Background: α-Conotoxins are small toxins produced by cone snails and antagonists of nicotinic acetylcholine receptors.
Results: Two mutants were created to investigate the unusual C terminus of a novel α-conotoxin from Conus longurionis.
Conclusion: We characterized an important residue for discrimination between neuronal and muscle subtype nicotinic acetylcholine receptors.
Significance: This opens perspectives for designing new ligands to affect brain disorders.
Keywords: Alzheimer Disease, Electrophysiology, Nicotinic Acetylcholine Receptors, NMR, Protein Engineering, Schizophrenia, Conotoxin, Conotoxin Structure-Function Relationship
Abstract
α-Conotoxins are peptide toxins found in the venom of marine cone snails and potent antagonists of various subtypes of nicotinic acetylcholine receptors (nAChRs). nAChRs are cholinergic receptors forming ligand-gated ion channels in the plasma membranes of certain neurons and the neuromuscular junction. Because nAChRs have an important role in regulating transmitter release, cell excitability, and neuronal integration, nAChR dysfunctions have been implicated in a variety of severe pathologies such as epilepsy, myasthenic syndromes, schizophrenia, Parkinson disease, and Alzheimer disease. To expand the knowledge concerning cone snail toxins, we examined the venom of Conus longurionis. We isolated an 18-amino acid peptide named α-conotoxin Lo1a, which is active on nAChRs. To the best of our knowledge, this is the first characterization of a conotoxin from this species. The peptide was characterized by electrophysiological screening against several types of cloned nAChRs expressed in Xenopus laevis oocytes. The three-dimensional solution structure of the α-conotoxin Lo1a was determined by NMR spectroscopy. Lo1a, a member of the α4/7 family, blocks the response to acetylcholine in oocytes expressing α7 nAChRs with an IC50 of 3.24 ± 0.7 μm. Furthermore, Lo1a shows a high selectivity for neuronal versus muscle subtype nAChRs. Because Lo1a has an unusual C terminus, we designed two mutants, Lo1a-ΔD and Lo1a-RRR, to investigate the influence of the C-terminal residue. Lo1a-ΔD has a C-terminal Asp deletion, whereas in Lo1a-RRR, a triple-Arg tail replaces the Asp. They blocked the neuronal nAChR α7 with a lower IC50 value, but remarkably, both adopted affinity for the muscle subtype α1β1δϵ.
Introduction
Nicotinic acetylcholine receptors (nAChRs)4 are expressed in the central and peripheral nervous systems where they are involved in many neuronal functions. These neuronal functions include differentiation and synaptic plasticity, which are the basis for learning and memory (1–3). The nicotinic acetylcholine receptor family is classified into two subtypes based on their primary sites of expression, namely neuronal and muscle subtype nAChRs. Both subtypes are pentameric integral membrane protein complexes classified as ligand-gated ion channels that open in response to binding of the neurotransmitter acetylcholine (ACh) (4, 5). The neuronal subtype nAChRs can include either exclusively α-subunits such as α7, α8, and α9, called homomeric ion channels or a combination of two or more different types of subunits, in a heteromeric assembly. These heteromeric channels are compiled of at least one α subunit (α2–α10) and one β subunit (β2–β4). The muscle subtype nAChRs are composed of four different types of subunits (α1, β1, δ, and ϵ/γ) (6–8).
One of the neuronal nAChRs, α7, has received much attention since its discovery thanks to the role of α7 in the central nervous system (CNS) (9). This is because α7 nAChRs are highly distributed in the brain, including regions involved in learning and memory, hippocampus, and cerebral cortex (10–12). Consequently, nAChR dysfunctions have been implicated in a variety of severe pathologies such as certain types of epilepsy, myasthenic syndromes, schizophrenia, Parkinson disease, and Alzheimer disease (13–15). Therefore, the discovery of new ligands binding with high affinity and selectivity to nAChR subtypes is of prime interest to study these receptors and to potentially discover new drugs for the treatment of these pathologies (16).
New ligands may be found in cone snail species, from which the so-called family of α-conotoxins is a group of potent nAChRs antagonists (17, 18). These α-conotoxins are a series of structurally and functionally related peptides found in the venom of cone snail species. They are classified into subfamilies based on the number of residues in their two “loops” between conserved Cys residues, with 3/5 (CCX3CX5C), 4/3 (CCX4CX3C), and 4/7 (CCX4CX7C) subfamilies the most common (19). The toxins of Conus sp. are usually potent, selective, and small (12–25 amino acids), which is an advantage for cost-effective synthesis (20). Moreover, they are shown to function as specific probes to investigate the structure-function relationship of nAChRs (17).
In this study, we report the isolation of a novel 18-amino acid α-conotoxin from the venom of the marine snail Conus longurionis and its electrophysiological screening against six different types of nAChRs. To the best of our knowledge, this is the first conotoxin to be characterized from this species found in the Indian Ocean near Tamil Nadu, India. The peptide, called Lo1a, has a W-shaped structural conformation with two loops that are reinforced by two disulfide bonds. The function of the peptide revealed that Lo1a was most active against neuronal homomeric α7 nAChRs. To further determine the structure-function relationship of Lo1a and its target α7, we engineered two synthetic analogues, namely Lo1a-ΔD and Lo1a-RRR, based on the protein sequence of Lo1a and its homology to other conotoxins from the α4/7 family. The first peptide, Lo1a-ΔD, has an Asp deletion at the C terminus, whereas in the second peptide, an Arg tail replaces this Asp. Both analogues were found to block the neuronal nAChR α7 with a lower IC50, but remarkably, they adopted affinity for the muscle subtype α1β1δϵ. These results revealed an unexpected role for the C terminus in determining subtype selectivity and efficacy. Consequently, our findings might be relevant in the context of designing novel therapeutic compounds with potential utility in diseases such as Alzheimer disease, schizophrenia, and attention deficit hyperactivity disorder because, as previously mentioned, α7 nAChRs are thought to play important roles in the brain (21).
EXPERIMENTAL PROCEDURES
Cone Snail Specimens and Venom Extraction
Specimens of C. longurionis (identified by Kiener in 1845 and classified by Tucker and Tenorio (22)) were collected from the Indian Ocean near Tamil Nadu, India. The venomous apparatuses (venom bulbs and venom ducts) were extracted from the specimens as previously described (23). The collected tissue was preserved in RNAlater solution (Ambion) and stored at −20 °C. The venomous apparatuses were used for total RNA extraction and peptide/protein extraction.
Peptide and Purification
Sample fractionation occurred by reversed phase HPLC (Gilson, Middleton, WI). Two steps were followed for the separation of the venom compounds. In the first step, the lyophilized crude venom powder was solubilized into 50% acetonitrile (ACN)/water, and aliquots were loaded on a gel filtration SuperdexTM peptide 10/300 GL column with 50% ACN/water as mobile phase (flow rate, 0.5 ml/min) to separate the peptides and proteins based on their size. Two sample collections were made that were stored overnight at −80 °C, freeze-dried and finally solubilized in 5% ACN/water. For the second step, an analytical Vydac C18 column (218MS54, 4.6 × 250 mm, 5-μm particle size; Grace, Deerfield, IL) with a two-solvent system was used: (A) 0.1% TFA/H2O and (B) 0.085% TFA/ACN. The sample was eluted at a constant flow rate of 1 ml min−1 with a 0–80% gradient of solvent B over 90 min (1% ACN per min after 10 min of solvent A). The HPLC column fractions were monitored by a UV/VIS-155 detector (Gilson) scanning both 214 and 280 nm.
Peptide Sequencing
Isolated Lo1a was collected and freeze-dried for direct peptide sequencing and molecular mass analysis (MALDI-TOF). A Protein Sequencer PPSQ-31A/33A (Shimadzu, Japan) was used to determine the amino acid sequence of the separated compound. In this Edman degradation method, the sample was loaded onto a polybrene-pretreated, precycled glass fiber disk and Edman sequenced for 24 residue cycles.
Peptide Synthesis and Folding
Lo1a was synthesized using Fmoc (N-(9-fluorenyl)methoxycarbonyl) chemistry by GeneCust (Luxemburg). Lo1a-ΔD and Lo1a-RRR were synthesized by GenicBio Limited (Shanghai, China). Formation of the two disulfide bridges was carried out by adopting the selective protection and deprotection strategy in vitro. The resulting bicyclic peptides were subsequently purified by HPLC and analyzed with ESI-MS, then freeze-dried, and stored at −20 °C until use.
Functional Characterization
Complementary DNA encoding the nAChR channels was subcloned into the corresponding vector: human α3/pcDNA3(XbaI), human α4/pGEM-HE(NheI), chick α7/pBlueScript(NotI), human β2/pSP64(PvuII), human β4/pcDNA3(XbaI), rat α1/pSP0oD(SalI), rat β1/pSP0oD(SalI), rat γ/pSP0oD(SalI), rat δ/pSP0oD(SalI), and rat ϵ/pSP0oD(SalI). The linearized plasmids (respective restriction enzymes are indicated in parentheses) were transcribed using the T7 (α3, α4, α7, or β4) or the SP6 (β2, α1, β1, γ, δ, or ϵ) mMESSAGE mMACHINE transcription kit (Ambion, Austin, TX).
The harvesting of stage V–VI oocytes from anesthetized female Xenopus laevis frogs was previously described (24). Oocytes were injected with 50–70 nl of cRNA at a concentration of 1–3 ng/nl using a micro-injector (Drummond Scientific, Broomall, PA). The oocytes were incubated in a ND-96 solution containing: 96 mm NaCl, 2 mm KCl, 1.8 mm CaCl2, 2 mm MgCl2, and 5 mm HEPES (pH 7.4), supplemented with 1.25 ml/liter gentamicin and 90 mg/liter theophylline. The oocytes were stored for 1–5 days at 16 °C until sufficient expression of nAChRs was achieved.
Whole cell currents from oocytes were recorded at room temperature (18–22 °C) by the two-electrode voltage clamp technique using a GeneClamp 500 amplifier (Molecular Devices, Sunnyvale, CA) controlled by a pClamp data acquisition system (Molecular Devices). The oocytes were placed in a bath containing ND-96 solution. Voltage and current electrodes were filled with 3 m KCl, and the resistances of both electrodes were between 0.5 and 1.5 mΩ. The elicited currents were sampled at 100 Hz and filtered at 50 Hz using a four-pole, low pass Bessel filter. To eliminate the effect of the voltage drop across the bath grounding electrode, the bath potential was actively controlled by a two-electrode bath clamp. During recordings, oocytes were continuously perfused with ND96-A (ND96 with 1 μm atropine; except for α7) at a rate of 2 ml/min, with the conopeptides applied for 30 s before ACh was added. ACh (200 μm for α7, 70 μm for α3β4, 50 μm for α4β2 and α4β4, and 10 μm for α1β1γδ and α1β1δϵ) was applied for 2 s at 2 ml/min, with 30-s washout periods between different ACh applications and 200 s after toxin application. The percentage response or percentage inhibition was obtained by averaging the peak amplitude of three control responses (two directly before exposure to the peptide and one after 200 s of washout).
Whole cell current traces were evoked from a holding potential of −90 mV. Concentration-response curves were constructed by application of different toxin concentrations to the nAChR-expressing oocytes. The percentage of nAChR blockade was plotted against the logarithm of the applied concentrations and fitted with the Hill equation: y = 100/[1 + (IC50/[toxin])h], where y is the amplitude of the toxin-induced effect, IC50 is the toxin concentration at half-maximal efficacy, [toxin] is the toxin concentration, and h is the Hill coefficient.
Comparison of two sample means was made using a paired Student's t test (p < 0.05). All data are presented as the means ± S.E. of at least three independent experiments (n ≥ 3). All data were analyzed using pClamp Clampfit 10.0 (Molecular Devices) and Origin 7.5 software (Originlab, Northampton, MA).
NMR Spectroscopy
NMR spectra were recorded with a 2.6 mm solution (200 μl, pH 5.9) of the folded conopeptide in D2O and in H2O:D2O (9:1) mixture at 5 °C on a 600 MHz Bruker Avance II spectrometer equipped with a 5-mm TCI HCN Z gradient cryoprobe. Spectra were processed using Topspin 2.1 (Bruker Biospin) and analyzed by using CARA program (version 1.8.4) (25).
In the one-dimensional and two-dimensional spectra that were recorded in H2O:D2O mixture, the water signal was suppressed by using excitation sculpting with gradients (26). The two-dimensional NOESY in H2O (mixing time, 150 and 300 ms) was recorded with a sweep width of 7210 Hz in both dimensions, 64 scans, 2048 data points in t2, and 1024 free induction decays in t1.
A two-dimensional total correlation spectroscopy (TOCSY) (27) in 90% H2O was recorded with DIPSI2 sequence for mixing (mixing time, 80 ms). A double quantum-filtered COSY (28) in H2O was acquired using excitation sculpting with gradients for water suppression with a sweep width of 7210 Hz in both dimensions, 64 scans, 2048 data points in t2, and 1024 free induction decays in t1. In the processing of two-dimensional spectra, the data were apodized with a shifted sine bell square function in both dimensions. Proton chemical shifts were calibrated by using residual HOD (water in which one of the protons has been exchanged with a deuteron) signal as reference (4.9745 ppm at 5 °C).
Natural abundance 1H,13C heteronuclear single quantum correlation (1H-13C HSQC) spectrum in D2O was recorded with sensitivity enhancement and gradient coherence selection optimized for selection of aliphatic CH groups (JCH = 135 Hz) using 64 scans, 1024/2048 complex data points, and 12,072/7210 Hz spectral widths in t1 and t2, respectively. For the selection of aromatic CH groups, 170 Hz was used for JCH along with 32 scans and 64/2048 complex data points. The two-dimensional HSQC-TOCSY spectrum was recorded with a pulse program consisted of an HSQC building block followed by a clean MLEV-17 TOCSY transfer step of 80 ms of mixing time just prior to the refocusing gradients with exactly the same spectral widths and number of points as in the aliphatic 1H-13C HSQC.
Distance restraints were derived from cross-peak volumes of the NOESY spectrum recorded with 150 ms of mixing time. Estimated interproton distances were derived using the isolated spin pair approximation, rij = rref (vref/vij)1/6, where rij is the estimated interproton distance, rref is the fixed internal reference distance, and vref and vij are the NOE cross-peak volumes of the reference and estimated cross-peaks, respectively. Average cross-peak volume of the geminal methylene proton pairs was used as reference volume, which corresponds to the fixed reference distance of 1.8 Å. Generally an experimental error of ± 20% on the calculated interproton distances was used as upper and lower bounds. The 3JHNHα coupling constants were measured from the one-dimensional proton spectrum recorded in H2O and then converted to dihedral restraints as follows: 3JHNHα > 8 Hz, ϕ = −120 ± 30°; 3JHNHα < 6 Hz; ϕ = −60 ± 30°; ω = 180 ± 30° to define the trans X-Pro7/14 conformation as confirmed by the observation of strong NOE interactions between Hα(n) and HD2, HD3 (n+1) Pro.
All structure calculations were performed by using Xplor-NIH program, version 2.25 (29). A set of 100 structures was generated by torsion angle molecular dynamics, starting from an extended strand and by using NMR-derived restraints. After the torsion angle molecular dynamics round (30), the majority of the structures had converged to very similar structures with similar total energies and having no violations of the NOE and dihedral restraints. The 15 lowest energy structures were used for further refinement during a “gentle molecular dynamics” round in explicit water (31). A box of water was constructed and optimized around selected structures obtained from the previous torsion angle dynamics step. The final stage of refinement commenced with a 20-ps constant temperature molecular dynamics simulation at 300 K (20,000 steps of 0.001 ps) and was followed by a 200-step conjugate gradient energy minimization of the average structure of the last 10 ps of the 20-ps simulation. Structures were analyzed by using PROCHECK (32). Visual representations of the molecule were created by using the University of California, San Francisco Chimera program (version 1.8rc).
RESULTS
Isolation of a Novel α-Conotoxin from C. longurionis Venom
N-terminal Edman degradation of the purified active peptide revealed a novel 18-residue α-conotoxin, called Lo1a, with the sequence H-EGCCSNPACRTNHPEVCD-NH2 (bridges Cys3–Cys9 and Cys4–Cys17) and a molecular mass of 1930.12 Da, determined by MALDI-TOF (4800 Analyzer; Applied Biosystems). To the best of our knowledge, Lo1a is the first conotoxin isolated and pharmacologically characterized from C. longurionis, a species of a (vermivorous) cone snail commonly found in the Indian Ocean in Tamil Nadu, India. It has highest sequence homology with Qc1.5 (81%; Fig. 1), which is isolated from Conus quercinus, another vermivorous cone snail (33).
FIGURE 1.
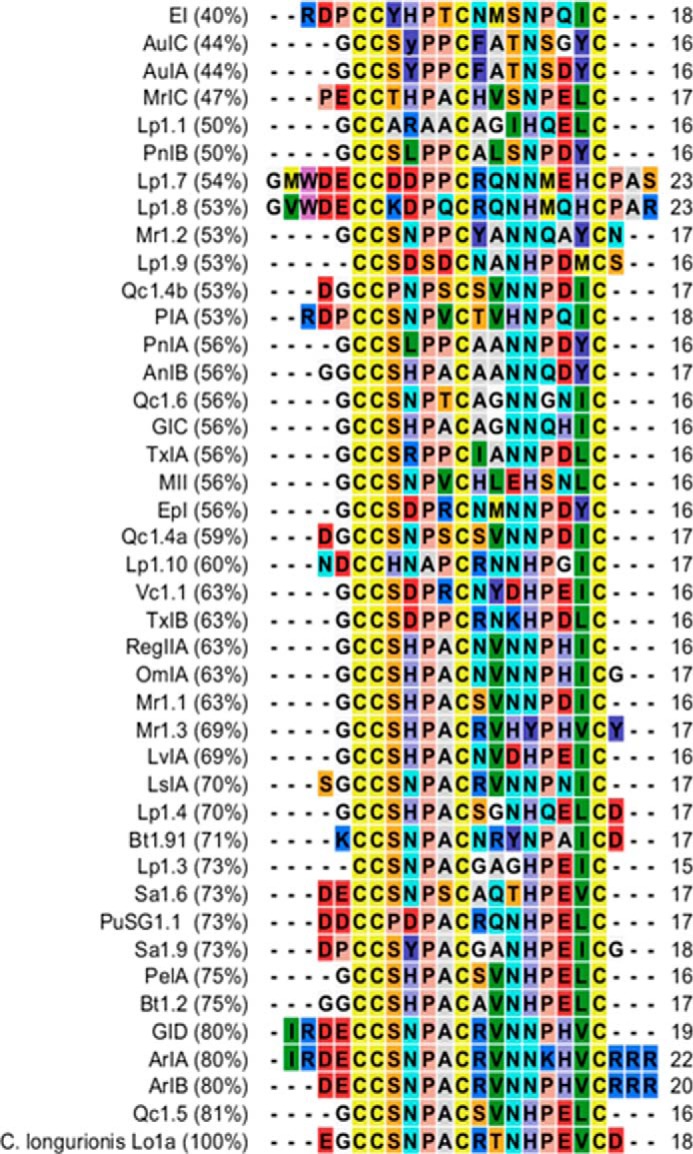
Alignment of 41 α4/7 conotoxins with Lo1a, sorted by percentage of identity (indicated in the left column) with Lo1a. References from top to bottom are as follows: EI (C. ermineus (63)), AuIC (C. aulicus (72)), AuIA (C. aulicus (72)), MrIC (C. marmoreus (73)), Lp1.1 (C. leopardus (74)), PnIB (C. pennaceus (75)), Lp1.7 (C. leopardus (35)), Lp1.8 (C. leopardus (35)), Mr1.2 (C. marmoreus (35)), Lp1.9 (C. leopardus (35)), Qc1.4b (C. quercinus (35)), PIA (C. purpurascens (62)), PnIA (C. pennaceus (75)), AnIB (C. anemone (49)), Qc1.6 (C. quercinus (35)), GIC (C. geographus (76)), TxIA (C. textile (45)), MII (C. magus (55)), EpI (C. episcopates (77)), Qc1.4a (C. quercinus (35)), Lp1.10 (C. leopardus (35)), Vc1.1 (C. victoriae (53)), TxIB (C. textile (35)), RegIIA (C. regius (52), OmIA (C. omaria (50)), Mr1.1 (C. marmoreus (34)), Mr1.3 (C. marmoreus (35)), LvIA (C. lividus (35)), LsIA (C. limpusi (53)), Lp1.4 (C. leopardus (34)), Bt1.91 (C. betulinus (35)), Lp1.3 (C. leopardus (35)), Sa1.6 (C. sanguinolentus (35)), PuSG1.1 (C. pulicarius (35)), Sa1.9 (C. sanguinolentus (35)), PeIA (C. pergrandis (18)), Bt1.2 (C. betulinus (35)), GID (C. geographus (51)), ArIA (C. arenatus (61)), ArIB (C. arenatus (61)), Qc1.5 (C. quercinus (35)), and Lo1a (C. longurionis, this work).
α-Conotoxin Lo1a Analogues
Typically, α-conotoxins end C-terminally with a Cys (Fig. 1). Interestingly, Lo1a ends with an Asp following this Cys. Only two more conotoxins having an Asp at the C terminus are described, namely Lp1.4 (34) and Bt1.91 (35). To explore the influence of the C-terminal Asp18 to the functional properties of Lo1a, we created two synthetic analogues: Lo1a-ΔD and Lo1a-RRR. The synthetic analogue Lo1a-ΔD (molecular mass, 1815.03 Da) is C-terminally truncated, whereas in Lo1a-RRR (molecular mass, 2283.60 Da), the negatively charged Asp is replaced by a positively charged Arg tail.
Lo1a Inhibition of Recombinant nAChR Subtypes
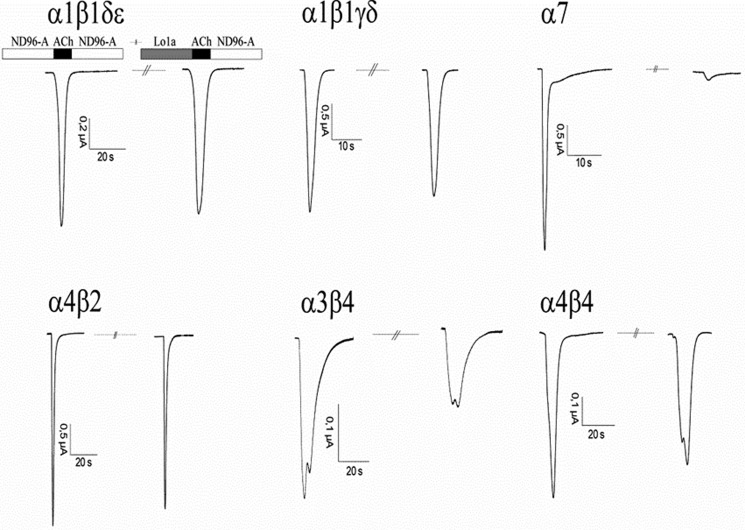
The potency and selectivity of Lo1a at neuronal nAChRs was determined by examining its effect on ACh-evoked currents mediated by different nAChR subunit combinations expressed in Xenopus oocytes. Lo1a (10 μm) inhibited ACh-evoked current amplitude mediated by α7 (85%), α3β4 (40%), α4β2 (19%), and α4β4 (13%) (Fig. 2). No remarkable effect was seen at the muscle subtype nAChRs α1β1γδ and α1β1δϵ (for concentrations up to 50 μm). Concentration-response curves for Lo1a inhibition of ACh-evoked currents at the α7 nAChR revealed an IC50 value of 3.24 ± 0.70 μm (Fig. 3).
FIGURE 2.
Electrophysiological profile of Lo1a. During recordings, oocytes were continuously perfused with ND96-A (ND96 with 1 μm atropine; except for oocytes expressing α7 nAChR) at a rate of 2 ml/min, with 10 μm Lo1a applied for 30 s before ACh was added. ACh (200 μm for α7, 70 μm for α3β4, 50 μm for α4β2 and α4β4, and 10 μm for α1β1γδ and α1β1δϵ) was applied for 2 s at 2 ml/min, with 30-s washout periods between different ACh applications and 200 s after toxin application.
FIGURE 3.
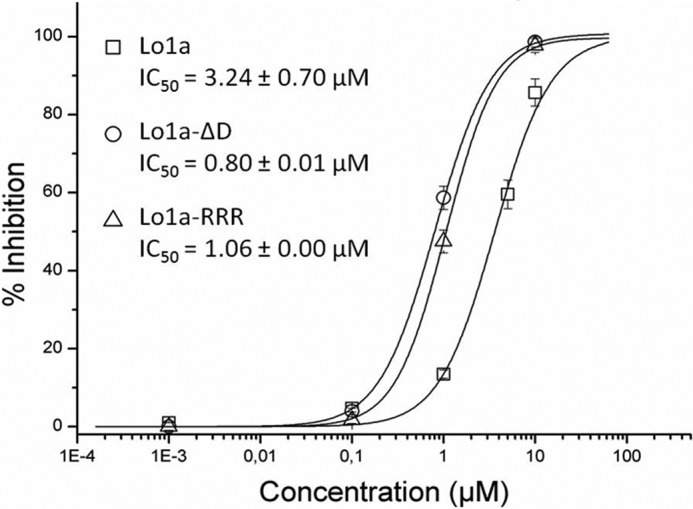
Concentration-response curve on α7 nAChR. Different concentration of Lo1a (□), Lo1a-ΔD (○), and Lo1a-RRR (▵) were tested on α7. The corresponding IC50 values are: 3.24 ± 0.70, 0.80 ± 0.01, and 1.06 ± 0.00 μm, respectively. Currents were obtained as described under “Experimental Procedures.” The percentage of block was plotted against the logarithm of the tested concentrations. The results were fit with the Hill equation.
Influence of C-terminal Truncation and Replacement of D by RRR
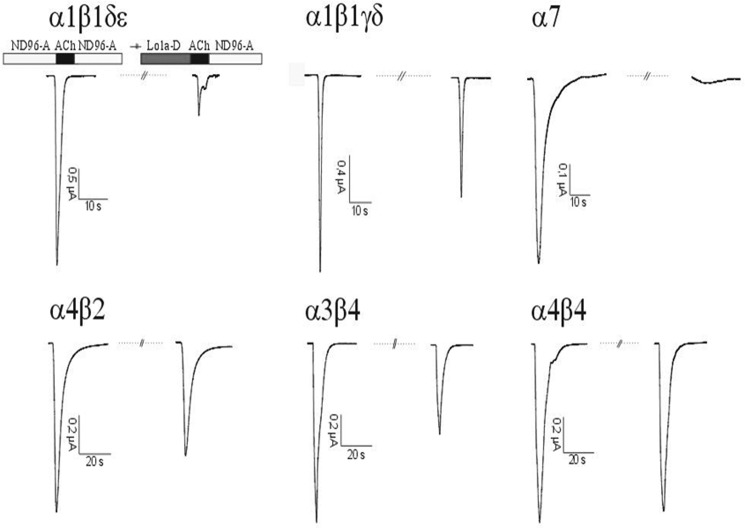
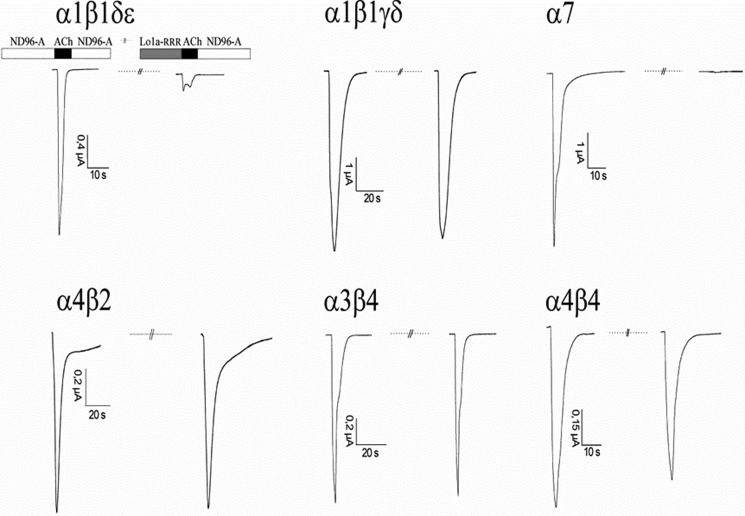
We synthesized a C-terminally truncated and a triple Arg-tailed analogue of Lo1a to examine its activity at different nAChR subtypes (Figs. 4 and 5). Interestingly, Lo1a-ΔD was more potent at the neuronal nAChR α7 (IC50 = 0.80 ± 0.01 nm) but surprisingly also adopted affinity for the muscle subtype α1β1δϵ (IC50 = 4.40 ± 0.18 μm) (Fig. 6). The same was noticed for Lo1a-RRR, where the IC50 for α7 was 1.06 μm, and an IC50 value of 1.47 ± 0.39 μm was determined for α1β1δϵ (Fig. 6). An overview of subtype selectivity for Lo1a and both mutants is given in Fig. 7.
FIGURE 4.
Electrophysiological profile of Lo1a-ΔD. During recordings, oocytes were continuously perfused with ND96-A (ND96 with 1 μm atropine; except for oocytes expressing α7 nAChR) at a rate of 2 ml/min, with 10 μm Lo1a-ΔD applied for 30 s before ACh was added. ACh (200 μm for α7, 70 μm for α3β4, 50 μm for α4β2 and α4β4, and 10 μm for α1β1γδ and α1β1δϵ) was applied for 2 s at 2 ml/min, with 30-s washout periods between different ACh applications and 200 s after toxin application.
FIGURE 5.
Electrophysiological profile of Lo1a-RRR. During recordings, oocytes were continuously perfused with ND96-A (ND96 with 1 μm atropine; except for oocytes expressing α7 nAChR) at a rate of 2 ml/min, with 10 μm Lo1a-RRR applied for 30 s before ACh was added. ACh (200 μm for α7, 70 μm for α3β4, 50 μm for α4β2 and α4β4, and 10 μm for α1β1γδ and α1β1δϵ) was applied for 2 s at 2 ml/min, with 30-s washout periods between different ACh applications and 200 s after toxin application.
FIGURE 6.
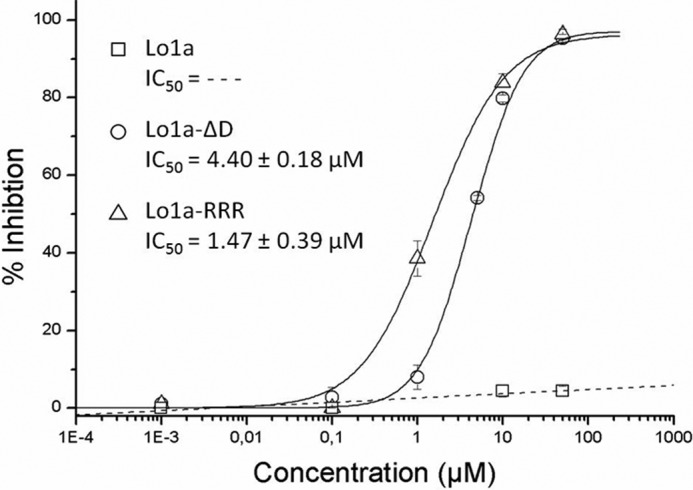
Concentration-response curve on α1β1δϵ nAChR. Different concentrations of Lo1a (□), Lo1a-ΔD (○), and Lo1a-RRR (▵) were tested on α1β1δϵ. The corresponding IC50 values for Lo1a-ΔD and Lo1a-RRR are: 4.40 ± 0.18 and 1.47 ± 0.39 μm, respectively. Currents were obtained as described under “Experimental Procedures.” The percentage of block was plotted against the logarithm of the tested concentrations. The results were fit with the Hill equation.
FIGURE 7.
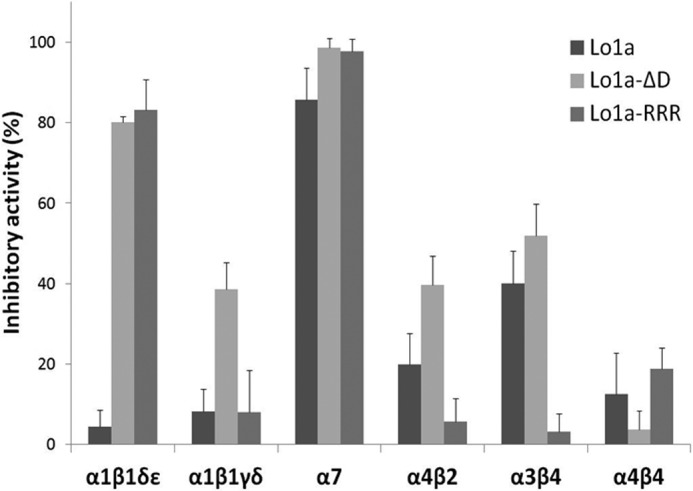
Overview of subtype selectivity. The three peptides, Lo1a, Lo1a-ΔD, and Lo1a-RRR were tested on six different nAChRs: α1β1δϵ, α1β1γδ, α7, α4β2, α3β4, and α4β4 at a concentration of 10 μm. The data are represented as the means ± S.E. (indicated by error bars); each experiment was performed at least three times (n ≥ 3).
NMR Spectroscopy
NMR spectral analysis shows the formation of a single set of resonances for the conopeptide, indicating that it adopts one type of structural form in solution. Resonance assignment was performed by following standard procedures as outlined by Wüthrich (36). Complete sequence specific proton assignments were achieved by analyzing homonuclear two-dimensional spectra (double quantum-filtered COSY, TOCSY, and NOESY). Initially the NH and Hα resonances of the individual spin systems (except Pro) were identified by analyzing the “fingerprint” region of the double quantum-filtered COSY and TOCSY spectra, and the remaining resonances of the spin systems were identified by following the “TOCSY tower.” Sequence specific assignments were achieved by linking individual spin system via sequential inter-residue Hαn-HN(n+1) cross-peaks in the fingerprint region of the NOESY spectrum. Carbon assignments were also performed by using 1H-13C HSQC spectra, which further reconfirmed most of the homonuclear proton assignments and clarified the Hβ and Hγ proton assignments of the two proline residues that were not resolved in the homonuclear two-dimensional spectra because of signal overlaps. The geminal methylene protons were not assigned stereo-specifically, and the NOE distance restraints involving these protons were used ambiguously during structure calculation in the Xplor-NIH program. Fig. 8 shows the observed short and medium range NOEs that were used for the resonance assignment, 3JHNHα, and chemical shift index along the amino acid sequences of α-conotoxin Lo1a. The chemical shift index values indicate the presence of a α-helix in the middle of the peptide from residue Pro7 to Asn12.
FIGURE 8.
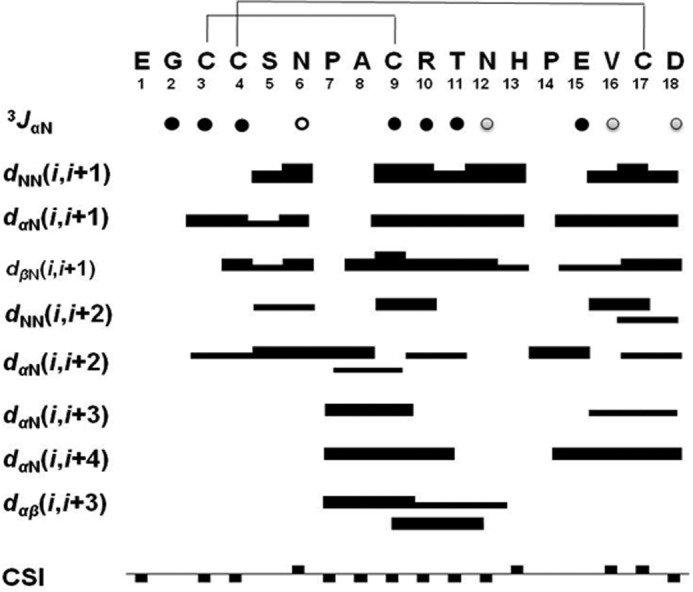
Sequence and disulfide connectivity of α-conotoxin Lo1a along with a summary of short and medium range NOEs, 3JHNHα (3JαN) couplings, and chemical shift index for the Hα protons. The thickness of each bar represents relative strength of NOEs (strong, medium, and weak). Filled circle, 3JHNHα < 6 Hz; shaded circle, 3JHNHα = 6–8 Hz; open circle, 3JHNHα > 8 Hz. The filled rectangles above and below the horizontal line represent chemical shift index values of +1 and −1, respectively. Note the presence of α-helix from Pro7 to Asn12, which is common to all α4/7 subfamily conotoxins.
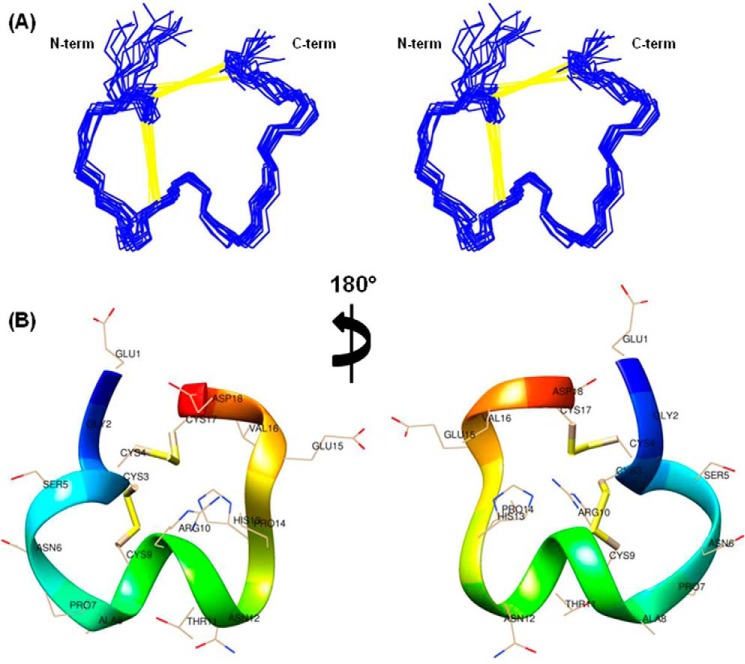
Fig. 9 displays the structural representations of the final ensemble of 15 superimposed structures (Fig. 9A) and the minimum energy closest to average structure (Fig. 9B) of α-conotoxin Lo1a. The peptide structures are well defined with backbone and heavy atom root mean square deviations of 0.43 and 0.83, respectively, over residues 3–17. The structural evaluation using PROCHECK demonstrates that all the resulted structures have no bad nonbonded contacts, and all the backbone dihedral angles are within the allowed regions of the Ramachandran plot (63% residues fall in the most favored region). Detailed structure determination statistics are provided in Table 1. The coordinates for 15 structures, NMR restraints, and chemical shifts have been deposited in the RCSB Protein Data Bank with RCSB identifier RCSB103496 and Protein Data Bank code 2MD6.
FIGURE 9.
NMR solution structure of α-conotoxin Lo1a. A, stereo views of a family of 15 final structures of α-conotoxin Lo1a with superimposed backbone heavy atoms (N, CA, C′). Disulfide bonds are shown in yellow as pseudo bonds. B, front and back ribbon views of the closest to average structure of α-conotoxin Lo1a. Side chain heavy atoms are displayed along with residue label. The two views are related by a 180° rotation around the vertical axis.
TABLE 1.
NMR structure determination statistics of α-conotoxin Lo1a for an ensemble of 15 structures
The values where applicable are the means ± S.D.
| Total NOE distance restraints | 190 |
|---|---|
| Intraresidue | 65 |
| Sequential (|i − j| = 1) | 66 |
| Medium range (2 ≤|i − j| ≤4) | 46 |
| Long range (|i − j| ≥ 5) | 13 |
| Dihedral angle restraints | 10 (ϕ = 8, ωX-Pro = 2) |
| RMSD from the average structure () | |
| Residues 3–17 | |
| Backbone atoms (N, Cα, C′) | 0.43 ± 0.09 |
| All heavy atoms | 0.83 ± 0.13 |
| Root mean square deviation from the idealized covalent geometry | |
| Bond () | 0.000008 ± 0.000000 |
| Angle (°) | 3.809 ± 0.074 |
| Improper (°) | 3.245 ± 0.412 |
| Ramachandran analysis (%) | |
| Residues in favored regions | 63 |
| Residues in additional allowed regions | 30.3 |
| Residues in generously allowed regions | 6.7 |
| Residues in disallowed regions | 0 |
A close look into the structure (Fig. 9) of the α-conotoxin Lo1a reveals that the peptide backbone adopts a compact W-shaped conformation having two loops that are reinforced by two disulfide bonds (Cys3–Cys9 and Cys4–Cys17). Residues from Pro7 to Asn12 at the bottom of the W shape formed a α-helix involving two turns. In addition to this, in the N-terminal part of the peptide structure, two overlapping β-turns (type IV) exist between Gly2 and Ser5 and between Cys4 and Pro7. In the C-terminal part of the peptide, residues from Pro14 to Cys17 formed a type I/IV θ β-turn. This type of overall W-shaped molecular topology/fold was previously identified in the reported structures of other α4/7 subfamily α-conotoxins (37, 38).
DISCUSSION
α-Conotoxins are a family of cysteine-rich peptides that behave pharmacologically as competitive antagonists of the nicotinic acetylcholine receptor (39). In general, there are two main nAChR subtypes, the neuronal and muscle subtype nAChRs. Considering the neuronal subtype nAChRs, and particularly their ACh-binding site, a high percentage of sequence identity exists among the known neuronal nAChR subunits (40, 41). Because of the high sequence conservation, it has been difficult to obtain subtype selective ligands, principally agonists that take action deep within the conserved ACh-binding pocket. However, recently, selective peptide antagonists from cone snail venom have shown to be highly selective pharmacological tools displaying the ability to discriminate among many of the different nAChR subunit combinations (41). This high selectivity toward a particular mammalian nAChR subtype is often established through specific interactions with particular residues located outside the conserved ACh-binding site (42). Nowadays, co-crystal structures of α-conotoxins binding to the ACh-binding protein (43), which is a protein model for the extracellular ligand-binding domain of nAChRs, can offer useful information about the molecular interactions of these small peptides (44, 45).
Lo1a Has a Typical α-Conotoxin Structure
In our study, we revealed the amino acid composition and the three-dimensional structure of a new α4/7-conotoxin from C. longurionis, called Lo1a. The peptide was investigated electrophysiologically, and it was found that Lo1a inhibits α7 nAChRs preferentially. Once the activity of Lo1a had been determined, we employed NMR-based techniques to elucidate the structure of this novel peptide. According to Marx et al. (46), NMR is the method of choice for determining conotoxin structures, because conotoxins are generally difficult to crystallize and are not amenable to x-ray methods, with a few exceptions such as PnIA and PnIB (47, 48). α-Conotoxin Lo1a was shown to be highly soluble, and NMR spectra were readily assigned and used to generate a high resolution structure. This structure demonstrated that Lo1a shares many of the structural and biochemical properties that define α-conotoxins, including the characteristic I–III, II–IV disulfide connectivity and the size of the first and the second loop of the peptide, i.e. four and seven residues, respectively. Several α-conotoxins including AnIB (49), OmIA (50), GID (51), RegIIA (52), and LsIA (5) have an SXPA motif similar to the first loop of Lo1a. The peptide also shares the common fold comprising a short disulfide bond stabilized helix and a conserved proline. This proline is shown to be the only highly conserved amino acid residue apart from the cysteines and is responsible for helix initiation by inducing the 310 helix turn in the peptide backbone (39). Dutertre and Lewis (39) revealed that this small α-helix structure appears to be a very important determinant for binding, even if the orientations and specific interactions differ significantly. Because the side chains in an α-helix protrude at 360°, this structure is likely suitable to allow multiple contacts on both sides of a binding pocket located at the interface of two subunits (39).
As indicated in the previous section, Lo1a was shown to be very selective for the neuronal versus the muscle nicotinic receptors. Other α-conotoxins having these characteristics are ImI, α4/3 conotoxin from Conus imperialis (54) and MII, α4/7 conotoxin from Conus magus (55). ImI targets α7 and α9 subunits with an IC50 of 220 nm and 1.8 μm, respectively (56). Its amino acid sequence, GCCSDPRCAWRC, shares three non-Cys amino acid residues with the sequence of Lo1a. These similar residues are Gly at the N terminus, Ser after the second Cys, and Pro between the second and the third Cys. The latter was identified as one of the determinants, in addition to Asp5, Arg7, and Trp10 to influence the potency of ImI at the α7 nAChR (57, 58). α-Conotoxin MII blocks α6-containing nAChRs and α3β2 with an IC50 of 0.39 nm (α6/α3β2β3) and 2.18 nm, respectively (59). If its amino acid sequence, GCCSNPVCHLEHSNLC, is aligned with Lo1a, 5 of 14 non-Cys residues of Lo1a are similar with MII, resulting in a homology percentage of 56 (Fig. 1). Everhart et al. (60) identified amino acid residues Asn5, Pro6, and His12 from MII as major determinants for potency on the α3β2 nAChR. Consequently, it is expected that Lo1a also targets α3β2 nAChRs.
Lo1a Has an Atypical C Terminus
The N-terminal amino acid residue of α-conotoxins is typically a glycine followed by the first two cysteine residues (Fig. 1). However, sequence alignment of Lo1a with other α-conotoxins reveals, in addition to a unique loop 2 sequence, an atypical terminal amino acid sequence, because both N and C termini contain negatively charged amino acid residues, i.e. glutamic acid and aspartic acid, respectively. Few conotoxins have other residues preceding the first cysteine. From these conotoxins, ArIA (61), ArIB (61), GID (51), PIA (62), EI (63), and LsIA (5) are the most investigated (Fig. 1). On the other hand, a negative residue at the C terminus of an α-conotoxin has only been observed in conotoxins Lp1.4 from Conus leopardus (33) and Bt1.91 from Conus betulinus (35). However, to the best of our knowledge, the influence of this negative C-terminal residue has never been investigated before.
C-terminal Charges Are Important for Neuronal/Muscle Subtype Selectivity and Efficacy
To explore the role of the C-terminal Asp of Lo1a, two analogues were synthesized: one C-terminally truncated peptide named Lo1a-ΔD and another named Lo1a-RRR with an extremely positively charged Arg tail at the C terminus. Deleting the C-terminal amino acid resulted in a more positively charged peptide, whereas the rationale for the Arg tailing peptide can be found in the sequences of ArIA and ArIB from Conus arenatus (Fig. 1). Both peptides, ArIA and ArIB, are potent inhibitors of the α7 nAChR having IC50 values in the nanomolar range (61). Whiteaker et al. (61) made a series of directed substitutions in ArIB to obtain a more selective α7 antagonist. The mutant ArIB[V11L,V16D] improved its selectivity on α7 more than 10,000-fold, suggesting the importance of Lys and Asp at positions 11 and 16, respectively. Via replacing the C-terminal Asp by a triple Arg tail, we aimed to investigate whether we could improve potency/selectivity on α7 with Lo1a-RRR.
The two synthetic C-terminally modified analogues revealed unexpected roles for the C terminus in determining subtype selectivity and efficacy. Both mutants demonstrated more potency toward the neuronal α7 AChR because both IC50 values decreased from 3.24 μm (Lo1a) to 0.80 μm (Lo1a-ΔD) and 1.06 μm (Lo1a-RRR). Because the difference in potency was not very astonishing, we can conclude that the C-terminal residue(s) does not play a crucial role in activity on α7. Remarkably, both mutants conferred affinity for the adult muscle subtype nAChR α1β1δϵ (IC50 values 4.40 ± 0.18 μm (Lo1a-ΔD) and 1.47 ± 0.39 μm (Lo1a-RRR)) but much less for the fetal muscle subtype nAChR α1β1γδ. Consequently, C-terminal charges seem to have an influence in making the distinction between neuronal or muscle subtype nAChRs and even among muscle subtype nAChRs.
α-Conotoxins that target selectively muscle subtype nAChRs typically have a 3/5 structure (64). An exception is the α-conotoxin EI (α4/7) from Conus ermineus that selectively targets the α/δ interface of muscle subtype nAChRs (63). Conotoxins that distinguish between the adult and the fetal muscle subtype nAChRs are rare. One example is ψ-conotoxin PrIIIE from Conus parius, characterized by Lluisma et al. (65), which showed higher inhibition potency against the adult subtype (IC50 of 245 nm) than the fetal subtype nAChR (IC50 of 3.24 μm). Another ψ-conotoxin PIIIE from Conus purpurascens shows an IC50 of 7.4 μm on the adult muscle subtype, but no inhibition on the fetal muscle subtype for concentrations up to 10 μm. Teichert et al. (66) reports αA-conotoxin OIVB from Conus obscurus, a unique selective inhibitor of the mammalian fetal muscle nAChR (IC50 of 56 nm), whereas affinity for the adult muscle nAChR is more than 1800-fold lower. According to Groebe et al. (67), many of the α-conotoxins bind with 10,000-fold higher affinity to the mammalian α/δ interface than the α/γ interface. The peptides Lo1a-ΔD and Lo1a-RRR, which are described in this work, apparently demonstrate higher affinity to the α/ϵ interface.
Considering the terminal charges of Lo1a and its analogues, it can be expected that the positively charged triple-Arg tail in Lo1a-RRR is more likely to adopt a conformation extended away from the negatively charged N terminus (as derived by torsion-angle molecular dynamics and energy minimization in the structural model). In contrast, the structure from Lo1a has a more “compact” conformation, with an inward-facing C terminus as derived from the solution state NMR structural data. Consequently, because the C and N terminus of Lo1a are in close proximity (less than 10 nm), the charges at both termini may interact, playing a role in making the distinction between neuronal versus muscle subtype nAChRs. Further structure-function studies combined with co-crystallization experiments are necessary to see whether this hypothesis is applicable for Lo1a-ΔD.
Muscle and Neuronal Subtype nAChR Binding Sites: Structural Receptor Elements for Binding of Lo1a and Its Homologous
The muscle subtype nAChR has a pentameric structure comprised of two α1 subunits, one β1, one δ, and, depending on whether the receptor is in an embryonic or adult stage, one γ or ϵ subunit, respectively. Each α1 subunit folds such that the primary binding site directly faces a neighboring subunit, which is either a γ/ϵ or a δ subunit. The γ subunit is believed to be the one that forms stable contacts being the lone subunit between the two α1 subunits, whereas the δ subunit pairs with the β subunit to form stable contacts between the α1 subunits on the opposite side. Because two α1 subunits are separated by at least one non α1 subunit, correct coupling between these subunits is required for cooperative binding of agonists (68). Agonists of the muscle subtype nAChR initiate channel opening and desensitization by binding to a site on each of these two α1 subunits (69). Moreover, Arias and Blanton (19) established that two adjacent cysteines (at positions 192 and 193 according to the sequence number of Torpedo AChR) in the α1 subunits are involved in the recognition and binding of cholinergic agonists and competitive antagonists. Later on, Sine (70) demonstrated that not only do the two α1 subunits form the binding sites of agonists and antagonists, but the γ/(ϵ) and δ subunits are also involved. Agonists and antagonists can specifically distinguish between the α1γ/(α1ϵ) and α1δ binding sites of the fetal/(adult) muscle acetylcholine receptor because of different contributions by the γ/(ϵ) and δ subunits where a minimum of four loops in both subunits is required to create the agonist binding site (71). In 1991, Sine and Claudio (68) showed that AChRs lacking either a γ or a δ subunit assemble into pentamers with subunit compositions, (α)2β(γ)2 and (α)2β(δ)2. Therefore, we tried to express (α)2β(γ)2 and (α)2β(δ)2 and (α)2β(ϵ)2 heterologously to determine which binding site is responsible for interactions causing the observed results. Unfortunately, because of poor heterologous expression in frog oocytes, these experiments could not provide sufficient information about the binding sites of Lo1a-ΔD and Lo1a-RRR.
Whereas the two ligand-binding sites of muscle AChRs are formed at interfaces between α1 and δ, ϵ, or γ subunits, binding sites of α7 neuronal AChRs are formed at interfaces between identical α7 subunits. Residues of the α face of the binding site, termed the (+) face, cluster in three well separated regions of the primary sequence, termed loops A, B, and C (71). The stabilization of this loop C conformation in an open state appears to be an essential element for antagonist activity. The stabilization in the ACh-binding protein is established on the basis of the loop C vicinal disulfide bonds where the α-conotoxin disulfide bond Cys I–III interacts, as is shown for conotoxins PnIA and ImI (39). This type of antagonist activity can also be expected for Lo1a and its analogues, because both conotoxins, PnIA and ImI, are structurally quite homologous to Lo1a. In summary, our study provides insight into α-conotoxin pharmacology and the molecular basis of nAChR selectivity, highlighting the influence and importance of C-terminal residues on conotoxin pharmacology.
Acknowledgments
We thank L. Sivilotti for sharing the hα3 and hβ4 cDNA. We are grateful to V. Witzemann for sharing the rα1, rβ1, rγ, rδ, rϵ1 clones and to J. Lindstrom for kindly providing the hα4 and hβ2 clones. We thank F. Clementi for sharing rα5 and hα7.
Footnotes
- nAChR
- nicotinic acetylcholine receptor
- ACh
- acetylcholine
- ACN
- acetonitrile
- TOCSY
- total correlation spectroscopy
- HSQC
- heteronuclear single quantum correlation.
REFERENCES
- 1. Alberola-Die A., Martinez-Pinna J., González-Ros J. M., Ivorra I., Morales A. (2011) Multiple inhibitory actions of lidocaine on Torpedo nicotinic acetylcholine receptors transplanted to Xenopus oocytes. J. Neurochem. 117, 1009–1019 [DOI] [PubMed] [Google Scholar]
- 2. Gold A. B., Lerman C. (2012) Pharmacogenetics of smoking cessation: role of nicotine target and metabolism genes. Hum. Genet. 131, 857–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rezvani A. H., Levin E. D. (2001) Cognitive effects of nicotine. Biol. Psychiatry 49, 258–267 [DOI] [PubMed] [Google Scholar]
- 4. Cooper E., Couturier S., Ballivet M. (1991) Pentameric structure and subunit stoichiometry of a neuronal nicotinic acetylcholine receptor. Nature 350, 235–238 [DOI] [PubMed] [Google Scholar]
- 5. Inserra M. C., Kompella S. N., Vetter I., Brust A., Daly N. L., Cuny H., Craik D. J., Alewood P. F., Adams D. J., Lewis R. J. (2013) Isolation and characterization of α-conotoxin LsIA with potent activity at nicotinic acetylcholine receptors. Biochem. Pharmacol. 86, 791–799 [DOI] [PubMed] [Google Scholar]
- 6. Colquhoun L. M., Patrick J. W. (1997) Pharmacology of neuronal nicotinic acetylcholine receptor subtypes. Adv. Pharmacol. 39, 191–220 [DOI] [PubMed] [Google Scholar]
- 7. Le Novère N., Changeux J. P. (1995) Molecular evolution of the nicotinic acetylcholine receptor: an example of multigene family in excitable cells. J. Mol. Evol. 40, 155–172 [DOI] [PubMed] [Google Scholar]
- 8. Gotti C., Fornasari D., Clementi F. (1997) Human neuronal nicotinic receptors. Prog. Neurobiol. 53, 199–237 [DOI] [PubMed] [Google Scholar]
- 9. Couturier S., Bertrand D., Matter J. M., Hernandez M. C., Bertrand S., Millar N., Valera S., Barkas T., Ballivet M. (1990) A neuronal nicotinic acetylcholine receptor subunit (α7) is developmentally regulated and forms a homo-oligomeric channel blocked by α-BTX. Neuron 5, 847–856 [DOI] [PubMed] [Google Scholar]
- 10. Rubboli F., Court J. A., Sala C., Morris C., Chini B., Perry E., Clementi F. (1994) Distribution of nicotinic receptors in the human hippocampus and thalamus. Eur. J. Neurosci. 6, 1596–1604 [DOI] [PubMed] [Google Scholar]
- 11. Wevers A., Jeske A., Lobron C., Birtsch C., Heinemann S., Maelicke A., Schröder R., Schröder H. (1994) Cellular distribution of nicotinic acetylcholine receptor subunit mRNAs in the human cerebral cortex as revealed by non-isotopic in situ hybridization. Brain Res. Mol. Brain Res. 25, 122–128 [DOI] [PubMed] [Google Scholar]
- 12. Breese C. R., Adams C., Logel J., Drebing C., Rollins Y., Barnhart M., Sullivan B., Demasters B. K., Freedman R., Leonard S. (1997) Comparison of the regional expression of nicotinic acetylcholine receptor α7 mRNA and [125I]α-bungarotoxin binding in human postmortem brain. J. Comp. Neurol. 387, 385–398 [DOI] [PubMed] [Google Scholar]
- 13. Gotti C., Clementi F. (2004) Neuronal nicotinic receptors: from structure to pathology. Prog. Neurobiol. 74, 363–396 [DOI] [PubMed] [Google Scholar]
- 14. Sacco K. A., Bannon K. L., George T. P. (2004) Nicotinic receptor mechanisms and cognition in normal states and neuropsychiatric disorders. J. Psychopharmacol. 18, 457–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Steinlein O. K., Bertrand D. (2010) Nicotinic receptor channelopathies and epilepsy. Pflugers Arch. 460, 495–503 [DOI] [PubMed] [Google Scholar]
- 16. Quinton L., Servent D., Girard E., Molgó J., Le Caer J. P., Malosse C., Haidar el A., Lecoq A., Gilles N., Chamot-Rooke J. (2013) Identification and functional characterization of a novel α-conotoxin (EIIA) from Conus ermineus. Anal. Bioanal. Chem. 405, 5341–5351 [DOI] [PubMed] [Google Scholar]
- 17. Sine S. M., Kreienkamp H. J., Bren N., Maeda R., Taylor P. (1995) Molecular dissection of subunit interfaces in the acetylcholine receptor: identification of determinants of α-conotoxin M1 selectivity. Neuron 15, 205–211 [DOI] [PubMed] [Google Scholar]
- 18. McIntosh J. M., Plazas P. V., Watkins M., Gomez-Casati M. E., Olivera B. M., Elgoyhen A. B. (2005) A novel α-conotoxin, PeIA, cloned from Conus pergrandis, discriminates between rat α9α10 and α7 nicotinic cholinergic receptors. J. Biol. Chem. 280, 30107–30112 [DOI] [PubMed] [Google Scholar]
- 19. Arias H. R., Blanton M. P. (2000) α-Conotoxins. Int. J. Biochem. Cell Biol. 32, 1017–1028 [DOI] [PubMed] [Google Scholar]
- 20. Olivera B. M., Rivier J., Clark C., Ramilo C. A., Corpuz G. P., Abogadie F. C., Mena E. E., Woodward S. R., Hillyard D. R., Cruz L. J. (1990) Diversity of Conus neuropeptides. Science 249, 257–263 [DOI] [PubMed] [Google Scholar]
- 21. Grønlien J. H., Håkerud M., Ween H., Thorin-Hagene K., Briggs C. A., Gopalakrishnan M., Malysz J. (2007) Distinct profiles of α7 nAChR positive allosteric modulation revealed by structurally diverse chemotypes. Mol. Pharmacol. 72, 715–724 [DOI] [PubMed] [Google Scholar]
- 22. Tucker J. K., Tenorio M. J. (2009) Systematic Classification of Recent and Fossil Conoidean Gastropods: With Keys to the Genera of Cone Shells, ConchBooks, Hackenheim, Germany [Google Scholar]
- 23. Olivera B. M., Gray W. R., Zeikus R., McIntosh J. M., Varga J., Rivier J., de Santos V., Cruz L. J. (1985) Peptide neurotoxins from fish-hunting cone snails. Science 230, 1338–1343 [DOI] [PubMed] [Google Scholar]
- 24. Van Der Haegen A., Peigneur S., Tytgat J. (2011) Importance of position 8 in mu-conotoxin KIIIA for voltage-gated sodium channel selectivity. FEBS J. 278, 3408–3418 [DOI] [PubMed] [Google Scholar]
- 25. Keller M., Steiger R. (2004) The pi plate: an implant for unstable extension fractures of the distal radius in patients with osteoporotic bone. Tech. Hand Up Extrem. Surg. 8, 212–218 [DOI] [PubMed] [Google Scholar]
- 26. Hwang T. L., Shaka A. J. (1998) Multiple-pulse mixing sequences that selectively enhance chemical exchange or cross-relaxation peaks in high-resolution NMR spectra. J. Magn. Reson. 135, 280–287 [DOI] [PubMed] [Google Scholar]
- 27. Shaka A. J., Lee C. J., Pines A. (1988) Iterative schemes for bilinear operators: application to spin decoupling. J. Magn. Reson. 77, 274–293 [Google Scholar]
- 28. Derome A. E., Williamson M. P. (1990) Rapid-pulsing artifacts in double-quantum-filtered COSY. J. Magn. Reson. 88, 177–185 [Google Scholar]
- 29. Schwieters C. D., Kuszewski J. J., Tjandra N., Clore G. M. (2003) The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 160, 65–73 [DOI] [PubMed] [Google Scholar]
- 30. Stein E. G., Rice L. M., Brünger A. T. (1997) Torsion-angle molecular dynamics as a new efficient tool for NMR structure calculation. J. Magn. Reson. 124, 154–164 [DOI] [PubMed] [Google Scholar]
- 31. Spronk C. A., Linge J. P., Hilbers C. W., Vuister G. W. (2002) Improving the quality of protein structures derived by NMR spectroscopy. J. Biomol. NMR 22, 281–289 [DOI] [PubMed] [Google Scholar]
- 32. Laskowski R. A., Macarthur M. W., Moss D. S., Thornton J. M. (1993) Procheck: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 33. Yuan D. D., Han Y. H., Wang C. G., Chi C. W. (2007) From the identification of gene organization of α conotoxins to the cloning of novel toxins. Toxicon 49, 1135–1149 [DOI] [PubMed] [Google Scholar]
- 34. Peng C., Chen W., Sanders T., Chew G., Liu J., Hawrot E., Chi C. (2010) Chemical synthesis and characterization of two α4/7-conotoxins. Acta Biochim. Biophys. Sin. 42, 745–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kaas Q., Yu R., Jin A. H., Dutertre S., Craik D. J. (2012) ConoServer: updated content, knowledge, and discovery tools in the conopeptide database. Nucleic Acids Res. 40, D325–D330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wüthrich K. (1986) NMR of Proteins and Nucleic Acids, John Wiley & Sons Inc., New York [Google Scholar]
- 37. Park K. H., Suk J. E., Jacobsen R., Gray W. R., McIntosh J. M., Han K. H. (2001) Solution conformation of α-conotoxin EI, a neuromuscular toxin specific for the α1/δ subunit interface of Torpedo nicotinic acetylcholine receptor. J. Biol. Chem. 276, 49028–49033 [DOI] [PubMed] [Google Scholar]
- 38. Chi S. W., Kim D. H., Olivera B. M., McIntosh J. M., Han K. H. (2004) Solution conformation of α-conotoxin GIC, a novel potent antagonist of α3β2 nicotinic acetylcholine receptors. Biochem. J. 380, 347–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dutertre S., Lewis R. J. (2006) Toxin insights into nicotinic acetylcholine receptors. Biochem. Pharmacol. 72, 661–670 [DOI] [PubMed] [Google Scholar]
- 40. Le Novère N., Corringer P. J., Changeux J. P. (2002) The diversity of subunit composition in nAChRs: evolutionary origins, physiologic and pharmacologic consequences. J. Neurobiol. 53, 447–456 [DOI] [PubMed] [Google Scholar]
- 41. Tsetlin V. I., Hucho F. (2004) Snake and snail toxins acting on nicotinic acetylcholine receptors: fundamental aspects and medical applications. FEBS Lett. 557, 9–13 [DOI] [PubMed] [Google Scholar]
- 42. Dutertre S., Lewis R. J. (2004) Computational approaches to understand α-conotoxin interactions at neuronal nicotinic receptors. Eur. J. Biochem. 271, 2327–2334 [DOI] [PubMed] [Google Scholar]
- 43. Brejc K., van Dijk W. J., Klaassen R. V., Schuurmans M., van Der Oost J., Smit A. B., Sixma T. K. (2001) Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature 411, 269–276 [DOI] [PubMed] [Google Scholar]
- 44. Armishaw C., Jensen A. A., Balle T., Clark R. J., Harpsøe K., Skonberg C., Liljefors T., Strømgaard K. (2009) Rational design of α-conotoxin analogues targeting α7 nicotinic acetylcholine receptors: improved antagonistic activity by incorporation of proline derivatives. J. Biol. Chem. 284, 9498–9512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dutertre S., Ulens C., Büttner R., Fish A., van Elk R., Kendel Y., Hopping G., Alewood P. F., Schroeder C., Nicke A., Smit A. B., Sixma T. K., Lewis R. J. (2007) AChBP-targeted α-conotoxin correlates distinct binding orientations with nAChR subtype selectivity. EMBO J. 26, 3858–3867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Marx U. C., Daly N. L., Craik D. J. (2006) NMR of conotoxins: structural features and an analysis of chemical shifts of post-translationally modified amino acids. Magn. Reson. Chem. 44, S41–S50 [DOI] [PubMed] [Google Scholar]
- 47. Hu S. H., Gehrmann J., Guddat L. W., Alewood P. F., Craik D. J., Martin J. L. (1996) The 1.1 A crystal structure of the neuronal acetylcholine receptor antagonist, α-conotoxin PnIA from Conus pennaceus. Structure 4, 417–423 [DOI] [PubMed] [Google Scholar]
- 48. Hu S. H., Gehrmann J., Alewood P. F., Craik D. J., Martin J. L. (1997) Crystal structure at 1.1 A resolution of α-conotoxin PnIB: comparison with α-conotoxins PnIA and GI. Biochemistry 36, 11323–11330 [DOI] [PubMed] [Google Scholar]
- 49. Loughnan M. L., Nicke A., Jones A., Adams D. J., Alewood P. F., Lewis R. J. (2004) Chemical and functional identification and characterization of novel sulfated α-conotoxins from the cone snail Conus anemone. J. Med. Chem. 47, 1234–1241 [DOI] [PubMed] [Google Scholar]
- 50. Talley T. T., Olivera B. M., Han K. H., Christensen S. B., Dowell C., Tsigelny I., Ho K. Y., Taylor P., McIntosh J. M. (2006) α-Conotoxin OmIA is a potent ligand for the acetylcholine-binding protein as well as α3β2 and α7 nicotinic acetylcholine receptors. J. Biol. Chem. 281, 24678–24686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nicke A., Loughnan M. L., Millard E. L., Alewood P. F., Adams D. J., Daly N. L., Craik D. J., Lewis R. J. (2003) Isolation, structure, and activity of GID, a novel α 4/7-conotoxin with an extended N-terminal sequence. J. Biol. Chem. 278, 3137–3144 [DOI] [PubMed] [Google Scholar]
- 52. Franco A., Kompella S. N., Akondi K. B., Melaun C., Daly N. L., Luetje C. W., Alewood P. F., Craik D. J., Adams D. J., Marí F. (2012) RegIIA: an α4/7-conotoxin from the venom of Conus regius that potently blocks α3β4 nAChRs. Biochem. Pharmacol. 83, 419–426 [DOI] [PubMed] [Google Scholar]
- 53. Sandall D. W., Satkunanathan N., Keays D. A., Polidano M. A., Liping X., Pham V., Down J. G., Khalil Z., Livett B. G., Gayler K. R. (2003) A novel α-conotoxin identified by gene sequencing is active in suppressing the vascular response to selective stimulation of sensory nerves in vivo. Biochemistry 42, 6904–6911 [DOI] [PubMed] [Google Scholar]
- 54. McIntosh J. M., Yoshikami D., Mahe E., Nielsen D. B., Rivier J. E., Gray W. R., Olivera B. M. (1994) A nicotinic acetylcholine receptor ligand of unique specificity, α-conotoxin ImI. J. Biol. Chem. 269, 16733–16739 [PubMed] [Google Scholar]
- 55. Cartier G. E., Yoshikami D., Gray W. R., Luo S., Olivera B. M., McIntosh J. M. (1996) A new α-conotoxin which targets α3β2 nicotinic acetylcholine receptors. J. Biol. Chem. 271, 7522–7528 [DOI] [PubMed] [Google Scholar]
- 56. Johnson D. S., Martinez J., Elgoyhen A. B., Heinemann S. F., McIntosh J. M. (1995) α-Conotoxin ImI exhibits subtype-specific nicotinic acetylcholine receptor blockade: preferential inhibition of homomeric α7 and α9 receptors. Mol. Pharmacol. 48, 194–199 [PubMed] [Google Scholar]
- 57. Quiram P. A., Sine S. M. (1998) Structural elements in α-conotoxin ImI essential for binding to neuronal α7 receptors. J. Biol. Chem. 273, 11007–11011 [DOI] [PubMed] [Google Scholar]
- 58. Quiram P. A., Sine S. M. (1998) Identification of residues in the neuronal α7 acetylcholine receptor that confer selectivity for conotoxin ImI. J. Biol. Chem. 273, 11001–11006 [DOI] [PubMed] [Google Scholar]
- 59. McIntosh J. M., Azam L., Staheli S., Dowell C., Lindstrom J. M., Kuryatov A., Garrett J. E., Marks M. J., Whiteaker P. (2004) Analogs of α-conotoxin MII are selective for α6-containing nicotinic acetylcholine receptors. Mol. Pharmacol. 65, 944–952 [DOI] [PubMed] [Google Scholar]
- 60. Everhart D., Cartier G. E., Malhotra A., Gomes A. V., McIntosh J. M., Luetje C. W. (2004) Determinants of potency on α-conotoxin MII, a peptide antagonist of neuronal nicotinic receptors. Biochemistry 43, 2732–2737 [DOI] [PubMed] [Google Scholar]
- 61. Whiteaker P., Christensen S., Yoshikami D., Dowell C., Watkins M., Gulyas J., Rivier J., Olivera B. M., McIntosh J. M. (2007) Discovery, synthesis, and structure activity of a highly selective α7 nicotinic acetylcholine receptor antagonist. Biochemistry 46, 6628–6638 [DOI] [PubMed] [Google Scholar]
- 62. Dowell C., Olivera B. M., Garrett J. E., Staheli S. T., Watkins M., Kuryatov A., Yoshikami D., Lindstrom J. M., McIntosh J. M. (2003) α-Conotoxin PIA is selective for α6 subunit-containing nicotinic acetylcholine receptors. J. Neurosci. 23, 8445–8452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Martinez J. S., Olivera B. M., Gray W. R., Craig A. G., Groebe D. R., Abramson S. N., McIntosh J. M. (1995) α-Conotoxin EI, a new nicotinic acetylcholine receptor antagonist with novel selectivity. Biochemistry 34, 14519–14526 [DOI] [PubMed] [Google Scholar]
- 64. Lewis R. J., Dutertre S., Vetter I., Christie M. J. (2012) Conus venom peptide pharmacology. Pharmacol. Rev. 64, 259–298 [DOI] [PubMed] [Google Scholar]
- 65. Lluisma A. O., López-Vera E., Bulaj G., Watkins M., Olivera B. M. (2008) Characterization of a novel psi-conotoxin from Conus parius Reeve. Toxicon 51, 174–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Teichert R. W., Rivier J., Torres J., Dykert J., Miller C., Olivera B. M. (2005) A uniquely selective inhibitor of the mammalian fetal neuromuscular nicotinic acetylcholine receptor. J. Neurosci. 25, 732–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Groebe D. R., Dumm J. M., Levitan E. S., Abramson S. N. (1995) α-Conotoxins selectively inhibit one of the two acetylcholine binding sites of nicotinic receptors. Mol. Pharmacol. 48, 105–111 [PubMed] [Google Scholar]
- 68. Sine S. M., Claudio T. (1991) γ- and δ-subunits regulate the affinity and the cooperativity of ligand binding to the acetylcholine receptor. J. Biol. Chem. 266, 19369–19377 [PubMed] [Google Scholar]
- 69. Changeux J. P. (1990) The TiPS lecture: the nicotinic acetylcholine receptor: an allosteric protein prototype of ligand-gated ion channels. Trends Pharmacol. Sci. 11, 485–492 [DOI] [PubMed] [Google Scholar]
- 70. Sine S. M. (1993) Molecular dissection of subunit interfaces in the acetylcholine receptor: identification of residues that determine curare selectivity. Proc. Natl. Acad. Sci. U.S.A. 90, 9436–9440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Prince R. J., Sine S. M. (1996) Molecular dissection of subunit interfaces in the acetylcholine receptor. Identification of residues that determine agonist selectivity. J. Biol. Chem. 271, 25770–25777 [DOI] [PubMed] [Google Scholar]
- 72. Luo S., Kulak J. M., Cartier G. E., Jacobsen R. B., Yoshikami D., Olivera B. M., McIntosh J. M. (1998) α-Conotoxin AuIB selectively blocks α3β4 nicotinic acetylcholine receptors and nicotine-evoked norepinephrine release. J. Neurosci. 18, 8571–8579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Jin A. H., Vetter I., Dutertre S., Abraham N., Emidio N. B., Inserra M., Murali S. S., Christie M. J., Alewood P. F., Lewis R. J. (2014) MrIC, a novel α-conotoxin agonist in the presence of PNU at endogenous α7 nicotinic acetylcholine receptors. Biochemistry 53, 1–3 [DOI] [PubMed] [Google Scholar]
- 74. Peng C., Han Y., Sanders T., Chew G., Liu J., Hawrot E., Chi C., Wang C. (2008) α4/7-Conotoxin Lp1.1 is a novel antagonist of neuronal nicotinic acetylcholine receptors. Peptides 29, 1700–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Luo S., Nguyen T. A., Cartier G. E., Olivera B. M., Yoshikami D., McIntosh J. M. (1999) Single-residue alteration in α-conotoxin PnIA switches its nAChR subtype selectivity. Biochemistry 38, 14542–14548 [DOI] [PubMed] [Google Scholar]
- 76. McIntosh J. M., Dowell C., Watkins M., Garrett J. E., Yoshikami D., Olivera B. M. (2002) α-Conotoxin GIC from Conus geographus, a novel peptide antagonist of nicotinic acetylcholine receptors. J. Biol. Chem. 277, 33610–33615 [DOI] [PubMed] [Google Scholar]
- 77. Loughnan M., Bond T., Atkins A., Cuevas J., Adams D. J., Broxton N. M., Livett B. G., Down J. G., Jones A., Alewood P. F., Lewis R. J. (1998) α-Conotoxin EpI, a novel sulfated peptide from Conus episcopatus that selectively targets neuronal nicotinic acetylcholine receptors. J. Biol. Chem. 273, 15667–15674 [DOI] [PubMed] [Google Scholar]