Background: The mechanism of ligand gating in physiologically important cyclic nucleotide-modulated channels is unknown.
Results: We constructed and purified a chimeric ion channel with activity modulated by cAMP and used it to measure ligand-binding energetics.
Conclusion: cAMP binds with high lipid-dependent affinity to the chimeric channel.
Significance: The availability of a good protein preparation enables assays that shed light on ligand gating.
Keywords: Electron Microscopy (EM), Energetics, Fluorescence, Gating, Ion Channels, Isothermal Titration Calorimetry, Nanodisc, Single-channel Recording
Abstract
Cyclic nucleotide-modulated ion channels play crucial roles in signal transduction in eukaryotes. The molecular mechanism by which ligand binding leads to channel opening remains poorly understood, due in part to the lack of a robust method for preparing sufficient amounts of purified, stable protein required for structural and biochemical characterization. To overcome this limitation, we designed a stable, highly expressed chimeric ion channel consisting of the transmembrane domains of the well characterized potassium channel KcsA and the cyclic nucleotide-binding domains of the prokaryotic cyclic nucleotide-modulated channel MloK1. This chimera demonstrates KcsA-like pH-sensitive activity which is modulated by cAMP, reminiscent of the dual modulation in hyperpolarization-activated and cyclic nucleotide-gated channels that display voltage-dependent activity that is also modulated by cAMP. Using this chimeric construct, we were able to measure for the first time the binding thermodynamics of cAMP to an intact cyclic nucleotide-modulated ion channel using isothermal titration calorimetry. The energetics of ligand binding to channels reconstituted in lipid bilayers are substantially different from those observed in detergent micelles, suggesting that the conformation of the chimera's transmembrane domain is sensitive to its (lipid or lipid-mimetic) environment and that ligand binding induces conformational changes in the transmembrane domain. Nevertheless, because cAMP on its own does not activate these chimeric channels, cAMP binding likely has a smaller energetic contribution to gating than proton binding suggesting that there is only a small difference in cAMP binding energy between the open and closed states of the channel.
Introduction
Cyclic nucleotide-gated (CNG)6 channels and hyperpolarization-activated and cyclic nucleotide-gated (HCN) channels are important in visual and olfactory signaling and pacemaker activity, respectively (1–3). Both belong to the S6 superfamily of voltage-gated ion channels (4, 5). In addition to the four-helix voltage sensor domain and two-helix pore domain of each monomeric subunit, these channels contain a cyclic nucleotide-binding domain (CNBD) at the C terminus, which is connected to the pore-forming S6 helix via a so-called C-linker. The CNBD and C-linkers are conserved within this subfamily of channels (6, 7). CNG channels are activated by cyclic nucleotides binding to the CNBD and do not respond to membrane voltage changes, despite having a 4-helix voltage sensor domain (1). In contrast, HCN channels are activated by hyperpolarization (unlike canonical voltage-gated ion channels, which are activated by depolarization (8, 9)), and their activity is only modulated by binding of cyclic nucleotides to the CNBD (2).
Binding of cAMP to the CNBD in both CNG and HCN channels is believed to induce a conformational change that promotes channel opening. Current understanding largely derives from electrophysiological experiments on cells heterologously expressing these channels (1, 10). Recent technical advances in confocal patch clamp fluorometry permit concomitant ligand-binding measurements and current recordings in the same membrane patch, allowing for detailed analysis of binding cooperativity and activation states triggered by partial ligand binding (11–14). For example, Kusch et al. (12) provide evidence that HCN2 channels possess alternating cooperative and noncooperative ligand binding behaviors (favoring two or four bound ligands rather than one or three) and are fully activated upon binding of two ligands. Yet the molecular mechanism by which ligand binding to the CNBD changes the closed-open equilibrium (opens the pore) remains unknown.
To acquire further insight into the mechanism(s) of HCN (and CNG) channel gating, we sought to probe the thermodynamics of ligand binding to the purified channel, isolated from other cellular components. This has proven difficult because heterologous expression of eukaryotic CNG and HCN channels so far has yielded insufficient functional protein for biochemical and biophysical studies. Only the cytoplasmic C termini (CNBDs with C-linker) from a few HCN channels have proven amenable to large scale purification and crystallization (15–19).
Many prokaryotic potassium channels such as KvAP, KcsA, and MthK have proven to be more manageable targets than their eukaryotic counterparts (20–23). MloK1, an HCN channel homolog from Mesorhizobium loti, remains the most promising candidate for biophysical characterization, displaying cyclic nucleotide-dependent ion channel activity when reconstituted into liposomes (24, 25). MloK1 contains the signature GYG selectivity filter sequence found in HCN channels, but the “voltage sensor domain” lacks the arginine repeats that allow voltage sensing (26). Crystal structures of the transmembrane and CNBD domains of MloK1 have been determined separately (24, 27–29). The transmembrane domain has a structure similar to that of other voltage-gated potassium channels, but the absence of the CNBDs precludes insight into ligand modulation of MloK1. The structures of the isolated MloK1 CNBDs, determined with and without ligand, show that cAMP binding leads to a clamping down of the C-terminal helix over the ligand-binding pocket and repositioning of the N-terminal helix, which in the full-length channel continues with the inner helix of the transmembrane domain (28). In addition, a 16-Å electron microscopy-derived structure of full-length MloK1 in the presence of ligand reveals a symmetrical arrangement of the CNBDs around the transmembrane domain (30). Currently, however, a high resolution structure of full-length MloK1 remains elusive, the protein detergent-solubilized is unstable, and binding and activity assays are challenging because of considerable difficulty in removing the ligand (cAMP) as well as protein aggregation at high concentrations. Finally, there have been no reports of successful electrical current measurements from this channel (24, 25).
To overcome these difficulties and to obtain the desired information about cyclic nucleotide-channel interactions, we constructed a chimeric channel by attaching the CNBDs of MloK1 to the transmembrane domain of KcsA (Fig. 1A). The design of a conceptually similar chimeric channel (see under “Discussion”), with some functional differences, was reported previously by Ohndorf and MacKinnon (31). Overexpression and purification of the chimera in this report allow for studies that have proven intractable with the full-length MloK1 protein. We show that the chimeric channel is active and that its activity is modulated by pH in a similar manner to KcsA. In addition, the chimera is modulated by cAMP, albeit only in the presence of protons, suggesting the chimera's response to cyclic nucleotides is similar to that of HCN channels. Using this chimera, we were able for the first time to measure the thermodynamics of cAMP binding to a full-length cyclic nucleotide-modulated channel using ITC. Furthermore, we show that the incorporation of the channel in nanodiscs (also termed nanoscale apolipoprotein-bound bilayers, or NABBs (32, 33)) leads to a 10-fold increase in the apparent cAMP affinity, as compared with the value obtained in detergent micelles. This affinity increase indicates that cAMP binding can induce a conformational change in the transmembrane domains, which are in direct contact with the lipid bilayer and remote from the CNBDs. At the same time, this conformational change is not sufficient by itself to open the channel, suggesting that the energetic contribution to channel opening from cAMP binding is much less than that from proton binding.
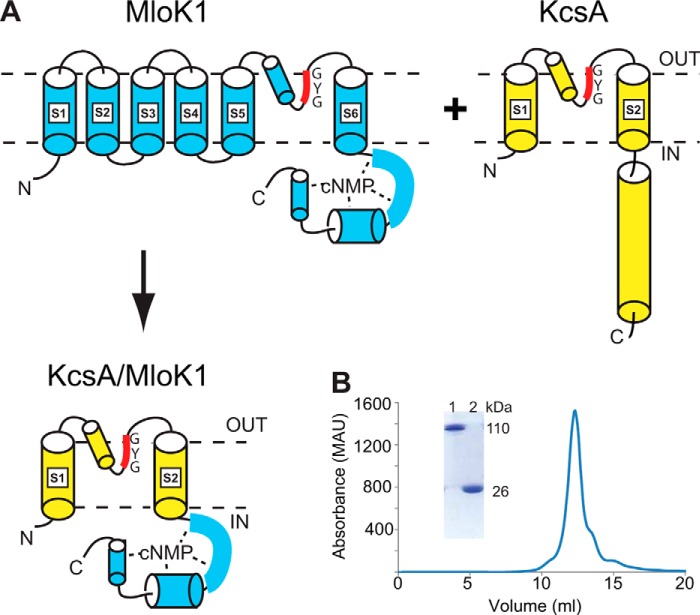
FIGURE 1.
KcsA/MloK1 chimera's design, expression, and purification. A, diagram detailing the expected arrangement of the chimera based on the structures of KcsA (yellow) and MloK1 (blue). The CNBD of MloK1 is directly attached to the bottom of the S6 helix of KcsA. Dashed lines represent the membrane. B, size exclusion chromatography profile of the chimera shows a single peak at a size corresponding to a tetrameric channel in a detergent micelle. Inset shows a portion of an SDS-acrylamide protein gel of unheated tetrameric chimera (lane 1) and heated monomeric chimera (lane 2).
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification
DNA for the chimeric gene was generated through ligation of KcsA and MloK1 segments (Fig. 1A). The region of overlap between the two segments consists of an RRG sequence contained in residues 121–123 in KcsA and residues 219–221 in MloK1. The chimeric gene was cloned into the pQE60 (Qiagen) plasmid and contains an N-terminal hexahistidine tag.
Protein expression was induced with isopropyl 1-thio-β-d-galactopyranoside at an optical density of 0.8 in transformed BL21 (DE3) cells and grown overnight in LB at 20 °C. Pelleted cells were resuspended in a solution of 50 mm Tris, pH 7.5, 100 mm KCl, 200 μm cAMP and sonicated in the presence of leupeptin (2 mg/ml), pepstatin (3 mg/ml), and PMSF (0.17 mg/ml) (Roche Applied Science). The resulting slurry was gently shaken for 2 h with 30 mm n-decyl β-d-maltopyranoside (DM). The lysate was spun down at 32,000 × g and the supernatant applied over a Ni2+ HiTrap chelating column (GE Healthcare). Protein was eluted with 300 mm imidazole in 20 mm Tris, 100 mm KCl, 5 mm DM, 200 μm cAMP, pH 7.6, and further purified over a Superdex 200 10/300 GL gel filtration column (GE Healthcare) using a solution of 20 mm Tris, 100 mm KCl, 5 mm DM, 200 μm cAMP, pH 7.6. The protein concentration was calculated using a factor of 0.476 mg/ml chimera for an absorbance unit of 1 at a wavelength of 280 nm as determined with the DC protein assay (Bio-Rad).
cAMP Removal
Tightly bound cAMP was competed off of the chimeric protein with a low affinity analog, as described previously for MloK1 (34). Protein was reloaded onto a Ni2+ HiTrap chelating column (GE Healthcare) and rinsed with a solution of 20 mm Tris, 100 mm KCl, 5 mm DM, pH 7.6 (Rinse Buffer), to remove free cAMP. The protein was then incubated with Rinse Buffer containing 1 mm 8-(4-chlorophenylthio)-guanosine 3′,5′-cyclic monophosphate (8-pCPT-cGMP) (Sigma) for 30 min, followed by another rinse step, an additional 30-min incubation with 1 mm 8-pCPT-cGMP, another rinse step, an overnight incubation in 10 mm 8-pCPT-cGMP, and a final extensive rinse step using 40 column volumes of rinse buffer. Protein was eluted with 300 mm imidazole and then dialyzed overnight in Rinse Buffer.
Bilayer Recordings
Chimeric protein for use in bilayer recordings included the construct described above (Fig. 1A) and a variant with an E71A mutation, which removes inactivation at the KcsA selectivity filter (35). Following gel filtration, protein was reconstituted into liposomes as reported previously (36, 37) by mixing a ratio of 5–10 μg of protein per mg of lipid with a 3:1 molar ratio of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine to 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol solubilized in reconstitution buffer (400 mm KCl, 10 mm HEPES, 5 mm N-methyl-d-glucamine, 200 μm cAMP) with 34 mm CHAPS, which was subsequently applied over a 20-ml G-50 Sephadex column to remove the detergent. Liposome aliquots in reconstitution buffer were frozen in liquid nitrogen and stored for future use. Thawed liposomes were briefly sonicated prior to use. Bilayers were assembled using a horizontal planar system with two chambers separated by a partition containing a 100-μm diameter hole (38). The bilayer was formed manually by applying 10 mg/ml 3:1 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine/1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol in decane over the hole in the partition, and the bilayer formation was monitored electrically using a cycling voltage ramp (39). The cis chamber was filled with 70 mm KCl, 30 mm KOH, 10 mm MOPS, 10 mm succinic acid, and 10 mm Tris, pH 7. The trans chamber was filled with an identical solution where pH varied between 4 and 5.5. Solutions of varying pH and cAMP concentrations were exchanged through the trans chamber. Currents were recorded in Clampex 10 with an Axopatch 200A, digitized with a Digidata 1322A, and analyzed in Clampfit (Molecular Devices).
Stopped Flow Ion Flux Assay
The thallium flux assay and data analysis were conducted as described previously (40, 41). The KcsA/MloK1 chimera used in the flux assay was dialyzed further to reduce the imidazole concentration to submillimolar levels and reconstituted into large unilamellar vesicles (LUVs) at a weight ratio of 17 μg of protein to 1 mg of lipid in a 3:1 molar ratio of DOPC to 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol (Avanti Polar Lipids) in 100 mm KNO3, 10 mm HEPES, 10 mm succinic acid, 25 mm aminonaphthalene-1,3,6-trisulfonate (ANTS) (Sigma), pH 7.0, with 14 mm CHAPS. The suspension was sonicated until completely clear, and the detergent was removed through a 2-h incubation with Bio-Beads (Bio-Rad) (1 g of Bio-Beads/35 mg of CHAPS). The protein/lipid mixture was sonicated briefly, siphoned from the Bio-Beads, and extruded 42 times through a 0.1-μm polycarbonate filter. The external ANTS was removed over a PD-10 desalting column (GE Healthcare), and the LUVs were eluted in a solution of 140 mm KNO3, 10 mm HEPES, and 10 mm succinic acid, pH 7.0. LUVs with different cAMP concentrations were prepared in parallel, and the appropriate cAMP concentrations were maintained in all solutions used during the reconstitution and functional characterization.
ANTS fluorescence quenching in response to thallium influx into the LUVs (through the chimeric channel) was measured using an Applied Photophysics SX.20 stopped-flow spectrophotometer with sequential mixing capability. The excitation wavelength was 352 nm, and the emitted light was recorded using a cutoff filter at 455 nm. Channel activity was determined using a sequential-push mixing sequence, where the LUVs were first rapidly mixed with a solution with a pH and cAMP concentration such that the extravesicular pH and [cAMP] in the mixing chamber had the desired value (40). After a 100-ms equilibration, the LUVs were mixed with a solution containing 140 mm KNO3, 10 mm HEPES, 10 mm succinic acid, and the appropriate cAMP concentration to determine the initial fluorescence level. Then the LUVs were mixed with the quench solution containing 50 mm TlNO3, 94 mm KNO3, 10 mm HEPES, and 10 mm succinic acid at the desired cAMP concentration and pH. The lipid bilayer has a very low Tl+ permeability, and Tl+ influx through the open channels caused quenching of the ANTS fluorescence. The fluorescence of the quenched samples was normalized to the initial fluorescence level and fit with a stretched exponential to the first 2–100 ms of the normalized fluorescence quench curve for each sample (41) as shown in Equation 1,
where F(t) is the fluorescence at time t; τ0 is a coefficient with units of time, and β (0 < β ≤1) is a measure of the dispersity of LUV size. The rate of fluorescence quenching at 2 ms was then calculated using Equation 2,
and changes in channel function were evaluated as the relative changes in k(2 ms).
Nanodisc Formation
Chimeric channels were incorporated into nanodiscs using a method identical to that previously described for KcsA (33). For the apolipoprotein, a variant of the pET28 ZAP1 construct, ZAP1NT, in which the N-terminal 50 residues were truncated and a Q22C (Gln-54 in the original ZAP1) mutation added, was used for these experiments as the truncation results in improved yields. The cysteine was introduced for labeling purposes (described below). ZAP1NT was expressed in BL21 (DE3) cells upon induction with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside at an optical density of 0.8 for 3 h at 37 °C in terrific broth media. Centrifuged cell pellets were resuspended in 50 mm Tris, pH 7.5, 1 mm DTT, and 300 mm KCl and then sonicated in the presence of protease inhibitors. 6 m guanidinium chloride was added to the sonicated slurry followed by agitation at room temperature (or 4 °C) for 30 min. The lysate was spun down at 32,000 × g, pH adjusted to 7.6 with KOH, and then combined with 4 ml of nickel-nitrilotriacetic acid resin (Invitrogen) and equilibrated with 20 mm HEPES, pH 6.9, 150 mm KCl, and 6 m guanidinium chloride. The resin was washed twice with an equilibration buffer containing 30 mm imidazole and then one more time in equilibration buffer without the imidazole. 0.5 mm tetramethylrhodamine-5-maleimide was added to the ZAP1NT nickel-nitrilotriacetic acid slurry and allowed to incubate at 4 °C overnight. The ratio of absorbance at 280 nm from the ZAP1NT to the absorbance at 550 nm due to the tag provides a means of separating the 280-nm absorbance signal belonging to the chimeric channel from that of ZAP1NT in the final channel-containing nanodisc, as described previously (32, 33). Following the overnight incubation, the reaction was stopped with 1 mm DTT, and the resin was rinsed with 10 column volumes of the equilibration buffer. The protein was then refolded using 5 column volumes of 3 m guanidinium chloride-containing buffer followed by 10 column volumes of guanidinium chloride-free buffer. The labeled ZAP1NT then was eluted with guanidinium chloride-free buffer containing 300 mm imidazole. Protein was further purified over a 16/600 Superdex 200 column (GE Healthcare), frozen in liquid nitrogen, and stored at −80 °C until needed.
Nanodiscs were formed by mixing the chimeric protein and labeled ZAP1NT with a solution of 100 mg/ml 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 100 mg/ml CHAPS, 20 mm HEPES, and 100 mm KCl, pH 7.5, in molar ratios of 2:1:45 and 1:1:15 (channel/ZAP1NT/1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine). The latter molar ratio yielded fewer liposomes, but both preparations gave the same ITC results. After incubation for 30 min at room temperature, the mixture was centrifuged for 1 min at 2000 × g, run over a detergent removal column (Pierce) equilibrated with 20 mm Tris, 100 mm KCl, pH 7.6, dialyzed in 10 mm HEPES, 100 mm KCl, pH 7.5, and further purified on a Superdex 200 10/300 GL (GE Healthcare) gel filtration column. The nanodiscs were further analyzed by application to a Superose 6 (GE Healthcare) column to confirm the absence of aggregates and liposomes. The concentration of chimeric channel protein was determined by multiplying the 550-nm absorbance of the chimera-containing nanodisc by the 280:550-nm absorbance ratio of the labeled ZAP1NT protein and subtracting this value from the 280-nm absorbance of the chimera-containing nanodisc.
Isothermal Titration Calorimetry
Data were collected using a MicroCal AutoITC200 (GE Healthcare). cAMP was removed from the chimera as described above, and either dialyzed in 10 mm HEPES, 100 mm KCl, 5 mm DM, pH 7.5, or incorporated into nanodiscs, as described above, and dialyzed in 10 mm HEPES, 100 mm KCl, pH 7.5. The cAMP ligand used in the ITC experiment was dissolved in dialysis buffer. Data were collected at 25 °C. The protein was loaded into the calorimeter at a concentration of 80–157 μm and titrated with 1–2-μl injections of 600–1200 μm cAMP in dialysis buffer until saturation occurred. Control experiments in which cAMP was injected into protein-free dialysis buffer or into dialysis buffer containing an equivalent concentration of empty nanodiscs were performed to ensure the recorded heats were channel-dependent. The results were analyzed using the Origin Software (MicroCal) from which the binding stoichiometry (n), enthalpy change (ΔH), and equilibrium dissociation constant (KD) were determined by fitting a one-site model to the results (unless stated otherwise). The reported results are the average and standard deviation of six separate ITC runs of the nanodisc preparation from six independent purifications and five separate runs of the detergent preparation from three independent purifications (Table 1).
TABLE 1.
Apparent affinities for cAMP binding and/or activation for CNBD-containing ion channels
Values are reported as averages with the corresponding standard deviation.
| Protein | n | KD | KA | ΔH | K½ | No. of experiments |
|---|---|---|---|---|---|---|
| μm | μm−1 | kcal/mol | μm | |||
| KcsA/MloK1 in detergent | 0.55 ± 0.01 | 2.14 ± 0.18 | 0.47 ± 0.04 | −11.27 ± 0.11 | 5 | |
| KcsA/MloK1 in nanodiscs | 0.73 ± 0.05 | 0.15 ± 0.02 | 6.89 ± 1.20 | −13.11 ± 0.58 | 6 | |
| MloK1a | 0.08 ± 0.02 | |||||
| KcsA/MloK1 in liposomes | 1.19 ± 0.49 | 4 | ||||
| MloK1 in liposomesb | 0.06 ± 0.02 | |||||
| CNBDa | 0.98 ± 0.12 | 0.10 ± 0.01 |
RESULTS
Construction of the KcsA/MloK1 Chimera
The KcsA/MloK1 chimera was designed to have a sequence overlap between the C-terminal end of transmembrane helix 2 of KcsA and helix A′ of the MloK1 CNBD consisting of the RRG sequence that is common to both KcsA and MloK1 (see under “Experimental Procedures”). Thus, in the final protein construct, the CNBDs of MloK1 replace the C-terminal cytoplasmic coiled-coil of KcsA, retaining residues previously found to be involved in H+ sensing (Fig. 1A) (42, 43). In the presence of cAMP, the purified protein elutes as a single peak on a gel filtration column at a retention volume of 12.1 ml (Fig. 1B), roughly corresponding to the molecular weight of the tetramer. Similar to KcsA, the chimeric channel remains a tetramer when assayed by SDS-PAGE in the absence of heating, suggesting that the CNBDs do not disrupt the robust KcsA tetramer assembly. After incubation at 95 °C for 5 min, the tetrameric assembly disassociates into monomers when assayed by SDS-PAGE (Fig. 1B).
KcsA/MloK1 Chimera Behaves Like KcsA in Single-channel Recordings
We first tested whether the KcsA/MloK1 chimera is a functional ion channel in lipid bilayers. One aspect of the rationale for designing the chimera to include the transmembrane scaffold of KcsA was the potential to assay protein function at the single-molecule level with electrophysiological recordings.
With this construct, we could indeed record KcsA-like K+ currents from chimera-reconstituted proteoliposomes incorporated in horizontal lipid bilayers (Fig. 2A). Channel openings were observed at an intracellular solution of pH 4 but not pH 7, consistent with the known proton-dependent activity of KcsA. Moreover, the channels display a single-channel current-voltage (I-V) curve similar to that for KcsA (Fig. 2C). The KcsA/MloK1 open probability is extremely low, however, suggesting that the chimera, like KcsA, is mostly inactivated at these steady-state recording conditions. This was confirmed upon mutation of a glutamate behind the selectivity filter to alanine (E71A), which removes inactivation in KcsA (35). This mutation also removes inactivation in the KcsA/MloK1 chimera (Fig. 2B). The apparent pKa of the noninactivating KcsA/MloK1 chimera is ∼5, similar to the noninactivating E71A KcsA (43). These results indicate that the KcsA/MloK1 chimeric channel has preserved the ion conduction, H+ activation, and inactivation properties of KcsA, despite the absence of the C-terminal coiled-coil domain of KcsA. This suggests that this domain has little effect on the H+-dependent activation and inactivation gates or on K+ ion conduction through the open channel.
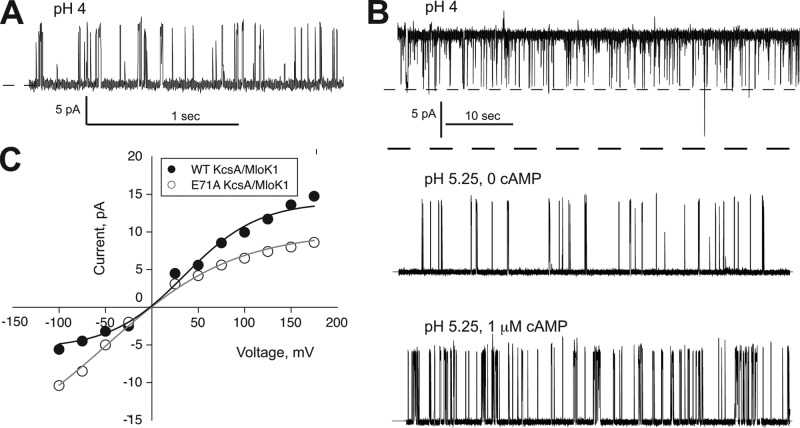
FIGURE 2.
KcsA/MloK1 possesses proton-activated KcsA-like single-channel current activity and is modulated by cAMP at the single-molecule level. A, single-channel recordings from KcsA/MloK1 WT; B, KcsA/MloK1 E71A (which is noninactivating). Single-channel recordings from a representative lipid bilayer containing KcsA/MloK1 E71A perfused from pH 4 (top trace, two channels open at the same time) to pH 5.25 (middle trace) and finally to pH 5.25 and 1 μm cAMP (bottom trace). Traces were obtained in 100 mm symmetrical K+ at 100 mV and filtered at 200 Hz for display. The dashed line corresponds to the closed state. At pH 4 (top trace), two channels are open concomitantly with a Po of ∼1. C, single-channel current-voltage plots from the experiments shown partially in A and B for KcsA/MloK1 WT (black circles) and KcsA/MloK1 E71A (open circles) channels.
Multiple perfusions destabilize the bilayer, complicating the study of the cAMP effect on the KcsA/MloK1 chimera at the single-channel level. An example of the effect of cAMP on the noninactivating KcsA/MloK1 channel using this method is shown in Fig. 2B. Addition of saturating concentrations of cAMP to the intracellular side of the noninactivating chimeric channels produces a 2–3-fold increase in channel open probability in ∼75% of the experiments, but the effect is not reversible. To remove the variability intrinsic to single-molecule measurements and to ascertain whether this chimera was indeed modulated by cAMP, we employed an ensemble-based channel activity assay.
KcsA/MloK1 Chimera Is a K+ Channel Modulated by pH and cAMP
To explore whether cAMP binding leads to channel activation, we used a stopped flow spectrofluorometric assay (40). Using this assay, we can measure the rate of fluorescence quenching of a water-soluble fluorophore (ANTS) trapped inside KcsA/MloK1-containing proteoliposomes by external Tl+ that enters the LUVs through open channels. Greater channel activity (higher time-averaged number of conducting channels in the LUV membrane) results in larger Tl+ flux through the open channels and faster rate of fluorescence quenching.
Using this assay, we found that the KcsA/MloK1 chimera is activated by H+ over a similarly acidic pH range as wild-type KcsA despite the truncation of the entire KcsA C-terminal cytoplasmic domain and its replacement with the MloK1 CNBD, similar to our results at the single-channel level (Figs. 2B and 3, A and C). Protein-free control LUVs display zero flux. The chimeric channel is closed above pH 5.5 (Fig. 3, A and C), again demonstrating that the cytoplasmic C terminus of KcsA is not required for pH sensing, which is in agreement with the conclusions in Refs. 43, 44 but not with those in Ref 45.
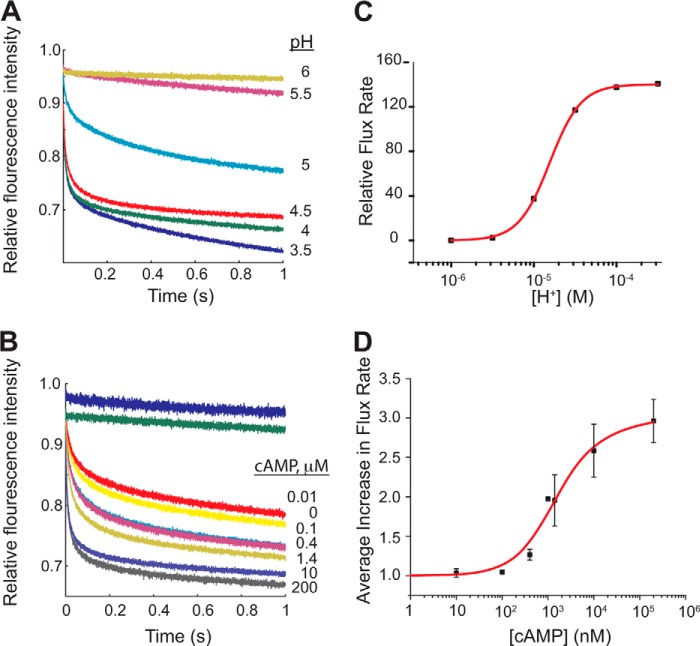
FIGURE 3.
Activity of KcsA/MloK1 is modulated by pH and cAMP. Representative experiments showing the normalized time course of ANTS fluorescence quenching due to Tl+ influx through KcsA/MloK1 channels reconstituted into LUVs at A, 200 μm cAMP and different extraliposomal pH values as follows: 6.0 (yellow), 5.5 (maroon), 5.0 (cyan), 4.5 (red), 4.0 (green), and 3.5 (blue); B, pH 4.0 and different cAMP concentrations as follows: 0 (yellow), 10 nm (red), 100 nm (cyan), 400 nm (pink), 1.4 μm (gold), 10 μm (gray), 200 μm (purple). The fluorescence time course in protein-free LUVs at pH 4.0 and channel-containing LUVs at 200 μm cAMP at pH 7.0 are shown in blue and green, respectively. C, changes in quench rate (Tl+ influx through the channels) in 200 μm cAMP as a function of pH. The red curve is a fit to a single-site curve with the following parameters: n = 2.3 ± 0.05 and pK½ = 4.7 ± 0.01 (mean ± error of the fit). D, average increase in flux through the channel as a function of cAMP (at pH 4). The relative flux increased ∼3-fold between 0 cAMP and 200 μm cAMP. Symbols are mean ± S.E. from four different experiments with protein from four different preparations (except for the experiment at 1 μm, which was done only once, and at 400 nm and 1.4 μm, which were done three times). Normalization of each data set was accomplished by dividing the relative flux rate at 0 cAMP from the flux at each represented cAMP concentration. The red curve is a fit to a modified Hill equation: v/R0 = ((Vmax/R0)(L)/(K½n + Ln)) + 1 from which the following parameters were derived: Vmax/R0 = 1.50 ± 0.69, n = 0.98 ± 0.04, and K½ = 1.19 ± 0.49 μm, where v is the flux rate; Vmax is the maximal flux rate; R0 is the rate in the absence of cAMP; L is the cAMP concentration; K½ is the cAMP concentration at half the maximal flux rate, and n is the Hill coefficient.
To examine how cAMP modulates KcsA/MloK1 function, we performed parallel experiments in which the chimera was reconstituted into LUVs in the presence of varying cAMP concentrations on both sides of the membrane, which were maintained in all subsequent manipulations. At all KcsA-activating pH values, cAMP (at saturating concentrations of 200 μm) increases the quench rate ∼3-fold, suggesting that cAMP increases the KcsA/Mlok1 channel activity (Fig. 3, B and D). This cAMP-dependent increase in channel activity is similar to the increase in Po we observe at the single-channel level (Fig. 2B), which was previously observed in flux assays with wild-type MloK1 (24, 25).
At pH 6 and 7, where no quenching is observed, the addition of cAMP does not increase the quench rate (data not shown). This indicates that the energetic contribution from cAMP binding is smaller than what is necessary to open the channel, i.e. cAMP only increases the open probability of H+-activated channels, rather than directly activating them. This is similar to the ligand modulation observed in eukaryotic HCN channels, where cAMP modulates channels that have been activated by voltage, but differs from CNG channels, which are directly activated by cAMP (7, 8, 46, 47). Plotting the quench rates as a function of cAMP concentration at pH 4 shows that cAMP enhances the KcsA/MloK1 activity with a K½ of ∼1.2 μm (Fig. 3D), which is an order of magnitude higher (meaning less affinity) than the K½ previously reported for MloK1 (25). The Hill coefficient of ∼1 is consistent with what was previously found for MloK1 (34, 37).
CNBD of the KcsA/MloK1 Chimera in Detergent Binds cAMP
We have so far established several lines of evidence that show that the KcsA/MloK1 chimera is a good biochemical model for investigating ligand gating in cyclic nucleotide-modulated, HCN-type ion channels. First, the purified protein is stable even at high concentrations in detergent micelles; second, the channel activity can be measured both at the single-channel level in lipid bilayers and with the ensemble-based fluorescence assay; third, the channel is activated by protons (like KcsA); and finally, the channel activity is increased by cAMP (like MloK1).
Given this, we investigated the energetics of cAMP binding to the KcsA/MloK1 chimera using ITC. Thermodynamic cAMP binding assays on full-length cyclic nucleotide-modulated channels using ITC have not been reported before (cyclic nucleotide binding assays with ITC were performed only on isolated CNBDs (with and without C-linkers); binding assays using a fluorescent cyclic nucleotide analog were performed on purified MloK1). Prior to performing these binding assays, it was necessary to completely remove cAMP from all binding sites, a considerably difficult task. As reported previously for the MloK1 CNBD (24, 34), dialysis is not sufficient to remove cAMP from these sites. ITC binding assays using dialyzed KcsA/MloK1 chimera in detergent yield a very low (n <0.1) apparent binding stoichiometry (molecules of cAMP per monomer). Because we expect each CNBD to bind one molecule of cAMP, the stoichiometry should be close to unity. The low stoichiometry suggests that either the CNBD was misfolded or that cAMP (or another ligand) remained bound to it. Indeed, it appears that this is not an issue restricted to prokaryotic channels as ITC studies of the HCN1 CNBD/C-linker demonstrate that the isolated domain also exhibits low stoichiometry for cAMP binding, leading the authors to propose that a subset of the domains retains their bound ligand (48). To address this problem, we repeatedly incubated the protein with a low affinity cyclic nucleotide analog (8-CPT-cGMP) to remove cAMP, followed by dialysis in ligand-free buffer to fully remove the analog, as described previously for MloK1 (34). This leads to an increased stoichiometry (n = 0.55), suggesting that the initial low n was due to slow dissociation of cAMP from the CNBD (Fig. 4A and Table 1). The measured KD was ∼2 μm, an order of magnitude larger than the dissociation constant previously found for binding of cAMP to the isolated MloK1 CNBD (as measured by ITC) as well as by binding a fluorescent cAMP analog to full-length MloK1 (as measured with fluorescence spectrometry). However, the enthalpy change was ∼11 kcal/mol, similar to that observed for the isolated CNBD (Table 1) (34). We discuss the possible reasons for the low apparent affinity below.
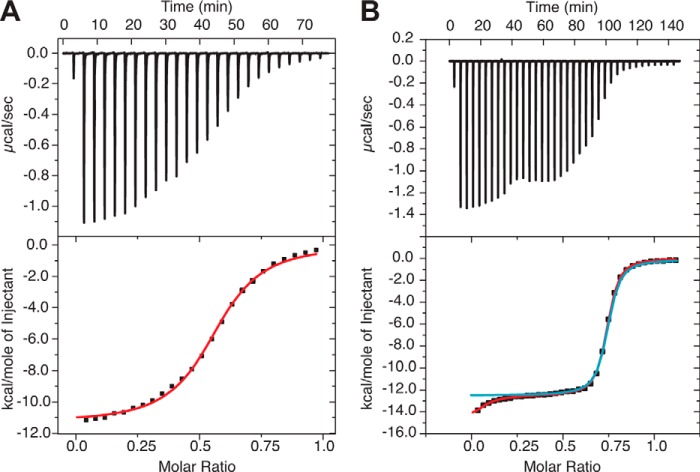
FIGURE 4.
KcsA/MloK1 binds cAMP with high affinity. ITC experiments with KcsA/MloK1 in detergent micelles (A) and nanodiscs (B). Top, heat is released upon successive injections of 2 μl of cAMP (from a 600 μm solution) to the KcsA/MloK1 protein in detergent (120 μm) (A) and 0.5 μl of cAMP (from an 800 μm solution) to the KcsA/MloK1 protein in nanodiscs (135 μm) (B). Bottom, binding curves obtained from the data on top. Protein buffer is 10 mm HEPES, 100 mm KCl, pH 7.5, with the addition of 5 mm DM in A. Red curve in A, bottom, is a least squares fit to a single-site binding model (KD = 2.0 ± 0.12 μm, n = 0.560 ± 0.001, ΔH = −11.3 ± 0.08 kcal/mol). Blue curve in B, bottom, is a least squares fit to a single-site binding model (KD = 165 ± 4.8 nm, n = 0.7 ± 0.01, ΔH = −12.4 kcal/mol). The use of a two-site model (red curve in B, bottom) gave values of KD = 16.0 ± 6.9 nm and n = 0.06 ± 0.03 for the first site and KD = 190 ± 21 nm and n = 0.67 ± 0.03 for the second site. The values are mean ± error of the fit. These values are for the individual experiments illustrated in this figure. Table 1 contains the mean ± S.D. of several experiments.
Incorporation of KcsA/MloK1 Chimera into Nanodiscs for ITC Assays
We next investigated whether the detergent might account for the reduced stoichiometry of cAMP binding, which could occur by the occlusion of the binding site (49, 50) or by incorrect folding of the CNBD resulting in a non-native conformation of the protein. To measure binding in the absence of detergent, we incorporated the channel into the lipid bilayer of nanodiscs.
Nanodiscs are small, ∼10 nm diameter, particles formed by two strands of apolipoprotein A1 (apoA1) that surround a lipid bilayer. Membrane proteins can be incorporated into this bilayer, recapitulating the native membrane condition (32). Nanodiscs provide two advantages over liposomes for these binding assays; there is access to both sides of the membrane, allowing a priori knowledge of the number of binding sites, and the nanodisc preparation is soluble and may thus be used to achieve higher protein concentrations compared with liposomes.
Using the zebrafish apoA1 construct (32) and bilayers composed of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine lipids, we incorporated the KcsA/MloK1 chimera into nanodiscs using our previously reported protocol for KcsA with minor modifications (33). Successful channel-containing nanodisc formation was verified by gel filtration and SDS-PAGE analysis (Fig. 5) and electron microscopy (Fig. 6). As determined by electron microscopy, both the empty and the channel-containing nanodiscs have an average diameter of 11–14 nm (Fig. 6A). Images of channel-containing nanodiscs show a 4-nm-long density protruding from the discs, as seen in side-view class averages (Fig. 6B), which is consistent with the tetrameric CNBD arrangement of the chimera. A three-dimensional single-particle reconstruction at 26 Å resolution suggests that the nanodiscs can accommodate a single tetrameric chimeric channel (Fig. 6C).
FIGURE 5.
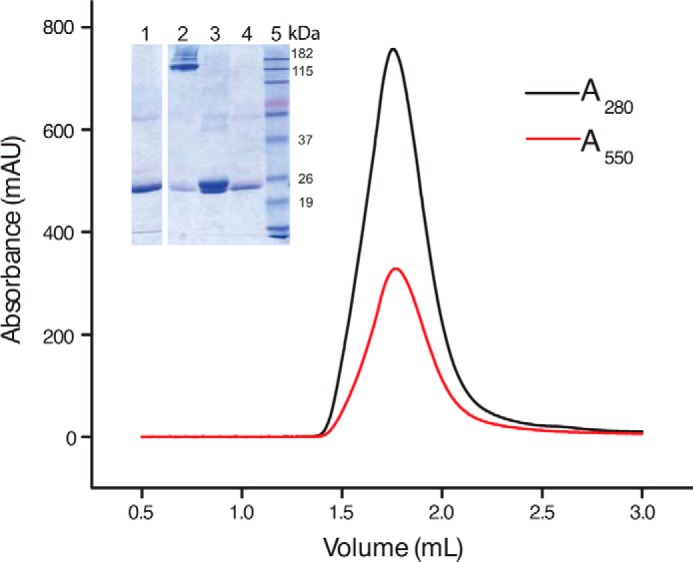
Size exclusion chromatography profile of the chimera in nanodiscs shows a single protein peak at 280 nm (black). Absorption at 550 nm (red) was simultaneously monitored and corresponds to the labeled Zap1NT protein. Inset shows an SDS-acrylamide protein gel of protein-free nanodisc (lane 1), unheated nanodisc containing chimera (lane 2), heated nanodisc containing chimera (lane 3), ZAP1NT (lane 4), and molecular weight marker (lane 5).
FIGURE 6.
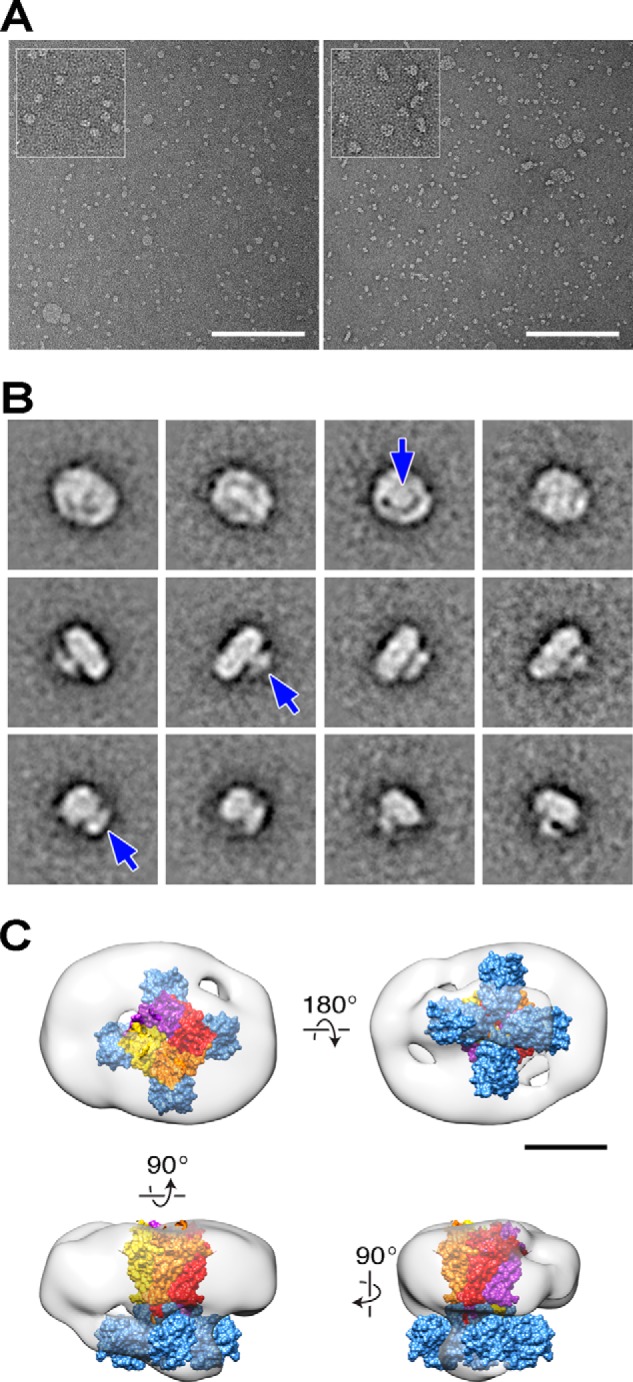
Transmission electron microscopy imaging of KcsA/MloK1 chimera incorporated into nanodiscs. A, TEM images comparing empty (left) and KcsA/MloK1-containing (right) nanodiscs. Scale bars, 200 nm. Insets show a 2-fold zoom of selected areas. B, representative class average images of KcsA/MloK1 nanodiscs. The top row shows averages interpreted as top views; the middle row shows side views, and the bottom row shows side views rotated by 90°. White density protruding from of the disc most likely corresponds to the CNBDs from a single tetramer (arrows). Each frame size is 35.5 × 35.5 nm. C, single particle three-dimensional reconstruction of the KcsA/MloK1 nanodisc, with an overlaid KcsA/MloK1 homology model docked into EM density (KcsA: yellow, orange, red, and purple, Protein Data Bank code 1K4C; CNBD: blue, Protein Data Bank code 3CL1). Scale bar, 5 nm.
CNBD of the KcsA/MloK1 Chimera in Nanodiscs Binds cAMP with High Affinity
The KcsA/MloK1 chimera in nanodiscs exhibits altered cAMP binding characteristics, as compared with the chimera in detergent (Fig. 4). First, the binding stoichiometry increases from 0.55 in detergent to 0.73 in nanodiscs. Second, the enthalpy change is larger (13 kcal/mol in nanodiscs compared with 11 kcal/mol in detergent), and the apparent affinity is about an order of magnitude higher than in detergent (KD is 0.15 μm for nanodiscs, as compared with 2 μm in detergent; Table 1). Third, although the binding curve for the channel in detergent is well described by a single-site model, ligand binding to the channel in nanodiscs shows an additional component. This additional component, which is observed in every experiment, is evident in the downward tail at very low molar ratios (Fig. 4B).
We attempted to fit the results in Fig. 4B with a two-site model. Although the first component was mostly unresolved, the fits suggest that n is ∼0.06 and that the KD is ∼16 nm. Given the lack of a starting baseline, these estimates have considerable uncertainty (Fig. 4B, legend). The second binding component is well resolved with n ≈ 0.67 and KD ≈ 0.2 μm. Reducing the cAMP concentration or altering the temperature in the ITC experiment does not provide an improvement in the resolution of the initial binding event. This additional event does not appear in control ITC experiments in which cAMP is injected into buffer without protein or into buffer containing empty lipid-filled nanodiscs (which do not contain chimeric channel). Additional incubation with the low affinity ligand 8-CPT-cGMP fails to increase the final binding stoichiometry beyond 0.73.
One explanation for this result is that the chimeric channels contain high affinity binding sites (approximately one-fourth of the total sites or one subunit per tetramer) rendering the removal of ligand very difficult. In this case, the cAMP would first bind to the small fraction of available high affinity sites of one CNBD, evident in the binding event in the initial injections in the ITC experiment (Fig. 4B). Upon saturation of these high affinity sites (or at higher concentrations of cAMP), cAMP would then bind to the low affinity sites of the remaining CNBDs, causing the binding event observed with subsequent injections of cAMP. This scenario is consistent with previously published ITC results for isolated CNBDs with C-linkers from HCN2 and HCN4 channels, which suggest that binding of the first ligand results in a negative cooperativity effect that reduces the binding affinity of the remaining three sites (48).
We cannot exclude the possibility that these initial heats could reflect a systematic effect (such as cAMP-induced minor rearrangement within the protein) that is not associated with the canonical cAMP-binding site. If this were true, it is still possible to fit the binding curve with a one-site model (blue curve in Fig. 4B). The binding parameters from this fit are very similar to those from the two-site fit (see Fig. 4 legend), suggesting that 70–75% of the sites in the channel are functional and bind cAMP with a similar affinity. It would then follow that the remaining 25% of the sites cannot bind cAMP, either because they are improperly folded or they have remained bound to the ligand.
Irrespective of which scenario is more accurate, ∼75% of the channels are well folded and bind cAMP with high affinity. Thus, incorporation of the channel into nanodiscs allows us to characterize the thermodynamics of ligand binding more completely than was possible with the detergent-solubilized channel. Importantly, when the channels are embedded in a more native environment such as a bilayer (lipid-filled nanodiscs), the CNBDs of the chimeric channel bind cAMP with high affinity (KD ≈ 150 nm), similar to the 100 nm affinity obtained for the isolated CNBD domains (34).
DISCUSSION
We have constructed a chimeric channel (KcsA/MloK1), which is composed of the transmembrane domain of KcsA and the cyclic nucleotide-binding domain of MloK1, and verified that it retains key properties of both parent channels. Like KcsA, the chimeric channel is activated by protons. Like MloK1, the activity of the chimera is modulated by cAMP. The chimeric channel, unlike MloK1, does not aggregate at concentrations necessary for ITC and is capable of conducting currents across bilayers. Thus, this chimera is well suited for both electrophysiological and biochemical assays, enabling examination of the thermodynamics of cAMP binding to a cyclic nucleotide-modulated ion channel using ITC, which has not been possible with either MloK1 or CNG/HCN channels.
Comparison between KcsA/MloK1 and the KcsA-CNG (Rhodopseudomonas palustris) Chimeras
A conceptually similar chimeric cAMP-modulated ion channel was reported previously by Ohndorf and MacKinnon (31). This chimera also contained the pore domain of KcsA, but the CNBD portion was from an R. palustris HCN homolog. Importantly, the R. palustris chimera possesses fewer residues from the KcsA bundle crossing and does not include the two glutamates (Glu-118 and Glu-120) that contribute to H+ sensing in KcsA (43). This likely explains why the R. palustris chimera has larger steady-state open probability and is less sensitive to H+, whereas our KcsA/MloK1 chimera displays H+-dependent gating similar to that of KcsA. In fact, because our KcsA/MloK1 chimera lacks the entire KcsA C-terminal domain and it is still H+-gated, our results demonstrate that this portion of the protein is not required for pH-induced activation, a result that directly addresses a controversy regarding the location of the pH sensor in KcsA channels (43–45).
Addition of saturating concentrations of cAMP produced a similar increase in open probability for both chimeras as follows: about 2-fold for the R. palustris and about 3-fold for the KcsA/MloK1 chimera, but K½ for the R. palustris chimera is about 10-fold less than that for the KcsA/MloK1 chimera (90 nm in Fig. 8 of Ref. 31 versus 1 μm in our Fig. 3D). This difference may reflect only the 33.3% shared identity between the CNBDs (residues 219–355 in M. loti and residues 255–410 in R. palustris) and their different oligomerization states (the isolated R. palustris CNBDs dimerize in solution, whereas the M. loti CNBDs are monomers (24)).
In addition to demonstrating cAMP modulation using flux assays, we also characterized the KcsA/MloK1 chimera using single-channel electrophysiology. Perfusion of cAMP to the CNBD-containing side of the chimera results in an increase in open probability in ∼75% of the channels tested, but this increase in open probability did not reverse when cAMP was removed. Neither the observed variability nor the irreversibility can be attributed to uncertainty in channel number and undetected gain of channels in the lipid bilayer during perfusions (51, 52) because we obtained similar results using the noninactivating E71A KcsA chimera (35). The apparent lack of reversibility could be due to a very slow off-rate of cAMP from the binding site, which is consistent with the difficulties encountered in removing the bound cAMP from any construct containing this CNBD (isolated CNBD, MloK1, and KcsA/MloK1 chimera). Ohndorf and MacKinnon (31) did not comment on the reversibility of the cAMP effect in the R. palustris chimera.
Effect of Lipids on cAMP Binding
Incorporation of the channel into lipid-containing nanodiscs increases the cAMP/subunit stoichiometry of binding from 0.55 to 0.73, reduces the measured KD 10-fold (from ∼2 μm to ∼150 nm), and increases the enthalpy change from −11 to −13 kcal/mol (Table 1). Interestingly, because the binding process itself occurs in a domain located outside of the membrane, the observed increase in cAMP binding affinity to lipid-embedded channels relative to detergent-solubilized channels suggests that binding induces a conformational change in the transmembrane portion of the protein, which is in direct contact with lipids. Thus, the nanodisc and detergent environments may favor different conformations of the channel with different cAMP affinities (14).
This may explain the observed differences in affinity but does not explain the difference in stoichiometry. The detergent might occlude the binding sites, either through direct interactions (49, 50) or by restructuring the protein in such a way as to impact cAMP binding (which can lead to both reduced binding stoichiometry and lower affinity in detergent micelles). Likely, both the removal of detergent and the addition of the lipids contribute to the measured differences. These results demonstrate the importance of the channel's lipid environment for its function, reflecting the energetic coupling between membrane proteins and the lipid bilayer (53).
cAMP Binding Curve Is Biphasic in Nanodiscs
A surprising but reproducible feature of cAMP binding to the channel in nanodiscs is the suggestion of a second partially resolved binding component with very high cAMP affinity. We could not resolve the entire binding curve, and we cannot exclude that it is unrelated to specific ligand binding events. Yet, given the cAMP/subunit stoichiometry of ∼0.75, this may mean that a fraction of binding sites (∼0.25) has very high cAMP affinity, such that they are likely to have a bound cAMP even in the nominal absence of cAMP. Our results thus suggest that empty CNBD tetramers in the full-length channel have very high affinity for cAMP and that binding the first cAMP molecule to a subunit in the tetramer decreases the cAMP affinity for the remaining three binding sites (corresponding to the 75% of the sites that we observe in the ITC experiments). If so, it would imply negative cooperativity in cAMP binding to MloK1.
Negative binding cooperativity has previously been suggested for full-length CNG and HCN eukaryotic channels based on patch clamp fluorometry experiments (11–14). Similarly, binding curves for cAMP to the isolated CNBDs of both HCN2 and HCN4 suggest negative cooperativity, with the first cAMP molecule binding with a 10-fold higher affinity than the subsequent molecules (48). Of particular interest to our studies is the report that ITC measurements performed on isolated CNBD/C-linkers from HCN1 channels yield a subunitary stoichiometry, leading the authors to hypothesize that a fraction of the cAMP-binding sites have constitutively bound cAMP (48), which is very similar to what we found for the chimera. In fact, because the experiments with the chimera were performed with the full-length channel rather than the isolated CNBDs, our results suggest that the findings of Chow et al. (48) may extend to the full-length HCN1 channel.
Previous measurements of cyclic nucleotide binding to the isolated CNBD of MloK1 revealed only a single population of noninteracting binding sites, with a stoichiometry close to 1; similarly, cyclic nucleotide binding to the full-length MloK1, using a fluorescent cAMP analog, revealed only a single binding affinity (34). The latter measurements were performed with detergent-solubilized protein, however, which may have obscured a high affinity component (cf. Fig. 4A). Moreover, although the equilibrium binding curve was consistent with a single population of binding sites, kinetic studies revealed two distinct values for the dissociation rate constant (0.07 and 0.47 s−1) (54), indicating two different classes of binding sites.
Apparent Affinities for cAMP Differ among Constructs and Assays
The cAMP equilibrium dissociation constant for the KcsA/MloK1 chimera in lipid bilayers/nanodiscs (∼150 nm) agrees well with that obtained for the isolated CNBD of MloK1 (107 nm) and for full-length MloK1 (82 nm using a fluorescent cAMP analog) (34). However, although our flux assays demonstrate an increase in KcsA/MloK1 chimeric activity in response to cAMP, the cAMP concentration for half-maximal activation was about 10-fold higher (∼1 μm) than the equilibrium dissociation constant for the chimera (150 nm) or the concentration for half-maximal activation that was observed previously for MloK1 (60 nm) (24, 25). This result differs from previous studies on MloK1 that found K½ and KD to be of the same order of magnitude, thus indicating that MloK1 could be significantly activated in partially liganded states (34); however, our results (Fig. 3) suggest that the chimeric channel activation occurs only after the full complement of cAMP has bound, perhaps reflecting the lack of voltage sensors in the chimera, which could compromise the coupling between CNBDs and channel gate.
KcsA/MloK1 Chimera, a Model for HCN Channel Gating
The dual modulation of the chimeric channel by protons and cAMP is reminiscent of the dual modulation of HCN channels by hyperpolarization and cAMP (2). Both HCN channels and the chimeric channels require a “strong” stimulus (hyperpolarization and proton binding, respectively) to open. Once the channels have been activated, however, their activity can be further modulated by cAMP. cAMP binding by itself therefore does not seem to provide sufficient energy to open either channel, meaning that the cAMP affinity cannot be highly state-dependent. Rather, cAMP binding to the CNBDs appears to cause local conformational changes that trigger further motion within the transmembrane domains to moderately increase the open probability of already active channels. This dual modulation of HCN channels may have evolved to enable stringent regulation of gating due to their importance in the heart and central nervous system. Furthermore, the chimera may exhibit negative cooperativity in cAMP binding, which was previously observed for the CNBDs of HCN channels. Therefore, by extension, the mechanistic details regarding binding may be consistent between isolated CNBDs with C-linkers and full-length HCN channels.
Conclusions
We constructed a KcsA/MloK1 chimeric channel that preserves the functional properties of both parent channels, thus allowing for investigations into how ligand binding modulates the gate of an ion channel in the context of a homogeneous, heterologously expressed protein, which is amenable to both binding and functional studies in a controlled environment. The channel's lipid environment is critical for cAMP binding even though the ligand-binding domain is located outside the membrane, indicating that the conformation of the transmembrane domains is different in micelles and lipid bilayers and that the transmembrane domains undergo conformational changes upon cAMP binding. These conformational changes, however, are not sufficiently large to open the channel, suggesting that the energetic contribution from cAMP binding is small relative to that from proton binding. This chimeric ion channel is ideal for examining the complex effects of two allosteric modulators, because the channel possesses both the proton gating machinery of KcsA and the cAMP modulation of MloK1 channels, making it a powerful tool for exploring the dual regulation observed in HCN channels.
This work was supported, in whole or in part, by National Institutes of Health Grants GM077560 (to C. M. N.) and GM021342 (to O. S. A.). This work was also supported by Swiss National Science Foundation Grant 315230 and National Center of Competence in Research TransCure (to H. S.).
- CNG
- cyclic nucleotide-gated
- HCN
- hyperpolarization-activated and cyclic nucleotide-gated
- CNBD
- cyclic nucleotide-binding domain
- DM
- n-decyl β-d-maltopyranoside
- LUV
- large unilamellar vesicle
- 8-pCPT-cGMP
- 8-(4-chlorophenylthio)-guanosine 3′,5′-cyclic monophosphate
- ANTS
- aminonaphthalene-1,3,6-trisulfonate
- ITC
- isothermal titration calorimetry.
REFERENCES
- 1. Kaupp U. B., Seifert R. (2002) Cyclic nucleotide-gated ion channels. Physiol. Rev. 82, 769–824 [DOI] [PubMed] [Google Scholar]
- 2. Kaupp U. B., Seifert R. (2001) Molecular diversity of pacemaker ion channels. Annu. Rev. Physiol. 63, 235–257 [DOI] [PubMed] [Google Scholar]
- 3. Robinson R. B., Siegelbaum S. A. (2003) Hyperpolarization-activated cation currents: from molecules to physiological function. Annu. Rev. Physiol. 65, 453–480 [DOI] [PubMed] [Google Scholar]
- 4. Yellen G. (2002) The voltage-gated potassium channels and their relatives. Nature 419, 35–42 [DOI] [PubMed] [Google Scholar]
- 5. Yu F. H., Catterall W. A. (2003) Overview of the voltage-gated sodium channel family. Genome Biol. 4, 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shabb J. B., Corbin J. D. (1992) Cyclic nucleotide-binding domains in proteins having diverse functions. J. Biol. Chem. 267, 5723–5726 [PubMed] [Google Scholar]
- 7. Zagotta W. N., Siegelbaum S. A. (1996) Structure and function of cyclic nucleotide-gated channels. Annu. Rev. Neurosci. 19, 235–263 [DOI] [PubMed] [Google Scholar]
- 8. Ludwig A., Zong X., Jeglitsch M., Hofmann F., Biel M. (1998) A family of hyperpolarization-activated mammalian cation channels. Nature 393, 587–591 [DOI] [PubMed] [Google Scholar]
- 9. Santoro B., Liu D. T., Yao H., Bartsch D., Kandel E. R., Siegelbaum S. A., Tibbs G. R. (1998) Identification of a gene encoding a hyperpolarization-activated pacemaker channel of brain. Cell 93, 717–729 [DOI] [PubMed] [Google Scholar]
- 10. Craven K. B., Zagotta W. N. (2006) CNG and HCN channels: two peas, one pod. Annu. Rev. Physiol. 68, 375–401 [DOI] [PubMed] [Google Scholar]
- 11. Kusch J., Biskup C., Thon S., Schulz E., Nache V., Zimmer T., Schwede F., Benndorf K. (2010) Interdependence of receptor activation and ligand binding in HCN2 pacemaker channels. Neuron 67, 75–85 [DOI] [PubMed] [Google Scholar]
- 12. Kusch J., Thon S., Schulz E., Biskup C., Nache V., Zimmer T., Seifert R., Schwede F., Benndorf K. (2012) How subunits cooperate in cAMP-induced activation of homotetrameric HCN2 channels. Nat. Chem. Biol. 8, 162–169 [DOI] [PubMed] [Google Scholar]
- 13. Nache V., Zimmer T., Wongsamitkul N., Schmauder R., Kusch J., Reinhardt L., Bönigk W., Seifert R., Biskup C., Schwede F., Benndorf K. (2012) Differential regulation by cyclic nucleotides of the CNGA4 and CNGB1b subunits in olfactory cyclic nucleotide-gated channels. Sci. Signal. 5, ra48. [DOI] [PubMed] [Google Scholar]
- 14. Wu S., Vysotskaya Z. V., Xu X., Xie C., Liu Q., Zhou L. (2011) State-dependent cAMP binding to functioning HCN channels studied by patch clamp fluorometry. Biophys. J. 100, 1226–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Flynn G. E., Black K. D., Islas L. D., Sankaran B., Zagotta W. N. (2007) Structure and rearrangements in the carboxy-terminal region of SpIH channels. Structure 15, 671–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lolicato M., Nardini M., Gazzarrini S., Möller S., Bertinetti D., Herberg F. W., Bolognesi M., Martin H., Fasolini M., Bertrand J. A., Arrigoni C., Thiel G., Moroni A. (2011) Tetramerization dynamics of C-terminal domain underlies isoform-specific cAMP gating in hyperpolarization-activated cyclic nucleotide-gated channels. J. Biol. Chem. 286, 44811–44820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Taraska J. W., Puljung M. C., Olivier N. B., Flynn G. E., Zagotta W. N. (2009) Mapping the structure and conformational movements of proteins with transition metal ion FRET. Nat. Methods 6, 532–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xu X., Vysotskaya Z. V., Liu Q., Zhou L. (2010) Structural basis for the cAMP-dependent gating in the human HCN4 channel. J. Biol. Chem. 285, 37082–37091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zagotta W. N., Olivier N. B., Black K. D., Young E. C., Olson R., Gouaux E. (2003) Structural basis for modulation and agonist specificity of HCN pacemaker channels. Nature 425, 200–205 [DOI] [PubMed] [Google Scholar]
- 20. Doyle D. A., Morais Cabral J., Pfuetzner R. A., Kuo A., Gulbis J. M., Cohen S. L., Chait B. T., MacKinnon R. (1998) The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science 280, 69–77 [DOI] [PubMed] [Google Scholar]
- 21. Jiang Y., Lee A., Chen J., Cadene M., Chait B. T., MacKinnon R. (2002) Crystal structure and mechanism of a calcium-gated potassium channel. Nature 417, 515–522 [DOI] [PubMed] [Google Scholar]
- 22. Jiang Y., Lee A., Chen J., Ruta V., Cadene M., Chait B. T., MacKinnon R. (2003) X-ray structure of a voltage-dependent K+ channel. Nature 423, 33–41 [DOI] [PubMed] [Google Scholar]
- 23. Schrempf H., Schmidt O., Kümmerlen R., Hinnah S., Müller D., Betzler M., Steinkamp T., Wagner R. (1995) A prokaryotic potassium ion channel with two predicted transmembrane segments from Streptomyces lividans. EMBO J. 14, 5170–5178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Clayton G. M., Silverman W. R., Heginbotham L., Morais-Cabral J. H. (2004) Structural basis of ligand activation in a cyclic nucleotide regulated potassium channel. Cell 119, 615–627 [DOI] [PubMed] [Google Scholar]
- 25. Nimigean C. M., Shane T., Miller C. (2004) A cyclic nucleotide modulated prokaryotic K+ channel. J. Gen. Physiol. 124, 203–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Aggarwal S. K., MacKinnon R. (1996) Contribution of the S4 segment to gating charge in the Shaker K+ channel. Neuron 16, 1169–1177 [DOI] [PubMed] [Google Scholar]
- 27. Clayton G. M., Altieri S., Heginbotham L., Unger V. M., Morais-Cabral J. H. (2008) Structure of the transmembrane regions of a bacterial cyclic nucleotide-regulated channel. Proc. Natl. Acad. Sci. U.S.A. 105, 1511–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schünke S., Stoldt M., Lecher J., Kaupp U. B., Willbold D. (2011) Structural insights into conformational changes of a cyclic nucleotide-binding domain in solution from Mesorhizobium loti K1 channel. Proc. Natl. Acad. Sci. U.S.A. 108, 6121–6126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schünke S., Stoldt M., Novak K., Kaupp U. B., Willbold D. (2009) Solution structure of the Mesorhizobium loti K1 channel cyclic nucleotide-binding domain in complex with cAMP. EMBO Rep. 10, 729–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chiu P. L., Pagel M. D., Evans J., Chou H. T., Zeng X., Gipson B., Stahlberg H., Nimigean C. M. (2007) The structure of the prokaryotic cyclic nucleotide-modulated potassium channel MloK1 at 16 Å resolution. Structure 15, 1053–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ohndorf U. M., MacKinnon R. (2005) Construction of a cyclic nucleotide-gated KcsA K+ channel. J. Mol. Biol. 350, 857–865 [DOI] [PubMed] [Google Scholar]
- 32. Banerjee S., Huber T., Sakmar T. P. (2008) Rapid incorporation of functional rhodopsin into nanoscale apolipoprotein bound bilayer (NABB) particles. J. Mol. Biol. 377, 1067–1081 [DOI] [PubMed] [Google Scholar]
- 33. Banerjee S., Nimigean C. M. (2011) Non-vesicular transfer of membrane proteins from nanoparticles to lipid bilayers. J. Gen. Physiol. 137, 217–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cukkemane A., Grüter B., Novak K., Gensch T., Bönigk W., Gerharz T., Kaupp U. B., Seifert R. (2007) Subunits act independently in a cyclic nucleotide-activated K+ channel. EMBO Rep. 8, 749–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cordero-Morales J. F., Cuello L. G., Zhao Y., Jogini V., Cortes D. M., Roux B., Perozo E. (2006) Molecular determinants of gating at the potassium-channel selectivity filter. Nat. Struct. Mol. Biol. 13, 311–318 [DOI] [PubMed] [Google Scholar]
- 36. Heginbotham L., Kolmakova-Partensky L., Miller C. (1998) Functional reconstitution of a prokaryotic K+ channel. J. Gen. Physiol. 111, 741–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nimigean C. M., Pagel M. D. (2007) Ligand binding and activation in a prokaryotic cyclic nucleotide-modulated channel. J. Mol. Biol. 371, 1325–1337 [DOI] [PubMed] [Google Scholar]
- 38. LeMasurier M., Heginbotham L., Miller C. (2001) KcsA: it's a potassium channel. J. Gen. Physiol. 118, 303–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chen T. Y., Miller C. (1996) Nonequilibrium gating and voltage dependence of the ClC-0 Cl− channel. J. Gen. Physiol. 108, 237–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rusinova R., Kim D. M., Nimigean C. M., Andersen O. S. (2014) Regulation of ion channel function by the host lipid bilayer examined by a stopped-flow spectroflurorimetric assay. Biophys. J. 106, 1070–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ingólfsson H. I., Andersen O. S. (2010) Screening for small molecules' bilayer-modifying potential using a gramicidin-based fluorescence assay. Assay Drug Dev. Technol. 8, 427–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Posson D. J., Thompson A. N., McCoy J. G., Nimigean C. M. (2013) Molecular interactions involved in proton-dependent gating in KcsA potassium channels. J. Gen. Physiol. 142, 613–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Thompson A. N., Posson D. J., Parsa P. V., Nimigean C. M. (2008) Molecular mechanism of pH sensing in KcsA potassium channels. Proc. Natl. Acad. Sci. U.S.A. 105, 6900–6905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cuello L. G., Cortes D. M., Jogini V., Sompornpisut A., Perozo E. (2010) A molecular mechanism for proton-dependent gating in KcsA. FEBS Lett. 584, 1126–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hirano M., Onishi Y., Yanagida T., Ide T. (2011) Role of the KcsA channel cytoplasmic domain in pH-dependent gating. Biophys. J. 101, 2157–2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gauss R., Seifert R., Kaupp U. B. (1998) Molecular identification of a hyperpolarization-activated channel in sea urchin sperm. Nature 393, 583–587 [DOI] [PubMed] [Google Scholar]
- 47. Seifert R., Scholten A., Gauss R., Mincheva A., Lichter P., Kaupp U. B. (1999) Molecular characterization of a slowly gating human hyperpolarization-activated channel predominantly expressed in thalamus, heart, and testis. Proc. Natl. Acad. Sci. U.S.A. 96, 9391–9396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chow S. S., Van Petegem F., Accili E. A. (2012) Energetics of cyclic AMP binding to HCN channel C terminus reveal negative cooperativity. J. Biol. Chem. 287, 600–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Quick M., Shi L., Zehnpfennig B., Weinstein H., Javitch J. A. (2012) Experimental conditions can obscure the second high affinity site in LeuT. Nat. Struct. Mol. Biol. 19, 207–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Quick M., Winther A. M., Shi L., Nissen P., Weinstein H., Javitch J. A. (2009) Binding of an octylglucoside detergent molecule in the second substrate (S2) site of LeuT establishes an inhibitor-bound conformation. Proc. Natl. Acad. Sci. U.S.A. 106, 5563–5568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bernèche S., Roux B. (2005) A gate in the selectivity filter of potassium channels. Structure 13, 591–600 [DOI] [PubMed] [Google Scholar]
- 52. Gao L., Mi X., Paajanen V., Wang K., Fan Z. (2005) Activation-coupled inactivation in the bacterial potassium channel KcsA. Proc. Natl. Acad. Sci. U.S.A. 102, 17630–17635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lundbaek J. A., Koeppe R. E., 2nd, Andersen O. S. (2010) Amphiphile regulation of ion channel function by changes in the bilayer spring constant. Proc. Natl. Acad. Sci. U.S.A. 107, 15427–15430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Peuker S., Cukkemane A., Held M., Noé F., Kaupp U. B., Seifert R. (2013) Kinetics of ligand-receptor interaction reveals an induced-fit mode of binding in a cyclic nucleotide-activated protein. Biophys. J. 104, 63–74 [DOI] [PMC free article] [PubMed] [Google Scholar]