Background: Striatins are novel regulatory subunits of PP2A in the striatin-interacting phosphatase and kinase (STRIPAK) complex.
Results: The striatin coiled coil forms a noncanonical dimer required for PP2A A subunit binding.
Conclusion: The coiled coil of striatins bind PP2A A subunits to form a 2:2 heterotetrameric core of the STRIPAK complex.
Significance: The current structural analysis provides insights into the assembly of the STRIPAK complex.
Keywords: Apoptosis, Phosphatase, PP2A, Protein Complexes, Protein Kinases, Protein Structure, Coiled Coil, Parallel Dimer, STRIPAK Complex, Striatin
Abstract
The protein phosphatase 2A (PP2A) and kinases such as germinal center kinase III (GCKIII) can interact with striatins to form a supramolecular complex called striatin-interacting phosphatase and kinase (STRIPAK) complex. Despite the fact that the STRIPAK complex regulates multiple cellular events, it remains only partially understood how this complex itself is assembled and regulated for differential biological functions. Our recent work revealed the activation mechanism of GCKIIIs by MO25, as well as how GCKIIIs heterodimerize with CCM3, a molecular bridge between GCKIII and striatins. Here we dissect the structural features of the coiled coil domain of striatin 3, a novel type of PP2A regulatory subunit that functions as a scaffold for the assembly of the STRIPAK complex. We have determined the crystal structure of a selenomethionine-labeled striatin 3 coiled coil domain, which shows it to assume a parallel dimeric but asymmetric conformation containing a large bend. This result combined with a number of biophysical analyses provide evidence that the coiled coil domain of striatin 3 and the PP2A A subunit form a stable core complex with a 2:2 stoichiometry. Structure-based mutational studies reveal that homodimerization of striatin 3 is essential for its interaction with PP2A and therefore assembly of the STRIPAK complex. Wild-type striatin 3 but not the mutants defective in PP2A binding strongly suppresses apoptosis of Jurkat cells induced by the GCKIII kinase MST3, most likely through a mechanism in which striatin recruits PP2A to negatively regulate the activation of MST3. Collectively, our work provides structural insights into the organization of the STRIPAK complex and will facilitate further functional studies.
Introduction
Striatin-interacting phosphatase and kinase (STRIPAK)4 is a recently identified supramolecular complex, in which striatins act as molecular scaffolds to recruit the protein phosphatase 2A (PP2A) and kinases including germinal center kinases (GCKs) (Fig. 1A) (1–3). PP2A holoenzyme, which consists of a scaffolding (A) subunit, a regulatory (B) subunit, and a catalytic (C) subunit, is a multifunctional serine/threonine phosphatase that regulates a variety of biological processes (4–6). There are four known families of PP2A regulatory subunits: B/B55/PR55, B′/B56/PR61, B″/PR48/PR72/PR130, and B‴/PR93/PR110. Striatins are a novel family of B‴ type regulatory subunits that directly interact with PP2A A subunits, and indirectly interact with some GCKs using adaptor molecules (Fig. 1A) (7–12). Because inhibition of PP2A using okadaic acid causes phosphorylation of MOB and hyperphosphorylation of striatins, PP2A may thus regulate striatins and the associated MOB proteins to modify the cytoskeleton and its interactions with membrane structures (10, 11).
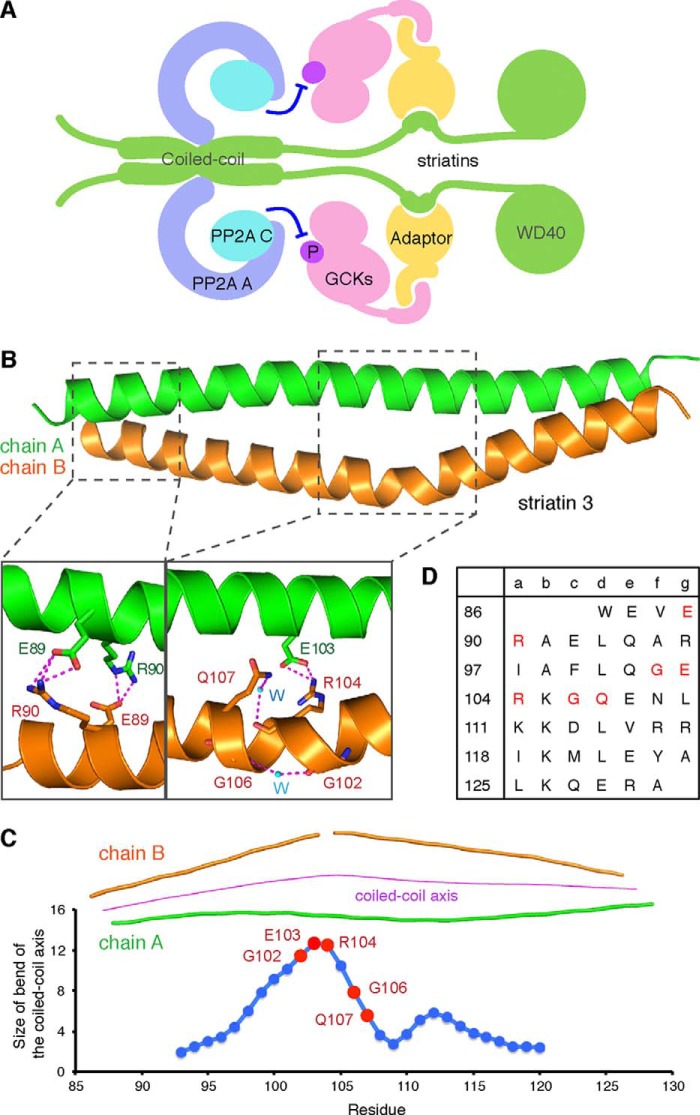
FIGURE 1.
Striatin 3 coiled coil domain forms an asymmetric parallel dimer. A, organization of the STRIPAK complex. Striatins are novel PP2A B‴ subunits that interact with PP2A A and C subunits via their coiled coil domains, and bind GCKs via adaptors. B, overall structure of the SeMet-labeled striatin 3 coiled coil domain. Chains A and B of striatin 3 are depicted as helical ribbons and colored green and orange, respectively. The coiled coil domain forms an asymmetric dimer, in which chain B contains a large bend. Noncanonical residues in the coiled coil result in formation of the asymmetric dimer. C, size of bend of the coiled coil axis. D, analysis of the heptad repeats of the striatin 3 coiled coil. Residues that may be involved in asymmetry of striatin 3 coiled coil are colored red.
Several GCKs have been identified as components of the STRIPAK complex, including MST1/2, MST3, MST4, STK25, and Misshapen-like kinase 1 (MINK) (1, 2, 9, 10, 13). Diverse biological functions have been documented for these kinases. For example, MST1/2 kinases are core components of the mammalian Hippo pathway, which is important for organ size control and normal development (14). MST4 may act downstream of the tumor suppressor liver kinase B1 (LKB1) to induce brush-border formation in intestinal epithelial cells (15). Multiple evidence including mass spectrometry, immunoprecipitation, and pulldown analysis indicate that kinases like MST3, MST4, or STK25 can bind striatins through an adaptor called CCM3 (also named PDCD10) to form the STRIPAK complex with PP2A A and C subunits (1, 2, 9, 10). It is thought that the PP2A holoenzyme within the STRIPAK complex acts as a negative regulator of the kinase components (10). Despite the important biological roles of these kinase components, the structural assembly and regulatory mechanism of the STRIPAK complex remain only partially understood.
The mammalian striatin proteins include striatin, striatin 3 (also named SG2NA), and striatin 4 (also named zinedin) (16–19). The members of the striatin family share four highly conserved domains: a caveolin-binding domain, a coiled coil domain, a calmodulin-binding domain, and a WD40 repeat domain. The caveolin-binding domain of striatins can directly bind caveolin-1 (20), a scaffolding protein within the caveolar membrane that can associate with TRAF2 (21), CD40 (22), and Gαq (23) to regulate T cell activation and proliferation. The coiled coil domain mediates homo- or hetero-oligomerization of striatins, which is important for not only the subcellular localization of striatins but also the assembly of STRIPAK (24). The calmodulin-binding domain of striatins binds to calmodulin in a Ca2+-dependent manner (25, 26). The WD40 repeat domain of striatins can interact with the armadillo repeat domain of adenomatous polyposis coli, a multifunctional tumor suppressor that negatively regulates the Wnt signaling pathway (27).
The striatin proteins are localized in dendritic spines of the striatum, which is associated with the motor system (25, 28, 29). Down-regulation of striatin impairs locomotor activity and inhibits the growth of dendrites (30). Localization of striatins to dendritic spines is regulated by cortactin-binding protein 2 (CTTNBP2) (31), which interacts with and regulates the mobility of cortactin to control dendritic spine formation (32). In addition to the aforementioned binding partners, striatins may associate with estrogen receptor α and Gαi to form a membrane signaling complex that facilitates rapid non-genomic signaling of estrogen receptor α (8, 33). Disruption of estrogen receptor-striatin interaction eliminates the ability of estrogen to stimulate cultured endothelial cell migration and to inhibit cultured vascular smooth muscle cell growth (34). Striatins were also found to interact with phocein, which has been implicated in vesicular trafficking, especially in the endocytic process (35). As striatins are highly concentrated in dendritic spines, their interactions with phocein might be involved in mediating synaptic plasticity through spine remodeling by endocytosis.
Previously we determined the structure of the STRIPAK component MST4 in complex with its adaptors MO25 (36) and CCM3 (37). To better understand the STRIPAK complex, we performed structural and functional studies of striatin. The striatin 3 coiled coil domain is shown to be sufficient for PP2A binding and to adopt a dimeric parallel but asymmetric conformation with a large bend. Mutational studies identified key amino acid residues important for striatin 3 homodimerization, and those mutants defective in homodimerization showed dramatically decreased binding to PP2A. The functional importance of striatin dimerization is further supported by our finding that overexpression of wild-type striatin 3, but not dimerization defective mutants, inhibited MST3-induced apoptosis in Jurkat cells.
EXPERIMENTAL PROCEDURES
Cloning, Protein Expression, and Purification
Residues 86–131 of human striatin 3 (which dimerizes to form the coiled coil domain) were cloned into pET28a with a His-SUMO tag and a tobacco etch virus protease cleavage site at the N terminus and expressed in Escherichia coli BL21 (DE3) CodonPlus cells (Stratagene). Protein expression was induced by 0.4 mm isopropyl β-d-thiogalactopyranoside at A600 = 0.8 in Terrific Broth medium and cells were cultured for an additional 18 h at 16 °C. Cells were then harvested by centrifugation and resuspended in lysis buffer (20 mm Hepes, 500 mm NaCl, 5% glycerol, 1 mm DTT, and 20 mm imidazole, pH 7.5). The cell homogenates were lysed by High Pressure Homogenizer (JNBIO3000 plus) at 1800 bar and cell debris was removed by centrifugation at 20,000 × g for 40 min. The soluble fraction was loaded to Ni-Sepharose pre-equilibrated with lysis buffer and the proteins were eluted with elution buffer (20 mm Hepes, 500 mm NaCl, 5% glycerol, 1 mm DTT, and 400 mm imidazole, pH 7.5). The eluted protein was desalted to lysis buffer and then cleaved by tobacco etch virus protease. The cleaved His-SUMO tag was removed by rebinding to Ni-Sepharose. Then the sample was concentrated and applied to a Superdex 75 column pre-equilibrated with 20 mm Hepes, 100 mm NaCl, 1 mm DTT, pH 7.5. The purity of proteins was evaluated by SDS-PAGE.
The selenomethionine (SeMet)-labeled striatin 3 coiled coil domain with Phe-99 and Tyr-123 mutated to methionines was expressed in E. coli BL21(DE3) CodonPlus cells cultured in M9 medium containing amino acid supplement (lysine, phenylalanine, and threonine to final concentration of 100 mg/liter, isoleucine, leucine, and valine to 50 mg/liter, and l-SeMet to 60 mg/liter). Note that these two mutations maintain the large apolar nature of the native side chains; they are also located at c and f heptad positions (Fig. 1D), which are highly exposed positions of α-helical coiled coils located away from the interhelical interface and that usually do not significantly influence the conformation of the coiled coil. SeMet-labeled protein was purified using the same procedure employed for the native protein.
For cross-linking experiments, wild-type or mutant striatin 3 coiled coil was cloned into pET28a with a MBP tag and a tobacco etch virus protease cleavage site at the N terminus. The proteins were expressed as described above and purified by amylose resin and size exclusive chromatography.
The human PP2A A subunit (amino acids 8–589) was cloned into pGEX4T-1 and expressed as described above. The protein was loaded to glutathione-Sepharose and eluted by reduced glutathione. The GST tag was cleaved by thrombin and removed by rebinding to glutathione-Sepharose. The sample was further purified by Mono Q and Superdex 200 column. For cell-based assays, Myc-tagged striatin 3, FLAG-tagged striatin 3, and HA-tagged PP2A A were cloned into pcDNA3.1.
Crystallization, Structure Determination, and Refinement
Crystallization trials were carried out at 18 °C by the sitting-drop vapor diffusion method. The sitting drops consisted of 1 μl of protein solution and 1 μl of reservoir solution and were equilibrated against 100 μl of reservoir solution. Crystals were grown in reservoir solution consisting of 0.1 m BisTris, pH 6.5, 0.2 m MgCl2, 23% PEG 3350. The crystals were then soaked in a cryoprotection solution composed of 0.1 m BisTris, pH 6.5, 0.2 m MgCl2, 23% PEG 3350, and 10% glycerol and flash frozen in liquid nitrogen.
Diffraction data of native and SeMet-derived crystals were collected on beamlines BL17U at Shanghai Synchrotron Radiation Facility (SSRF) and 1W2B at Beijing Synchrotron Radiation Facility (BSRF), respectively, and processed using HKL2000 (38). The structure was solved by the single-wavelength anomalous diffraction method from a SeMet derivative with the program AutoSol in Phenix (39). The structure was refined using phenix.refine and model building was performed in Coot (40, 41).
Cross-linking Experiment
Purified wild-type or mutant MBP-tagged STRN3 was diluted to 1 mg/ml and then incubated with 0.01% (v/v) glutaraldehyde in conjugation buffer (20 mm Hepes, 100 mm NaCl, 1 mm DTT, pH 7.5) at room temperature for 1 h. The reaction was quenched by 50 mm Tris-Cl, pH 8.0, for 30 min, and the cross-linked samples were analyzed on 10% SDS-PAGE.
Immunoprecipitation
HEK293 cells were transiently transfected with the indicated constructs for 24 h. All of the following steps were performed at 4 °C. Cells were harvested and washed three times using phosphate-buffered saline (PBS), and centrifuged at 500 × g for 5 min. Then the pelleted cells were suspended in lysis buffer of RIPA (50 mm Tris, pH 7.5, 150 mm NaCl, 10% glycerol, 0.1% Triton X-100, 0.5 mg/ml of BSA, and protease inhibitor mixture), and lysed by rotating for 30 min. The lysate was centrifuged at 14,000 × g for 30 min, and the supernatants were incubated with antibody and protein A/G-agarose beads (Santa Cruz) overnight. The immune complexes were washed three times with lysis buffer, then subjected to SDS-PAGE, and analyzed by Western blot.
GST Pulldown Assay
GST-fused proteins coupled on glutathione-Sepharose were mixed with different prey proteins at 4 °C for 2 h in 20 mm Hepes, 200 mm NaCl, 1 mm DTT, pH 7.5, and then washed three times. The input and output samples were loaded to SDS-PAGE and detected by Coomassie Blue staining.
Dynamic Light Scattering
Purified striatin 3 and PP2A A subunit were mixed at 4 °C for 1 h at a molar ratio of 2:1 and then loaded on a Superdex 200 column to remove excess striatin 3. The weighted distribution and homogeneity of striatin 3 and PP2A A subunit complex were determined by dynamic light scattering at 25 °C. Scattered light was measured at a 90° angle at a wavelength of 658 nm.
Bio-layer Interferometry Experiment
Bio-layer interferometry (BLI) was performed using Octet Red 96 (ForteBio). The PP2A A subunit was labeled by biotin in 20 mm Hepes, pH 7.5, 100 mm NaCl, 1 mm DTT, and biotinylated proteins were immobilized on streptavidin biosensors and then incubated with various concentrations of striatin 3 in kinetics buffer (137 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 2 mm KH2PO4, pH 7.4, 0.01% BSA, and 0.002% Tween 20). All binding experiments were performed in kinetics buffer at 25 °C. The experiments contained 5 steps: 1) baseline acquisition; 2) biotinylated proteins loading onto SA biosensor; 3) second baseline acquisition; 4) association of interacting protein for kon measurement; and 5) dissociation of interacting protein for koff measurement. Data were analyzed using Octet Data Analysis Software 7.0 (ForteBio). Equilibrium dissociation constants (Kd) were calculated by the ratio of kon to koff.
Flow Cytometry
Jurkat cells were cultured in RPMI 1640 medium and then transfected with different constructs encoding FLAG-tagged MST3 and FLAG-tagged striatin 3. Cells were incubated with annexin V and propidium iodide in PBS with 0.2 mg/ml of RNase A and 0.1% Triton X-100, and analyzed by flow cytometer.
Luciferase Assay
HEK293T cells were transfected with NF-κB-Luc and different constructs of MST3 or striatin 3 in the absence or presence of TNFα. Luciferase activities were determined using the Dual Luciferase Assay System (Promega), and then calculated as the ratio of luciferase/Renilla activity.
RESULTS
The Coiled Coil Domain of Striatin 3 Forms an Asymmetric Parallel Dimer
Previous studies identified the α-helical coiled coil domain of striatin 3 as necessary for its interaction with the PP2A A subunit (9, 10). We expressed and purified recombinant proteins of the coiled coil domain of human striatin 3, and crystallized it in a buffer containing 0.1 m BisTris, pH 6.5, 0.2 m MgCl2, and 23% PEG 3350. The structure of this domain was determined to 2.0-Å resolution by single-wavelength anomalous diffraction from a SeMet derivative crystal. Each asymmetric unit of the crystal contains two α-helical chains, which interact with each other to form a dimeric coiled coil. The structural model was refined to an Rwork of 0.204 and Rfree of 0.252. The details of the refinement statistics are summarized in Table 1.
TABLE 1.
Data collection and refinement statistics
Statistics for the highest-resolution shell are shown in parentheses.
| Native | SeMet | |
|---|---|---|
| Data collection | ||
| Wavelength (Å) | 0.9789 | 0.9793 |
| Resolution range (Å) | 50–2.15 (2.19–2.15) | 50–2.00 (2.03–2.00) |
| Space group | P622 | P3221 |
| Unit cell (Å, °) | a = b = 32.49, c = 181.88 | a = b = 34.06, c = 161.39 |
| α = β = 90, γ = 120 | α = β = 90, γ = 120 | |
| Total reflections | 51,993 | 130,110 |
| Unique reflections | 3,586 | 7,957 |
| Multiplicity | 14.5 (8.9) | 16.2 (10.0) |
| Completeness (%) | 96.6 (99.4) | 99.94 (99.73) |
| Mean I/σ(I) | 26.8 (3.8) | 43.11 (24.19) |
| Rmerge | 0.109 (0.627) | 0.041 (0.083) |
| Refinement | ||
| Rwork | 0.2035 | |
| Rfree | 0.2521 | |
| Number of atoms | 904 | |
| Protein | 790 | |
| Water | 114 | |
| Root mean square bonds) (Å) | 0.008 | |
| Root mean square (angles) (°) | 0.924 | |
| Ramachandran | ||
| Favored (%) | 100 | |
| Outliers (%) | 0 | |
| Average B-factor (Å2) | 19.20 | |
The α-helical coiled coil of human striatin 3 contains a number of features that are unusual for this class of helical structures. In the crystal, the two α-helical chains (named chain A and chain B) form a highly asymmetric parallel dimer, burying a total of 1287.4 Å2 surface area (Fig. 1B). Chain B actually consists of two distinct α-helices, because the main chain H-bond between residues 102 and 106 is broken and these residues are instead connected to each other by an intervening water molecule. As a result, the coiled coil axis contains a very large ∼12.7° bend in this vicinity (calculated using CCBENDS (42)), with chain B tending to wind around the fully intact α-helix of chain A (Fig. 1C).
This central portion of the structure contains a number of residues that are relatively unfavorable for canonical α-helical coiled coil formation but that instead may help promote and/or stabilize the large bend. Polar Arg-104 and Gln-107, respectively, occupy core a and d positions of the heptad repeat, wherein canonical coiled coils more hydrophobic residues are usually found (Fig. 1D). Note that a water molecule contacts both the Arg main chain and the Gln side chain (Fig. 1B). Arg-104 from chain B also forms an H-bonded salt link with Glu-103 (g position) from chain A (Fig. 1B). Moreover, glycines, which are generally underrepresented in the middle of α-helices (43), are found at positions 102 and 106, surrounding the location of the break of the α-helix in chain B. Collectively, these noncanonical residues in the middle of the striatin 3 coiled coil domain result in what we predict is a flexible bend, and help explain the highly asymmetric structure.
In contrast to the presence of such noncanonical features in the center of the coiled coil domain, typical hydrophobic interactions dominate the association between chains A and B elsewhere; these interactions include residues Trp-86, Leu-93, Ile-97, Leu-100, Leu-110, Lys-111, Leu-114, Val-115, Arg-117, Ile-118, Leu-121, Ala-124, and Leu-125 (Fig. 2, A and B). All of these residues are highly conserved in different species from fungi to mammal (Fig. 2C). The coiled coil of striatin 3 is further stabilized by salt bridges involving residues Glu-89, Arg-90, Arg-117, Glu-121, Glu-128, and Arg-129. Note that, in the N terminus, the a-position Arg-90 from each chain forms a salt bridge with the g-position Glu-89 from the other chain (Figs. 1B and 2A), similar to the one g-a′ salt link found in the central portion described above. Additionally, Glu-89 forms an indirect connection with Trp-86 by way of a bridging H-bonding water molecule.
FIGURE 2.
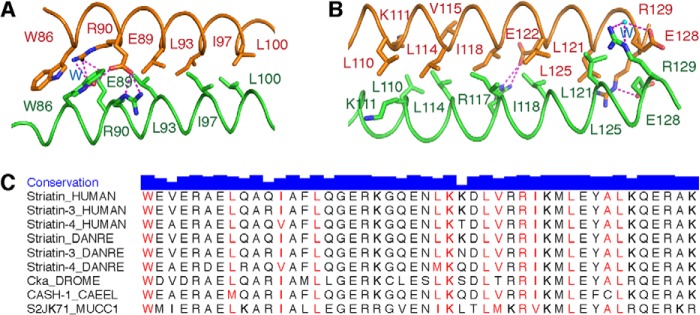
Detailed interface of striatin 3 coiled coil domain. A and B, hydrophobic interactions dominate the association of chains A and B. Salt bridges further stabilize their interaction. C, sequence alignment of striatin 3 from several species, including Homo sapiens (HUMAN), Danio rerio (DANRE), Drosophila melanogaster (DROME), Caenorhabditis elegans (CAEEL), and Mucor circinelloides f. circinelloides (MUCC1). Residue conservation is indicated by the height of the blue bar and residues in the interface are colored red.
Defining Key Residues for Striatin 3 Homodimerization
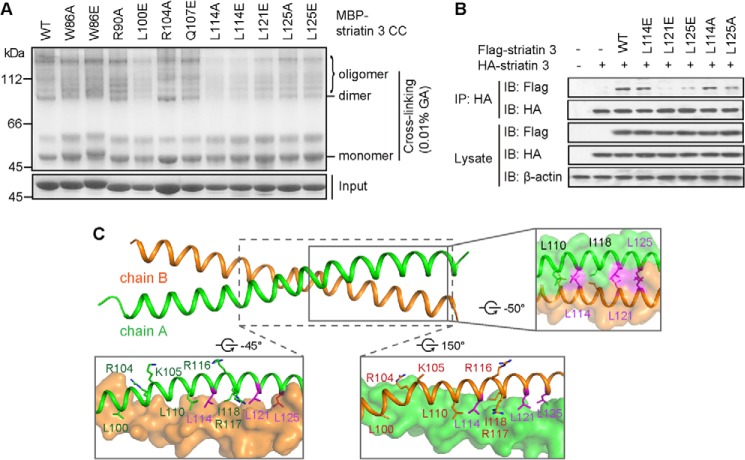
To evaluate the contribution of individual interface residues for striatin 3 dimerization, we constructed a series of striatin 3 mutants based on the structural analysis. MBP-tagged wild-type and mutant striatin 3 coiled coil proteins were purified and used for cross-linking assays. The α-helical chains of the coiled coil domain of wild-type striatin 3 can form dimers and higher-order oligomers. However, mutations W86A, W86E, L100E, L114A, L114E, L121E, L125A, and L125E, but not R90A and R104A, showed increased percentages of monomer (i.e. no coiled coil formation) and decreased percentages of oligomer, indicating impaired homooligomerization, including impaired homodimerization of striatin 3 caused by these mutations (Fig. 3A). Mutations L114A, L114E, L121E, L125A, and L125E were chosen for subsequent biochemical and cell-based studies. We further performed immunoprecipitation assays using the full-length striatin 3 in HEK293 cells. As shown in Fig. 3B, wild-type FLAG-striatin 3 was co-immunoprecipitated with HA-striatin 3, whereas FLAG-striatin 3 mutations L121E, L125E, and L125A exhibited disrupted or abolished homodimerization with HA-striatin 3. In contrast, mutations L114E and L114A of full-length striatin 3 did not show significant effects on homodimerization using this in vivo assay (Fig. 3B), whereas they did using the above in vitro assay, reflecting the fact that fragments may not function in the same way as they do in the full-length state. These results together suggest that hydrophobic residues Leu-121 and Leu-125 are critical for striatin 3 homodimerization (Fig. 3C).
FIGURE 3.
Mutations in the interface of the striatin 3 coiled coil domain impair its dimerization. A, wild-type or mutant MBP-tagged striatin 3 coiled coil (CC) was incubated with 0.01% glutaraldehyde (GA) at room temperature for 1 h. The cross-linked samples were analyzed on 10% SDS-PAGE. B, the indicated constructs of FLAG-tagged and HA-tagged striatin 3 were co-transfected into HEK293 cells. Cell lysate were immunoprecipitated (IP) with anti-HA antibody and subjected to immunoblot (IB) analysis. C, key residues in the interface of striatin 3 coiled coil. Chains A and B are shown as schematic or surface, and key residues are shown as sticks.
The Coiled Coil Domain of Striatin 3 Interacts with the N terminus of the PP2A A Subunit
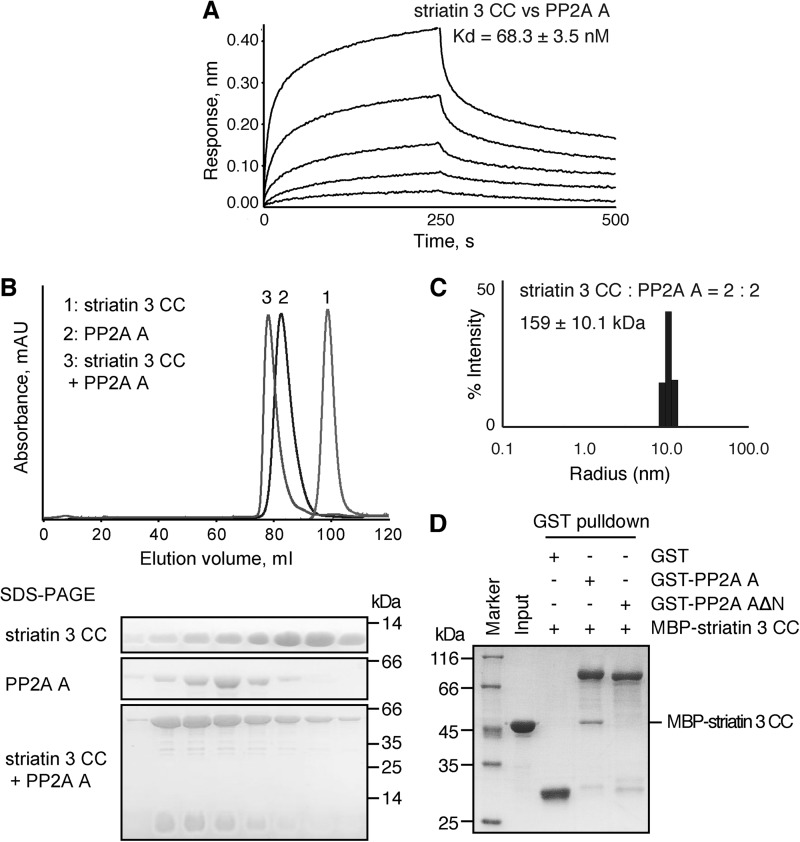
Consistent with previous studies on the association between striatin 3 and PP2A (9, 10), our bio-layer interferometry results showed that the striatin 3 coiled coil binds the PP2A A subunit with a dissociation constant (Kd) of 68.3 ± 3.5 nm (Fig. 4A), indicating a direct interaction between striatin 3 and PP2A A subunit. Furthermore, on size exclusion chromatography, the striatin 3 coiled coil domain and the PP2A A subunit, when mixed together, were eluted from the same peak, which shifted to higher molecular weight as compared with striatin 3 alone and PP2A A subunit alone (Fig. 4B). These results indicate that the coiled coil domain of striatin 3 is sufficient to form a stable complex with the PP2A A subunit. Given that striatin 3 is a homodimer, we speculate that the PP2A A subunit may interact with striatin 3 with a 2:2 stoichiometry. Dynamic light scattering in fact shows that the striatin 3 coiled coil and the PP2A A subunit form a complex in solution with an apparent molecular mass of 159 ± 10.1 kDa (Rh = 5.196 ± 0.171 nm) (Fig. 4C), which is consistent with a predominantly 2:2 heterotetramer of a striatin 3 coiled coil chain and PP2A A subunit whose theoretical molecular masses are 6 and 65 kDa, respectively.
FIGURE 4.
The interaction between striatin 3 and PP2A A subunit. A, binding affinity of the striatin 3 coiled coil with PP2A A subunit was determined by BLI. Biotinylated-PP2A A subunit was immobilized on streptavidin biosensors and incubated with the striatin 3 coiled coil at various concentrations (62.5–1000 nm). Curves are the experimental trace obtained from the BLI experiments. B, striatin 3 coiled coil and/or PP2A A subunit were loaded to a Superdex 200 column; absorbance curves are aligned. Proteins were loaded to SDS-PAGE and detected by Coomassie Blue staining. C, dynamic light scattering analysis of the striatin 3 coiled coil/PP2A A subunit complex. The corresponding molecular weight was calculated to be 159 kDa. D, GST or GST-tagged PP2A A subunit coupled on glutathione-Sepharose was mixed with MBP-tagged striatin 3 coiled coil. The input and output samples were loaded to SDS-PAGE and detected by Coomassie Blue staining.
Several structures of PP2A holoenzyme so far have been reported, showing that PP2A regulatory subunits, such as B/B55/PR55 (PDB code 3DW8), B′/B56/PR61 (PDB code 3FGA), and B″/CDC6 subunits (PDB codes 4I5L and 4I5N), interact with the N terminus of the PP2A A subunit, especially the first three α-helices (44, 45). We then asked whether the first three α-helices of the PP2A A subunit are also important for striatin 3 binding. We constructed a fragment of the PP2A A subunit with the N-terminal three helices truncated. GST pulldown experiments showed that the full-length PP2A A subunit, but not the N-terminal deletion mutant, could pull down the striatin 3 coiled coil, indicating that the N terminus of the PP2A A subunit is crucial for its interaction with striatin 3 (Fig. 4D).
Homodimerization of the Striatin 3 Coiled Coil Is Required for PP2A A Subunit Binding
The dynamic light scattering results described above indicated that striatin 3 and the PP2A A subunit exist predominantly as a 2:2 heterotetramer. To investigate whether striatin 3 homodimerization is required for the formation of the striatin-PP2A complex, we performed the pulldown assay using striatin 3 mutants to examine their interactions with the PP2A A subunit. As shown in Fig. 5A, wild-type striatin 3 and mutations L114E and L114A, which did not affect homodimerization, could pull down the PP2A A subunit. By contrast, mutations L121E, L125E, and L125A of striatin 3, which exhibited impaired homodimerization, showed decreased interactions with the PP2A A subunit (Fig. 5A). Bio-layer interferometry analysis also gave rise to a similar result, except for L114E and L114A, which showed impaired PP2A binding (Fig. 5B) probably due to the sensitivity of this method. Further immunoprecipitation assays were performed in HEK293 cells. Consistent with the results of GST pulldown, wild-type striatin 3, as well as mutations L114E and L114A, were shown to associate with the PP2A A subunit in vivo, whereas mutations L121E, L125E, and L125A did not immunoprecipitate with the PP2A A subunit (Fig. 5C). Together, these results indicate that homodimerization of the striatin 3 coiled coil chains is essential for PP2A A subunit binding.
FIGURE 5.
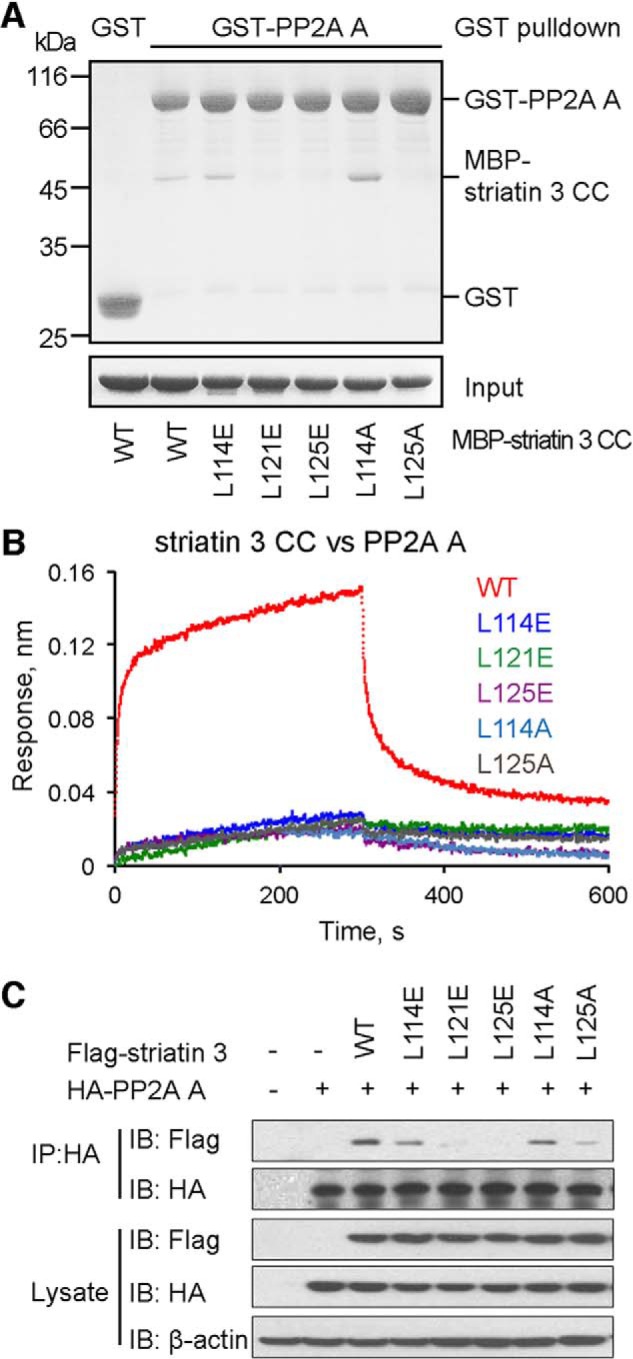
Mutations disrupting the dimerization of striatin 3 impair PP2A A subunit binding. A, GST or GST-tagged PP2A A subunit coupled on glutathione-Sepharose was mixed with wild-type or mutant MBP-tagged striatin 3 coiled coil. The input and output samples were loaded to SDS-PAGE and detected by Coomassie Blue staining. B, binding affinity of wild-type or mutant striatin 3 coiled coil with PP2A A subunit was determined by BLI. Biotinylated-PP2A A subunit was immobilized on streptavidin biosensors and incubated with wild-type or mutant striatin 3 coiled coil. Curves are the experimental trace obtained from the BLI experiments. C, the indicated constructs of FLAG-tagged striatin 3 and HA-tagged PP2A A subunit were co-transfected into HEK293 cells. Cell lysates were immunoprecipitated (IP) with anti-HA antibody and subjected to immunoblot (IB) analysis.
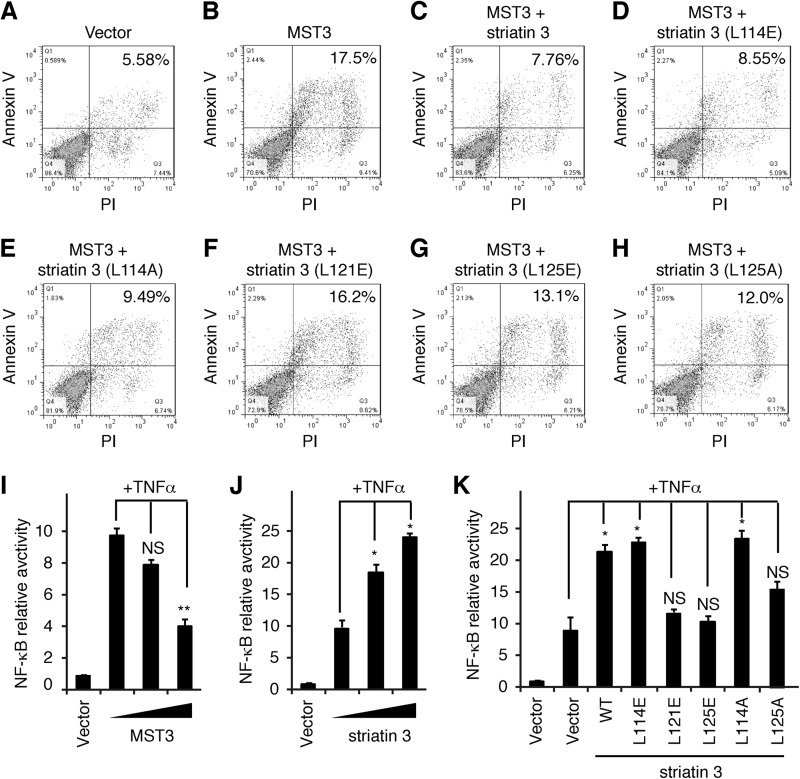
Striatin 3 Inhibits MST3-induced Apoptosis of Jurkat Cells
Previous studies showed that MST3 is involved in both caspase-dependent and -independent apoptosis (46–48). Regardless of these context-dependent differential mechanisms, the kinase activity of MST3 is important for its function in apoptosis. Given that PP2A may negatively regulate the phosphorylation and activation of MST3 by using striatin 3 as a scaffold that recruits both phosphatase and kinase (10), we asked whether striatin 3 could inhibit MST3-induced apoptosis. Annexin V and propidium iodide staining showed that compared with the empty vector, transfection of MST3 promoted apoptosis in Jurkat cells (Fig. 6, A and B), whereas co-expression of MST3 and striatin 3 significantly suppressed this apoptotic effect (Fig. 6C). Striatin 3 mutants L114E and L114A, which did not impair striatin-PP2A interaction, retained the inhibitory effect on MST3-induced apoptosis of Jurkat cells (Fig. 6, D and E). By contrast, striatin 3 mutants L121E, L125E, and L125A, which are defective in binding PP2A A subunit, do not inhibit MST3-induced apoptosis (Fig. 6, F–H).
FIGURE 6.
Striatin 3 inhibits MST3-induced apoptosis in Jurkat cells. A-H, Jurkat cells were transfected with the indicated constructs encoding FLAG-tagged MST3 and FLAG-tagged striatin 3. Cells were stained with annexin V and propidium iodide (PI) and analyzed by flow cytometer. The numbers are the percentage of apoptosis in each sample. I and J, HEK293T cells were transfected with NF-κB-Luc and different doses of MST3 (I) or striatin 3 (J) in the absence or presence of TNFα, and then the luciferase assay was performed. K, HEK293T cells were transfected with NF-κB-Luc and wild-type or mutant striatin 3 in the absence or presence of TNFα, and then the luciferase assay was performed.
Furthermore, we used TNFα as a stimulator to detect the relationship between the apoptosis-related signaling pathway and MST3-orientated STRIPAK complex. As shown in Fig. 6, I and J, MST3 could significantly decrease TNFα-induced NF-κB transactivation, whereas striatin 3 was able to increase NF-κB activity. Consistently, striatin 3 mutants L121E, L125E, and L125A, which are defective in binding PP2Aα, did not promote NF-κB transactivity (Fig. 6K). These observations suggest that MST3 promotes apoptosis at least in part through NF-κB signaling and striatin 3 acts as a regulatory subunit of the PP2A holoenzyme to down-regulate MST3-promoted apoptosis in cells.
DISCUSSION
As an important regulator in multiple signaling pathways, PP2A plays crucial roles in cell death, cell proliferation, cell mobility, cell invasion, and cytoskeleton dynamics (49–51). It also functions as an important tumor suppressor (52). PP2A dephosphorylates various substrates by recruiting distinct regulatory subunits, and thus conferring differential specificities. Striatins were recently identified as a novel group of PP2A B‴ subunits and implicated in both signaling and trafficking (18). More importantly, the PP2A-striatin holoenzyme is found as part of a supramolecular complex called STRIPAK, in which its kinase and phosphatase components could regulate each other and their respective substrates.
In this study, we determined the crystal structure of a SeMet-labeled striatin 3 coiled coil domain and found that it forms an asymmetric parallel dimer. To our knowledge, this is the first indication that the two striatin chains associate in a parallel manner. It is therefore reasonable to suggest that the entire dimeric STRIPAK complex is also organized in an overall parallel fashion (Fig. 1A), given the role of striatin as a scaffold. Moreover, the coiled coil dimer assumes an asymmetric conformation with a large bend, in which one of the two α-helices is nearly broken. Considering the likely central location of the coiled coil in STRIPAK, it is possible, albeit currently speculative, that the asymmetry could propagate from the coiled coil to other components of the dimeric STRIPAK complex. Such propagation does occur in other cases such as caspase-9 (53, 54). Future research could be directed toward using electron microscopy (EM) and small angle x-ray scattering in combination with x-ray crystallography and nuclear magnetic resonance (NMR) to ultimately resolve the structure of the entire STRIPAK complex, which would be important to determine the overall extent of conformational asymmetry.
Structural analysis combined with biochemical assays defined key residues important for striatin 3 homodimerization and function. Our results not only confirmed the direct interaction between striatin 3 and the PP2A A subunit and mapped the interacting regions on each protein, but also demonstrated that the coiled coil-mediated dimerization of striatin is required for its association with PP2A A subunit. The coiled coil domain of striatin 3 binds the N terminus of the PP2A A subunit, forming a 2:2 heterotetramer. A previous study by Gordon et al. (10) introduced mutations in the coiled coil domain of striatin, including R88S/K89E (Arg-104/Lys-105 in striatin 3), R100S/R101E (Arg-116/Arg-117 in striatin 3), L84A/L94A/I102A (Leu-100/Leu-110/Ile-118 in striatin 3), and L84A/L94A/L105A (Leu-100/Leu-110/Leu-121 in striatin 3), all of which reduced association between striatin and the PP2A C subunit, but did not disrupt striatin self-association in an immunoprecipitation assay. From the current structure, residues Lys-105 and Arg-116 lie on the lateral surface of the coiled coil, whereas residues Arg-104 and Arg-117 are located on the surface formed by homodimerization of striatin 3 (Fig. 3C). All four residues are well exposed for potential direct contacts with PP2A, mutation of which could therefore affect its interaction with PP2A. By contrast, residues Leu-100, Leu-110, Ile-118, and Leu-121 are mainly located on the homodimeric interface of the striatin coiled coil, mutation of which would most likely disrupt the homodimerization of striatin or at least disturb the dimeric conformation. Consistent with these structural observations, we found that mutations L121E, L125E, and L125A of striatin 3 not only disrupted its homodimerization, but also impaired its direct interaction with the PP2A A subunit. Meanwhile, our results revealed that some (d-position) leucines (e.g. Leu-114) are less important for homodimerization than others in terms of stabilizing the striatin 3 coiled coil structure. Taken together, these results indicate that striatin 3 and PP2A exist as a heterotetramer, in which two PP2A molecules bind to two broad surfaces of the striatin 3 coiled coil, respectively (Fig. 7A). Given that PP2A A and C subunits constitute a 1:1 heterodimeric core enzyme (4), our results thus suggest that PP2A A and C subunits would form a 2:2:2 heterohexamer with striatins (Figs. 1A and 7B), which is different from the canonical 1:1:1 heterotrimeric holoenzymes with regulatory subunits from other families. The specific assembly and regulatory mechanisms of the holoenzyme consisting of PP2A A and C subunits, and striatins warrant further structural and functional investigation.
FIGURE 7.
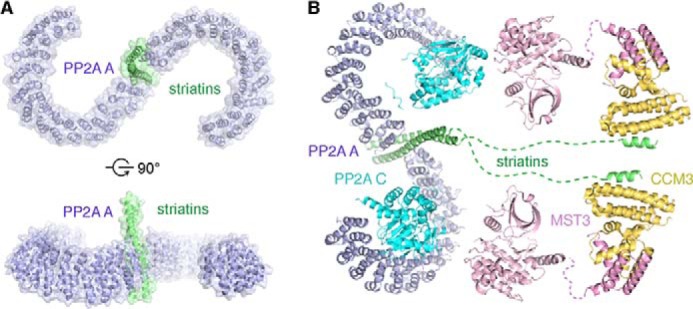
Model of the STRIPAK complex. A, a model of the striatin 3 coiled coil-PP2A A subunit complex. The structure of PP2A A subunit is from the Protein Data Bank (PDB code 3FGA). The location on the coiled coil to which PP2A is bound and the relative orientation of the molecules are not known but one possible example is displayed here according to mutational studies described in this article and those previously reported (10). B, model of the STRIPAK complex. The structures used in the model are from the PDB: PP2A A and C subunits (PDB code 3FGA), MST3 and CCM3 (PDB codes 3A7F and 4GEH).
MST3 has been demonstrated to be involved in apoptosis (46–48). Here, we showed that MST3 promotes apoptosis in Jurkat cells through affecting NF-κB signaling, whereas striatin 3 inhibits this apoptotic effect. Given that PP2A inhibits phosphorylation and activation of MST3 via CCM3 and striatins (10), our results support previous findings that the kinase activity of MST3 is required to induce apoptosis, and further suggest that striatin 3 suppresses MST3-induced apoptosis by recruiting PP2A phosphatase to negatively regulate MST3 activation (Fig. 6). Disrupting the homodimerization of striatin 3 and therefore its interaction with PP2A impairs its inhibitory effect on MST3-induced apoptosis, providing further support that the homodimer-mediated striatin interaction with the PP2A A subunit is indispensable for PP2A functions.
The STRIPAK complex appears to be evolutionarily conserved and play essential roles in multiple biological processes, indicating a general importance of this complex. For example, a complex similar to that in humans has been identified in Drosophila. The Drosophila homolog of striatins Cka, a core component of Drosophila STRIPAK (dSTRIPAK), interacts directly with PP2A and associates with Hippo/dMST (Drosophila homolog of mammalian GCK member MST1/2) via Drosophila Ras association family member (dRASSF) to negatively regulate the Hippo/dMST signaling pathway (12). In summary, the function and regulatory mechanism of the STRIPAK complex remains to be explored and fully defined. Our work thus provides a structural and biochemical basis for further studies on the STRIPAK complex.
CONCLUSION
As a novel type of PP2A regulatory subunits, the coiled coil of striatin forms noncanonical asymmetric parallel homodimers, in which one chain contains a large bend. The noncanonical coiled coil directly interacts with PP2A A subunits to form a 2:2 heterotetrameric core of the STRIPAK complex, which is required for inhibition of MST3-induced apoptosis. These structural features could propagate to the overall conformation of the STRIPAK complex and therefore determine its regulatory nature.
Acknowledgments
We thank Dr. Pei Gang for providing the plasmid encoding PP2A A subunit, Dr. Zhou Jinqiu for helpful discussion, and the staff at beamlines BL17U of Shanghai Synchrotron Radiation Facility (SSRF) and 1W2B at Beijing Synchrotron Radiation Facility (BSRF) for helping with data collection. We are grateful to Dr. Andrei Lupas at the Max Planck Institute for Developmental Biology and Dr. Frauke Gräter at the Partner Institute for Computational Biology and Heidelberg Institute for Theoretical Studies for assisting with the structural analysis.
This work was supported by grants from the 973 program of the Ministry of Science and Technology of China (2010CB529701 and 2012CB910204), the National Natural Science Foundation of China (31270808 and 31300734), the Science and Technology Commission of Shanghai Municipality (11JC14140000 and 13ZR1446400), and the “Cross and cooperation in science and technology innovation team” project of the Chinese Academy of Sciences.
The atomic coordinates and structure factors (code 4N6J) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- STRIPAK
- striatin interacting phosphatase and kinase
- GCK
- germinal center kinase
- SeMet
- selenomethionine
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- BLI
- bio-layer interferometry
- MBP
- myelin basic protein
- PDB
- Protein Data Bank
- MST
- mammalian STE20-like protein kinase.
REFERENCES
- 1. Goudreault M., D'Ambrosio L. M., Kean M. J., Mullin M. J., Larsen B. G., Sanchez A., Chaudhry S., Chen G. I., Sicheri F., Nesvizhskii A. I., Aebersold R., Raught B., Gingras A. C. (2009) A PP2A phosphatase high density interaction network identifies a novel striatin-interacting phosphatase and kinase complex linked to the cerebral cavernous malformation 3 (CCM3) protein. Mol. Cell. Proteomics 8, 157–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Glatter T., Wepf A., Aebersold R., Gstaiger M. (2009) An integrated workflow for charting the human interaction proteome: insights into the PP2A system. Mol. Syst. Biol. 5, 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sugden P. H., McGuffin L. J., Clerk A. (2013) SOcK, MiSTs, MASK and STicKs: the GCKIII (germinal centre kinase III) kinases and their heterologous protein-protein interactions. Biochem. J. 454, 13–30 [DOI] [PubMed] [Google Scholar]
- 4. Shi Y. (2009) Serine/threonine phosphatases: mechanism through structure. Cell 139, 468–484 [DOI] [PubMed] [Google Scholar]
- 5. Wlodarchak N., Guo F., Satyshur K. A., Jiang L., Jeffrey P. D., Sun T., Stanevich V., Mumby M. C., Xing Y. (2013) Structure of the Ca2+-dependent PP2A heterotrimer and insights into Cdc6 dephosphorylation. Cell Res. 23, 931–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Castermans D., Somers I., Kriel J., Louwet W., Wera S., Versele M., Janssens V., Thevelein J. M. (2012) Glucose-induced posttranslational activation of protein phosphatases PP2A and PP1 in yeast. Cell Res. 22, 1058–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Moreno C. S., Park S., Nelson K., Ashby D., Hubalek F., Lane W. S., Pallas D. C. (2000) WD40 repeat proteins striatin and S/G(2) nuclear autoantigen are members of a novel family of calmodulin-binding proteins that associate with protein phosphatase 2A. J. Biol. Chem. 275, 5257–5263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tan B., Long X., Nakshatri H., Nephew K. P., Bigsby R. M. (2008) Striatin-3γ inhibits estrogen receptor activity by recruiting a protein phosphatase. J. Mol. Endocrinol. 40, 199–210 [DOI] [PubMed] [Google Scholar]
- 9. Kean M. J., Ceccarelli D. F., Goudreault M., Sanches M., Tate S., Larsen B., Gibson L. C., Derry W. B., Scott I. C., Pelletier L., Baillie G. S., Sicheri F., Gingras A. C. (2011) Structure-function analysis of core STRIPAK proteins: a signaling complex implicated in Golgi polarization. J. Biol. Chem. 286, 25065–25075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gordon J., Hwang J., Carrier K. J., Jones C. A., Kern Q. L., Moreno C. S., Karas R. H., Pallas D. C. (2011) Protein phosphatase 2a (PP2A) binds within the oligomerization domain of striatin and regulates the phosphorylation and activation of the mammalian Ste20-like kinase Mst3. BMC Biochem. 12, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Moreno C. S., Lane W. S., Pallas D. C. (2001) A mammalian homolog of yeast MOB1 is both a member and a putative substrate of striatin family-protein phosphatase 2A complexes. J. Biol. Chem. 276, 24253–24260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ribeiro P. S., Josué F., Wepf A., Wehr M. C., Rinner O., Kelly G., Tapon N., Gstaiger M. (2010) Combined functional genomic and proteomic approaches identify a PP2A complex as a negative regulator of Hippo signaling. Mol. Cell 39, 521–534 [DOI] [PubMed] [Google Scholar]
- 13. Hyodo T., Ito S., Hasegawa H., Asano E., Maeda M., Urano T., Takahashi M., Hamaguchi M., Senga T. (2012) Misshapen-like kinase 1 (MINK1) is a novel component of striatin-interacting phosphatase and kinase (STRIPAK) and is required for the completion of cytokinesis. J. Biol. Chem. 287, 25019–25029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yu F. X., Guan K. L. (2013) The Hippo pathway: regulators and regulations. Genes Dev. 27, 355–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. ten Klooster J. P., Jansen M., Yuan J., Oorschot V., Begthel H., Di Giacomo V., Colland F., de Koning J., Maurice M. M., Hornbeck P., Clevers H. (2009) Mst4 and Ezrin induce brush borders downstream of the Lkb1/Strad/Mo25 polarization complex. Dev. Cell 16, 551–562 [DOI] [PubMed] [Google Scholar]
- 16. Landberg G., Tan E. M. (1994) Characterization of a DNA-binding nuclear autoantigen mainly associated with S phase and G2 cells. Exp. Cell Res. 212, 255–261 [DOI] [PubMed] [Google Scholar]
- 17. Muro Y., Chan E. K., Landberg G., Tan E. M. (1995) A cell-cycle nuclear autoantigen containing WD-40 motifs expressed mainly in S and G2 phase cells. Biochem. Biophys. Res. Commun. 207, 1029–1037 [DOI] [PubMed] [Google Scholar]
- 18. Benoist M., Gaillard S., Castets F. (2006) The striatin family: a new signaling platform in dendritic spines. J. Physiol. Paris 99, 146–153 [DOI] [PubMed] [Google Scholar]
- 19. Castets F., Rakitina T., Gaillard S., Moqrich A., Mattei M. G., Monneron A. (2000) Zinedin, SG2NA, and striatin are calmodulin-binding, WD repeat proteins principally expressed in the brain. J. Biol. Chem. 275, 19970–19977 [DOI] [PubMed] [Google Scholar]
- 20. Gaillard S., Bartoli M., Castets F., Monneron A. (2001) Striatin, a calmodulin-dependent scaffolding protein, directly binds caveolin-1. FEBS Lett. 508, 49–52 [DOI] [PubMed] [Google Scholar]
- 21. Feng X., Gaeta M. L., Madge L. A., Yang J. H., Bradley J. R., Pober J. S. (2001) Caveolin-1 associates with TRAF2 to form a complex that is recruited to tumor necrosis factor receptors. J. Biol. Chem. 276, 8341–8349 [DOI] [PubMed] [Google Scholar]
- 22. Li H., Nord E. P. (2004) Functional caveolae are a prerequisite for CD40 signaling in human renal proximal tubule cells. Am. J. Physiol. Renal Physiol. 286, F711–719 [DOI] [PubMed] [Google Scholar]
- 23. Sengupta P., Philip F., Scarlata S. (2008) Caveolin-1 alters Ca2+ signal duration through specific interaction with the Gαq family of G proteins. J. Cell Sci. 121, 1363–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gaillard S., Bailly Y., Benoist M., Rakitina T., Kessler J. P., Fronzaroli-Molinières L., Dargent B., Castets F. (2006) Targeting of proteins of the striatin family to dendritic spines: role of the coiled-coil domain. Traffic 7, 74–84 [DOI] [PubMed] [Google Scholar]
- 25. Castets F., Bartoli M., Barnier J. V., Baillat G., Salin P., Moqrich A., Bourgeois J. P., Denizot F., Rougon G., Calothy G., Monneron A. (1996) A novel calmodulin-binding protein, belonging to the WD-repeat family, is localized in dendrites of a subset of CNS neurons. J. Cell Biol. 134, 1051–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bartoli M., Monneron A., Ladant D. (1998) Interaction of calmodulin with striatin, a WD-repeat protein present in neuronal dendritic spines. J. Biol. Chem. 273, 22248–22253 [DOI] [PubMed] [Google Scholar]
- 27. Breitman M., Zilberberg A., Caspi M., Rosin-Arbesfeld R. (2008) The armadillo repeat domain of the APC tumor suppressor protein interacts with Striatin family members. Biochim. Biophys. Acta 1783, 1792–1802 [DOI] [PubMed] [Google Scholar]
- 28. Salin P., Kachidian P., Bartoli M., Castets F. (1998) Distribution of striatin, a newly identified calmodulin-binding protein in the rat brain: an in situ hybridization and immunocytochemical study. J. Comp. Neurol. 397, 41–59 [PubMed] [Google Scholar]
- 29. Moqrich A., Mattei M. G., Bartoli M., Rakitina T., Baillat G., Monneron A., Castets F. (1998) Cloning of human striatin cDNA (STRN), gene mapping to 2p22-p21, and preferential expression in brain. Genomics 51, 136–139 [DOI] [PubMed] [Google Scholar]
- 30. Bartoli M., Ternaux J. P., Forni C., Portalier P., Salin P., Amalric M., Monneron A. (1999) Down-regulation of striatin, a neuronal calmodulin-binding protein, impairs rat locomotor activity. J. Neurobiol. 40, 234–243 [PubMed] [Google Scholar]
- 31. Chen Y. K., Chen C. Y., Hu H. T., Hsueh Y. P. (2012) CTTNBP2, but not CTTNBP2NL, regulates dendritic spinogenesis and synaptic distribution of the striatin-PP2A complex. Mol. Biol. Cell 23, 4383–4392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang H., Ma X., Deng X., Chen Y., Mo X., Zhang Y., Zhao H., Ma D. (2012) PDCD10 interacts with STK25 to accelerate cell apoptosis under oxidative stress. Front. Biosci. 17, 2295–2305 [DOI] [PubMed] [Google Scholar]
- 33. Lu Q., Pallas D. C., Surks H. K., Baur W. E., Mendelsohn M. E., Karas R. H. (2004) Striatin assembles a membrane signaling complex necessary for rapid, nongenomic activation of endothelial NO synthase by estrogen receptor α. Proc. Natl. Acad. Sci. U.S.A. 101, 17126–17131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bernelot Moens S. J., Schnitzler G. R., Nickerson M., Guo H., Ueda K., Lu Q., Aronovitz M. J., Nickerson H., Baur W. E., Hansen U., Iyer L. K., Karas R. H. (2012) Rapid estrogen receptor signaling is essential for the protective effects of estrogen against vascular injury. Circulation 126, 1993–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bailly Y. J., Castets F. (2007) Phocein: a potential actor in vesicular trafficking at Purkinje cell dendritic spines. Cerebellum 1–9 [DOI] [PubMed] [Google Scholar]
- 36. Shi Z., Jiao S., Zhang Z., Ma M., Zhang Z., Chen C., Wang K., Wang H., Wang W., Zhang L., Zhao Y., Zhou Z. (2013) Structure of the MST4 in complex with MO25 provides insights into its activation mechanism. Structure 21, 449–461 [DOI] [PubMed] [Google Scholar]
- 37. Zhang M., Dong L., Shi Z., Jiao S., Zhang Z., Zhang W., Liu G., Chen C., Feng M., Hao Q., Wang W., Yin M., Zhao Y., Zhang L., Zhou Z. (2013) Structural mechanism of CCM3 heterodimerization with GCKIII kinases. Structure 21, 680–688 [DOI] [PubMed] [Google Scholar]
- 38. Otwinowski Z., Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 39. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Brown J. H. (2010) How sequence directs bending in tropomyosin and other two-stranded α-helical coiled coils. Protein Sci. 19, 1366–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Aurora R., Rose G. D. (1998) Helix capping. Protein Sci. 7, 21–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Xu Y., Chen Y., Zhang P., Jeffrey P. D., Shi Y. (2008) Structure of a protein phosphatase 2A holoenzyme: insights into B55-mediated Tau dephosphorylation. Mol. Cell 31, 873–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Xu Z., Cetin B., Anger M., Cho U. S., Helmhart W., Nasmyth K., Xu W. (2009) Structure and function of the PP2A-shugoshin interaction. Mol. Cell 35, 426–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Huang C. Y., Wu Y. M., Hsu C. Y., Lee W. S., Lai M. D., Lu T. J., Huang C. L., Leu T. H., Shih H. M., Fang H. I., Robinson D. R., Kung H. J., Yuan C. J. (2002) Caspase activation of mammalian sterile 20-like kinase 3 (Mst3). Nuclear translocation and induction of apoptosis. J. Biol. Chem. 277, 34367–34374 [DOI] [PubMed] [Google Scholar]
- 47. Wu H. Y., Lin C. Y., Lin T. Y., Chen T. C., Yuan C. J. (2008) Mammalian Ste20-like protein kinase 3 mediates trophoblast apoptosis in spontaneous delivery. Apoptosis 13, 283–294 [DOI] [PubMed] [Google Scholar]
- 48. Lin C. Y., Wu H. Y., Wang P. L., Yuan C. J. (2010) Mammalian Ste20-like protein kinase 3 induces a caspase-independent apoptotic pathway. Int. J. Biochem. Cell Biol. 42, 98–105 [DOI] [PubMed] [Google Scholar]
- 49. Lechward K., Awotunde O. S., Swiatek W., Muszyńska G. (2001) Protein phosphatase 2A: variety of forms and diversity of functions. Acta Biochim. Pol. 48, 921–933 [PubMed] [Google Scholar]
- 50. Sontag E. (2001) Protein phosphatase 2A: the Trojan Horse of cellular signaling. Cell. Signal. 13, 7–16 [DOI] [PubMed] [Google Scholar]
- 51. Janssens V., Goris J. (2001) Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem. J. 353, 417–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Perrotti D., Neviani P. (2013) Protein phosphatase 2A: a target for anticancer therapy. Lancet Oncol. 14, e229–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Renatus M., Stennicke H. R., Scott F. L., Liddington R. C., Salvesen G. S. (2001) Dimer formation drives the activation of the cell death protease caspase 9. Proc. Natl. Acad. Sci. U.S.A. 98, 14250–14255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Brown J. H. (2006) Breaking symmetry in protein dimers: designs and functions. Protein Sci. 15, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]