0DXF0). The model was created using MODELLER (23, 24). The N and C termini and the C-, F-, P-, and M-loops are indicated. All structure figures were generated using PyMOL (25). B, electrostatic surfaces for the structures in A in an equivalent orientation. Surfaces were generated using PyMOL (25).
FIGURE 2.
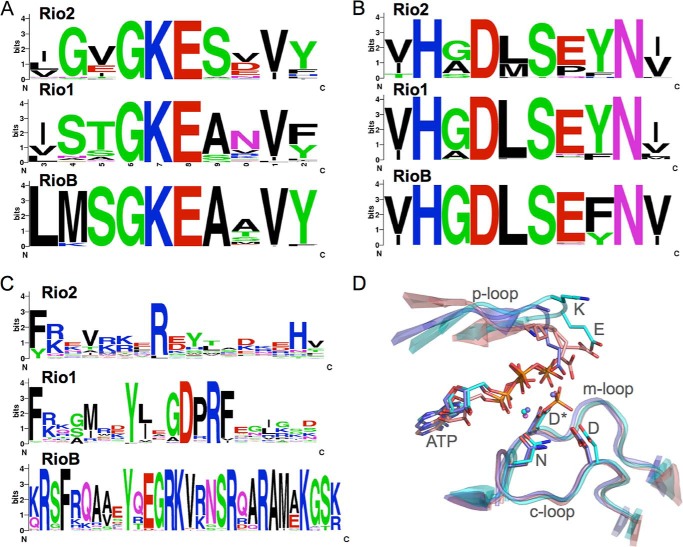
Sequences and structures of the conserved loops of the prokaryotic RIO kinases. Sequence logos (26) for the P-loop (A), C-loop (B), and F-loop (C) of the prokaryotic RIO kinases. WebLogo (27) was used to generate sequence logos that indicate the level of conservation of amino acids based on multiple sequence alignments generated using ClustalW (28). The colors indicate chemical properties: red, negatively charged; blue, positively charged; black, hydrophobic; green, polar or non-hydrophobic; magenta, amides. D, alignment of the C-, M-, and P-loops of A. fulgidus Rio1 (cyan), Rio2 (blue), and C. thermophilum Rio2 (pink; Protein Data Bank ID 4GYI) in their ATP-bound state. Invariant residues are indicated by their single-letter codes. The asterisk indicates the phosphoaspartate intermediate observed in eukaryotic Rio2.
The Search for RIO Kinase Substrates
For several years, there has been an ongoing search for substrates for the RIO kinases. This is, of course, with the goal of elucidating function for these ancient protein kinases. All RIO kinases that have been purified and biochemically characterized have shown autophosphorylation activity (8–10, 15–17). For at least one RIO kinase, Rio1 from A. fulgidus, this has been shown to occur in trans, i.e. one molecule phosphorylates another. Despite years of study in this area, the identification of other validated protein substrates for these enzymes has been elusive. In fact, incubation of Rio2-containing pre-40 S particles with radiolabeled ATP in yeast resulted in no labeling of the pre-40 S subunit, suggesting that there are no substrates for the Rio2 protein on the pre-40 S particle (17). In the haloarcheon Haloferax volcanii, a subunit of the proteasome (α1) is phosphorylated by Rio1 in vitro, and mutation of the phosphorylation sites of α1 resulted in a functional defect in vivo (18). Given the paucity of Ser/Thr/Tyr protein kinases in archaeal genomes, this result could indicate that Rio1 does phosphorylate this site in vivo. However, there have been no additional data to suggest that this is a conserved function of Rio1.
Known Functions of the RIO Kinases
In eukaryotes, it is well established that the functional role of the RIO kinases, especially Rio1 and Rio2, is the maturation of the small subunit of the ribosome. In Saccharomyces cerevisiae (budding yeast), in which the first RIO kinase was discovered, both RIO kinases are essential (6, 7, 15). Deletion of either one is lethal, and depletion of either one results in the accumulation of an immature pre-rRNA species (6, 7). The species that accumulates (20 S rRNA) is the same for depletion of either Rio1 or Rio2, indicating that they are involved in the same maturation step. For both genes, kinase activity is important to the function, and loss of kinase activity results in significant growth defects and cold sensitivity (7, 17). RIO kinase activity was shown to be necessary for 40 S maturation in human cells as well (19).
Although the mechanisms by which RIO kinases function in eukaryotes are poorly defined, recent data have emerged that yield important clues. A recent crystal structure of a eukaryotic Rio2 kinase from Chaetomium thermophilum revealed that ADP and a phosphoaspartate intermediate state were generated after incubation with ATP and magnesium ions (17). This led to the observation of robust, although modest, ATPase activity for this enzyme. The ATPase activity, which had a measured turnover rate of 0.9 min−1, is higher than the autophosphorylation activity by 50–100-fold. These data, along with the observation that the Rio2 kinase likely binds the pre-40 S particle with its active site inaccessible to protein substrate, led to the conclusion that Rio2 functions as an ATPase in the maturation steps of the pre-40 S particle (17).
All eukaryotes contain at least two RIO kinases, Rio1 and Rio2. Most archaeal organisms also contain one of each. A review containing a bioinformatics analysis of archaeal genomes has shown that some actually have duplicate copies of Rio2 and/or a combination of Rio2 and RioB instead of Rio2 and Rio1 (4). At this point, nothing is known about RioB, as there is no structural, genetic, or biochemical data addressing this subfamily. Neighborhood analysis has strongly implicated a role for RIO kinases in interaction with other proteins such as the archaeal initiation factor aIF1A (4). Therefore, what is the role of the RIO kinases in microbes? To fully answer this question, we will need more direct study of the RIO kinases of these organisms and some characterization of the RioB family. However, evidence is presented below to suggest that RIO kinases, at least Rio2, are also involved in ribosome biogenesis in archaea.
Archaeal RIO Kinases as Ribosome-processing Factors
In the first line of evidence, comparisons of the structures of the archaeal RIO kinases with the recently solved structures of eukaryotic RIO kinases have shown remarkable similarity in the RIO domain. In all RIO domains from both Rio1 and Rio2 that have been structurally characterized, the surface surrounding the entrance to the active site is positively charged. A recent structural and functional analysis of a eukaryotic Rio2 protein indicated that this positively charged surface is involved in interaction with rRNA when Rio2 is bound to the pre-ribosome (17). The conservation of this positive charge distribution in archaeal Rio2, which extends to the wHTH domain in Rio2, supports the premise that Rio2 proteins in archaea are also involved in interactions with the rRNA. As Rio1 proteins also display a positively charged surface surrounding the active site, Rio1 may also bind the pre-40 S particle in an orientation similar to that observed for Rio2. Fig. 1B shows the surface electrostatic distribution for A. fulgidus Rio1 and Rio2 and a homology model of RioB for comparison.
In addition, the conserved lysine residue of the P-loop (Rio1 STGKEA, Rio2 GXGKES, Rio3 STGKES, and RioB SGKEA) (Fig. 2, A and D) has no interaction with nucleotide in the active site in the known crystal structures (10, 13, 17). However, mutation of this residue to glutamate in yeast results in loss of binding to the pre-40 S particle and loss of cell viability (17). Therefore, it was concluded that this conserved lysine is important for pre-40 S interaction. If that is indeed the case, the conservation of this residue in all RIO kinases strongly suggests that it plays a similar role, namely rRNA binding, in archaea. The conserved glutamate residue next to the lysine is also a clue. In structures of the RIO kinases, it appears to be ideally located to play a role in hydrolysis of the phosphoaspartate intermediate (Fig. 2D). If that is indeed the case, the presence of this residue in all RIO kinases suggests that ATPase activity is common to all RIO kinases. However, further studies are required to support this possibility.
Other structural evidence has demonstrated that the F-loops of the RIO kinases have an overall net positive charge. The distribution of positive charge in these loops is in agreement with data in support of a major role in interaction with the pre-ribosome (17). Based on structural analysis, the F-loop is predicted to interact near helix 31 of the eukaryotic pre-40 S rRNA, and mutation of positively charges residues of yeast Rio2 to neutral residues results in loss of binding to the pre-40 S particle and loss of Rio2 function (17, 20). The conservation of positively charged residues in this loop in all RIO kinases strongly suggests that they all participate in a similar mode of rRNA binding. Fig. 2C shows the consensus sequences for the F-loops in the microbial RIO kinases. Even RioB contains an extended loop with conserved positively charged residues in this region.
In a less direct line of evidence, RIO kinases, in particular the activities of Rio2 proteins, are required in eukaryotes for the efficient release of several late pre-40 S particle-processing factors that are also present in some archaea. Nob1, Enp1, Ltv1, and Rio2 all accumulate on pre-40 S particles in the absence of Rio2 catalytic activity (17). Both Nob1 and Ltv1 have identifiable homologs in Euryarchaeota.
RIO Kinases in Microbes
Several unanswered questions still remain regarding the role of microbial RIO kinases. As some of the most ancient proteins with a kinase-like fold, one of the major questions is the nature of the reaction catalyzed by these enzymes in prokaryotes. Although it has been shown that RIO kinases of the Rio1 and Rio2 subfamilies are capable of measurable ATPase activity, no activity has been documented for the RioB family at this point. Arguably representative of the oldest RIO kinase, determining the nature of RioB activity will inform us on the question of the original activity of the RIO kinases. In addition, it will help elucidate the sparse distribution of RioB proteins in some Gram-negative bacteria.
More compelling, perhaps, is the idea that RIO kinases are ATPases rather than kinases. If so, were the first RIO kinases really ancient ATPases, and therefore, did kinases originate from ATPases? The reaction itself is not that different and involves the transfer of phosphate from ATP to a protein Ser, Thr, or Tyr for the kinases instead of to water for the ATPases. However, the mechanism that is proposed for phosphoryl transfer catalyzed by the kinase domain (the C-loop Asp-catalyzed nucleophilic attack by the hydroxyl-containing side chain on the γ-phosphate of ATP) is quite distinct from that for the transfer of the γ-phosphate to the M-loop Asp in an intermediate step prior to hydrolysis of the resulting phosphoaspartate proposed for the ATPase reaction of Rio2 (21). This is with all of the catalytic residues remaining invariant between RIO kinases and ePKs (Fig. 2B). It will be an interesting challenge to address that will benefit from knowledge of the function and activity of the microbial RIO kinases.
In addition, do the RIO kinases function as kinases with functional substrates in bacteria and archaea? Certainly, the identification of the α1 subunit of the haloarchaeal proteasome is suggestive of that possibility (18). However, studies of the activity of the RIO kinases have shown that they are very poor kinases compared with known kinases both on widely used substrates such as myelin basic protein and in autophosphorylation. This could mean that the proper substrate has not been identified, but coupled with the lack of the expected substrate-binding surfaces, it seems likely that the role of the RIO proteins is not to phosphorylate other proteins but simply to couple conformational change to ATP hydrolysis. If this is the case, why do RIO kinases all show this ability to autophosphorylate? Rio1 kinases from A. fulgidus and humans also oligomerize, and autophosphorylation reduces the amount of oligomerization (16). Therefore, autophosphorylation may provide a means by which the proteins are regulated, assuming that their activity is different in the oligomeric form. This scenario is plausible, as the proteins likely bind to the pre-ribosome as monomers. However, other possible roles for autophosphorylation of the RIO kinases cannot be ruled out, especially because Rio2 proteins have not been shown to oligomerize. Future work should focus on the identification of binding partners of RIO kinases in prokaryotes, including direct testing of their ability to bind RNA in the cell, and biochemical characterization of activity.
Development of Inhibitors Using Microbial RIO Kinases
Although the role of RIO kinases in microbes is far from clear, they are essential in yeast and are present in many harmful bacterial organisms. Research is ongoing in the pursuit of specific inhibitors of RIO activity, and a few starting molecules for the inhibition of Rio1 have already been identified. These molecules, which include a previously studied cytotoxic molecule (toyocamycin) and a group of caffeic acid benzyl derivatives, were discovered through virtual screening and structure-based drug design using the structures of the archaeal RIO kinases (16, 22). These molecules bind in the ATP-binding pocket of Rio1, and due to the high level of structural conservation between the active sites of archaeal and eukaryotic RIO kinases (see Fig. 2D for a comparison of active site loops), they are capable of interacting with human Rio1 kinase as well (16).
Conclusions
The RIO kinases continue to be an intriguing group of enzymes. They are essential in eukaryotes and have been around since before the divergence of archaea and eubacteria. Despite several years of study, their function and enzymatic activity remain unclear. In addition, although they bear strong resemblance to the well studied group of ePKs, they show enough divergence to perhaps function via a different catalytic mechanism. Furthermore, the demonstration of ATPase activity in the eukaryotic RIO kinases suggests that the origins of protein kinase activity may lie in ancient ATPases. To elucidate the functions of this group of atypical protein kinases and to determine how we might be served by modulation of their activity, the microbial RIO kinases must be studied to provide molecular level detail of their role.
This is the fourth article in the Thematic Minireview Series “Protein Serine/Threonine and Tyrosine Phosphorylation in Prokaryotes.” This work was supported in part by National Science Foundation Career Award Grant MCB0953493.
- ePK
- eukaryotic protein kinase
- C-loop
- catalytic loop
- M-loop
- metal-binding loop
- P-loop
- phosphate-binding loop
- F-loop
- flexible loop
- wHTH
- winged helix-turn-helix.
REFERENCES
- 1. Manning G., Whyte D. B., Martinez R., Hunter T., Sudarsanam S. (2002) The protein kinase complement of the human genome. Science 298, 1912–1934 [DOI] [PubMed] [Google Scholar]
- 2. Kannan N., Taylor S. S., Zhai Y., Venter J. C., Manning G. (2007) Structural and functional diversity of the microbial kinome. PLoS Biol. 5, e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. LaRonde-LeBlanc N., Wlodawer A. (2005) A family portrait of the RIO kinases. J. Biol. Chem. 280, 37297–37300 [DOI] [PubMed] [Google Scholar]
- 4. Esser D., Siebers B. (2013) Atypical protein kinases of the RIO family in archaea. Biochem. Soc. Trans. 41, 399–404 [DOI] [PubMed] [Google Scholar]
- 5. Leonard C. J., Aravind L., Koonin E. V. (1998) Novel families of putative protein kinases in bacteria and archaea: evolution of the “eukaryotic” protein kinase superfamily. Genome Res. 8, 1038–1047 [DOI] [PubMed] [Google Scholar]
- 6. Vanrobays E., Gelugne J. P., Gleizes P. E., Caizergues-Ferrer M. (2003) Late cytoplasmic maturation of the small ribosomal subunit requires RIO proteins in Saccharomyces cerevisiae. Mol. Cell. Biol. 23, 2083–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vanrobays E., Gleizes P. E., Bousquet-Antonelli C., Noaillac-Depeyre J., Caizergues-Ferrer M., Gélugne J. P. (2001) Processing of 20 S pre-rRNA to 18 S ribosomal RNA in yeast requires Rrp10p, an essential non-ribosomal cytoplasmic protein. EMBO J. 20, 4204–4213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Geerlings T. H., Faber A. W., Bister M. D., Vos J. C., Raué H. A. (2003) Rio2p, an evolutionarily conserved, low abundant protein kinase essential for processing of 20 S pre-rRNA in Saccharomyces cerevisiae. J. Biol. Chem. 278, 22537–22545 [DOI] [PubMed] [Google Scholar]
- 9. LaRonde-LeBlanc N., Wlodawer A. (2004) Crystal structure of A. fulgidus Rio2 defines a new family of serine protein kinases. Structure 12, 1585–1594 [DOI] [PubMed] [Google Scholar]
- 10. Laronde-Leblanc N., Guszczynski T., Copeland T., Wlodawer A. (2005) Structure and activity of the atypical serine kinase Rio1. FEBS J. 272, 3698–3713 [DOI] [PubMed] [Google Scholar]
- 11. Hanks S. K., Hunter T. (1995) Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 9, 576–596 [PubMed] [Google Scholar]
- 12. Knighton D. R., Zheng J. H., Ten Eyck L. F., Ashford V. A., Xuong N. H., Taylor S. S., Sowadski J. M. (1991) Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science 253, 407–414 [DOI] [PubMed] [Google Scholar]
- 13. LaRonde-LeBlanc N., Guszczynski T., Copeland T., Wlodawer A. (2005) Autophosphorylation of Archaeoglobus fulgidus Rio2 and crystal structures of its nucleotide-metal ion complexes. FEBS J. 272, 2800–2810 [DOI] [PubMed] [Google Scholar]
- 14. Fenner B. J., Scannell M., Prehn J. H. (2009) Identification of polyubiquitin binding proteins involved in NF-κB signaling using protein arrays. Biochim. Biophys. Acta 1794, 1010–1016 [DOI] [PubMed] [Google Scholar]
- 15. Angermayr M., Bandlow W. (2002) RIO1, an extraordinary novel protein kinase. FEBS Lett. 524, 31–36 [DOI] [PubMed] [Google Scholar]
- 16. Kiburu I. N., LaRonde-LeBlanc N. (2012) Interaction of Rio1 kinase with toyocamycin reveals a conformational switch that controls oligomeric state and catalytic activity. PLoS ONE 7, e37371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ferreira-Cerca S., Sagar V., Schäfer T., Diop M., Wesseling A. M., Lu H., Chai E., Hurt E., LaRonde-LeBlanc N. (2012) ATPase-dependent role of the atypical kinase Rio2 on the evolving pre-40 S ribosomal subunit. Nat. Struct. Mol. Biol. 19, 1316–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Humbard M. A., Reuter C. J., Zuobi-Hasona K., Zhou G., Maupin-Furlow J. A. (2010) Phosphorylation and methylation of proteasomal proteins of the haloarcheon Haloferax volcanii. Archaea 2010, 481725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Widmann B., Wandrey F., Badertscher L., Wyler E., Pfannstiel J., Zemp I., Kutay U. (2012) The kinase activity of human Rio1 is required for final steps of cytoplasmic maturation of 40 S subunits. Mol. Biol. Cell 23, 22–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Strunk B. S., Loucks C. R., Su M., Vashisth H., Cheng S., Schilling J., Brooks C. L., 3rd, Karbstein K., Skiniotis G. (2011) Ribosome assembly factors prevent premature translation initiation by 40 S assembly intermediates. Science 333, 1449–1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schwartz P. A., Murray B. W. (2011) Protein kinase biochemistry and drug discovery. Bioorg. Chem. 39, 192–210 [DOI] [PubMed] [Google Scholar]
- 22. Mielecki M., Krawiec K., Kiburu I., Grzelak K., Zagórski W., Kierdaszuk B., Kowa K., Fokt I., Szymanski S., Swierk P., Szeja W., Priebe W., Lesyng B., LaRonde-LeBlanc N. (2013) Development of novel molecular probes of the Rio1 atypical protein kinase. Biochim. Biophys. Acta 1834, 1292–1301 [DOI] [PubMed] [Google Scholar]
- 23. Eswar N., Eramian D., Webb B., Shen M. Y., Sali A. (2008) Protein structure modeling with MODELLER. Methods Mol. Biol. 426, 145–159 [DOI] [PubMed] [Google Scholar]
- 24. Eswar N., Webb B., Marti-Renom M. A., Madhusudhan M. S., Eramian D., Shen M., Pieper U., Sali A. (2007) Comparative protein structure modeling using MODELLER. Curr. Protoc. Protein Sci. Chapter 2, Unit 2.9, 10.1002/0471140864.ps0209s50 [DOI] [PubMed] [Google Scholar]
- 25. DeLano W. L. (2010) The PyMOL Molecular Graphics System, Version 1.3r1, Schrödinger, LLC, New York [Google Scholar]
- 26. Schneider T. D., Stephens R. M. (1990) Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18, 6097–6100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Crooks G. E., Hon G., Chandonia J. M., Brenner S. E. (2004) WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]