Background: The molecular composition of native TRPC4 channels is unknown.
Results: Tarbp2, like Ago2 and Dicer, a protein of the RNA-induced silencing complex, binds to and functionally interacts with TRPC4.
Conclusion: Upon TRPC4 binding, Tarbp2 modulates Ca2+ entry and promotes Ca2+-dependent proteolytic activation of Dicer.
Significance: Mechanistic insight into coupling TRPC4 activity and Dicer-dependent formation of noncoding RNAs is provided.
Keywords: Calcium Signaling, Cell Signaling, Dicer, Protein-Protein Interactions, TRP Channels
Abstract
TRPC4 proteins function as Ca2+ conducting, non-selective cation channels in endothelial, smooth muscle, and neuronal cells. To further characterize the roles of TRPC4 in vivo, detailed information about the molecular composition of native channel complexes and their association with cellular signaling networks is needed. Therefore, a mouse brain cDNA library was searched for novel TRPC4-interacting proteins using a modified yeast two-hybrid assay. This screen identified Trans-activation Response RNA-binding protein 2 (Tarpb2), a protein that recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Tarbp2 was found to bind to the C terminus of TRPC4 and TRPC5 and to modulate agonist-dependent TRPC4-induced Ca2+ entry. A stretch of basic residues within the Tarbp2 protein is required for these actions. Tarbp2 binding to and modulation of TRPC4 occurs in the presence of endogenously expressed Dicer but is no longer detectable when the Dicer cDNA is overexpressed. Dicer activity in crude cell lysates is increased in the presence of Ca2+, most probably by Ca2+-dependent proteolytic activation of Dicer. Apparently, Tarbp2 binding to TRPC4 promotes changes of cytosolic Ca2+ and, thereby, leads to a dynamic regulation of Dicer activity, essentially at low endogenous Dicer concentrations.
Introduction
The mammalian TRPC4 protein is a member of the transient receptor potential canonical subfamily of transient receptor potential (TRP)5 proteins, and its cDNA was identified initially (1) by its sequence similarity with the Drosophila TRP protein (40% sequence identity), the founding member of the TRP superfamily of proteins. TRPC4 proteins form non-selective Ca2+-conducting cation channels that are activated upon stimulation of receptors coupling to Gαi/Gαo- and Gαq/Gα11-dependent signaling pathways or receptor tyrosine kinases (2–4). TRPC4 is expressed in a broad range of tissues, including neurons (5, 6), macrovascular endothelial cells (7, 8), and intestinal smooth muscle cells (9). In ileal smooth muscle cells, TRPC4 channels are gated by muscarinic acetylcholine receptor types 2 and 3 and contribute more than 80% to the muscarinic receptor-induced cation current (9). In these cells, TRPC4 channels couple muscarinic receptors to smooth muscle cell depolarization, voltage-activated Ca2+ influx, and contraction, and thereby accelerate small intestinal motility. TRPC6 channels are responsible for the remaining <20% of muscarinic receptor-induced cation current, indicating that the simultaneous activation of two independent channels contribute to this current (9). In the brain, functional TRPC4 channels might be homotetramers or heterotetramers formed by TRPC4, TRPC5, and maybe TRPC1 (6, 10). These channels have been implicated in mediating neuronal excitation induced by stimulating metabotropic receptors, including group 1 metabotropic glutamate receptors (6). The mammalian TRPC4 and TRPC5 proteins share 65%, and the TRPC1, TRPC4, and TRPC5 proteins share 41% sequence identity, but little insight is available into the structural requirements of heteromerization and into the molecular composition of endogenous TRPC channels.
Several proteins have been implicated to interact with TRPC4 and modulate TRPC4 function, including various TRPCs, calmodulin (11, 12), the inositol 1,4,5-trisphosphate receptor (11), STIM1 (13), αII spectrin (14), protein 4.1 (15), Na+/H+ exchange regulatory factor/EBP50 (16), and the phospholipid-binding protein SESTD1 (17). In most cases it remains to be elucidated how the different proteins work together to modulate channel function. Moreover, little is known about the auxiliary proteins that regulate TRPC4 in diverse cell types.
Our experiments focused on the identification of components of TRPC4 channels. Tarbp2, or TRBP2 (18, 19), which was one of the first proteins involved in the identification of double-stranded RNA-binding proteins and which, in mammals, is part of the RNA-induced silencing complex, including Argonaute and Dicer (20–23), was found to interact with TRPC4 and TRPC5 and to modulate receptor-activated, TRPC4-dependent Ca2+ entry as well as constitutive Ca2+ entry accomplished by TRPC4 and TRPC5 gain-of-function channel mutants. In addition, Dicer activity is shown to be increased in the presence of Ca2+, most probably by Ca2+-dependent proteolytic activation of the enzyme. Apparently, Tarbp2-TRPC4/TRPC5 interaction might couple TRPC-dependent Ca2+ entry and Ca2+-dependent proteolytic activation of Dicer.
EXPERIMENTAL PROCEDURES
Yeast Two-hybrid Screening
To identify TRPC4-interacting proteins, we screened a mouse brain cDNA library using the CytoTrapTM two-hybrid system (Stratagene) relying on the restoration of the Ras signal transduction pathway, which allows screening for interactions in the cytoplasm. The cDNAs of the cytosolic TRPC4 N terminus (nt 1–986) and C terminus (nt 1862–2922) (GenBankTM accession number NM_016984.3) were amplified by RT-PCR (primer pairs C terminus, 5′-TAG GAT CCA TAA TTC TTA CCA ACT AAT TGC C-3′ and 5′-TAA TTG TCG ACT CAC AAT CTT GTG GTC ACA TAA-3′; N terminus, 5-GTA GGA TCC AAA TGG CTC AGT TCT ATT ACA AA-3′ and 5′-TTG GAT CCC TAC TTC ACC GCC CAG TGT CT-3′) and subcloned into the BamHI site of the pSos bait plasmid. Altogether, ∼105 and ∼106 clones were screened using the N terminus and C terminus as baits, respectively, according to the protocol of the manufacturer. Prey plasmids from positive clones were recovered and sequenced. Two independent partial cDNA clones of murine Tarbp2 (synonym TRBP2, GenBankTM accession number NM_009319) were identified (Fig. 1C) and recovered. The specificity of the yeast two-hybrid interaction was verified by retransformation of the prey and bait plasmids. In the case of the TRPC4 N terminus, only false positive clones were found (Fig. 1D). The Tarbp2 full-length cDNA was obtained from imaGenes GmbH (Berlin, Germany), sequenced, and subcloned into the EcoRV site of the N-terminal p3xFLAG-CMV vector (Sigma-Aldrich), into the EcoRV site of bicistronic pcAGGS-IRES-GFP vector, and into the HindIII/BamHI sites of the pcDNA3TagRFP-T (24) vector. All final cDNAs and vector constructs were sequenced.
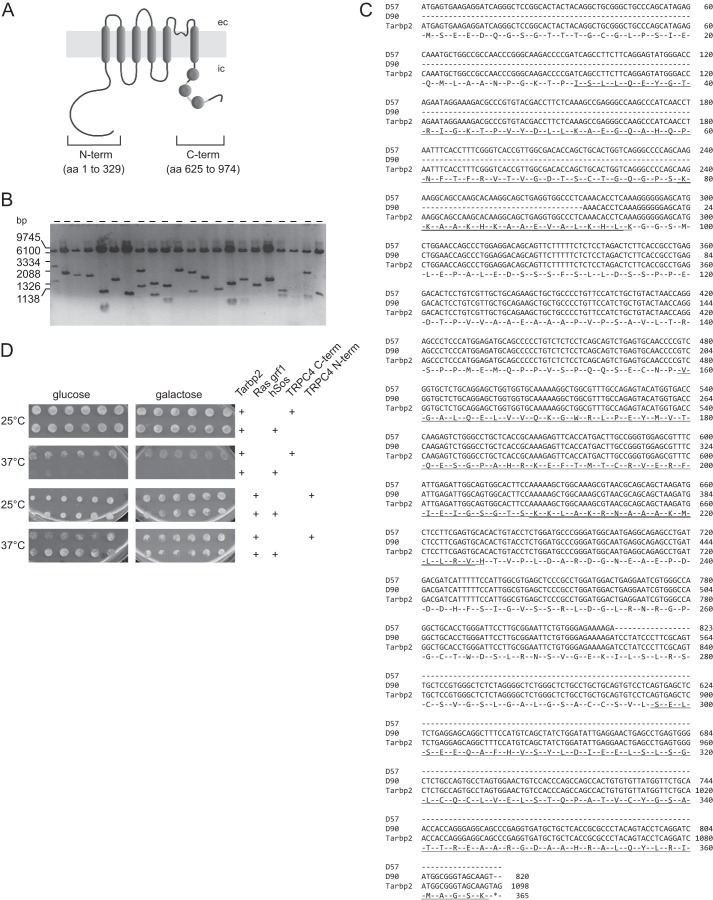
FIGURE 1.
Yeast two-hybrid screen to identify protein binding partners of TRPC4. A, the TRPC4 protein comprises six predicted transmembrane domains. The cDNAs encoding the cytosolic domains of the TRPC4 N- and C terminus were used as prey. Disks within the C terminus indicate calmodulin binding domains 1, 2, and 3. ec, extracellular; ic, intracellular. B, control of insert sizes of the mouse brain cDNA library used as bait in the screen. Twenty-two randomly selected cDNA clones were run on agarose gel (6.1-kb fragment, pMyr vector without insert after EcoRI/XhoI cut). C, alignment of the two independent cDNA clones D57 and D90 of Tarbp2 identified in the screen (Tarbp2 GenBankTM accession number NM_009319). Underlined are dsRBD1 (aa 32 to 96), dsRBD2 (aa153 to 225), and a third domain bearing structural homology to the dsRBDs (aa 289 to 386); double underlined are the KR-helix motifs within dsRBD1 and dsRBD2. D, results of retransformation to check the interaction of the bait and prey proteins. Plasmids were retransformed into the temperature-sensitive yeast strain cdc25H. The expression of the bait proteins is under the control of the GAL1 promoter, so the expression is repressed on glucose and activated by galactose. The yeast strain is temperature-sensitive, and the interaction of bait and prey protein rescue this phenotype, which leads to growth at the restrictive temperature of 37 °C. As a control, the yeast strain cdc25H was cotransformed with the bait plasmids and the pSos vector. Six clones of each transformation were plated on dropout medium containing glucose or galactose, respectively. The clone coding for Ras grf1 was found with the TRPC4 N terminus. The retransformation showed that the Ras grf1 clone was able to grow on galactose at 37 °C without the prey protein (TRPC4 N terminus), indicating a false positive interaction. The Tarbp2 clone exhibited a positive interaction with the TRPC4 C terminus (growth on galactose at 37 °C but not with pSos alone).
Cell Culture, Transfected cDNAs, and Transfection
HEK293 cells (ATCC, CRL 1573) were grown in 3-cm-diameter culture dishes until 80% confluence and then transiently transfected with 2 μg of the respective cDNA constructs in 5 μl of PolyFectTM reagent (Qiagen, Hilden, Germany). For Fura-2 measurements, cells transfected with the pcAGGS-IRES-GFP or pcAGGS-IRES-RFP vector were used as controls. Coverslips with transfected cells were used for Ca2+ imaging experiments 24–48 h after transfection. The bicistronic expression plasmids pdiTRPC4 and pdiTRPC5 and the stable inducible Flp-InTM TRPC4G503S and TRPC5G504S cell lines have been described recently (25). For patch clamping and Ca2+ imaging, wild-type, stably M2R-expressing HEK293 cells (26) and stably M2R TRPC4α-expressing HEK293 cells (26) (provided by Dr. M. X. Zhu, University of Texas, Houston, Texas) were plated on glass coverslips 24 h after transfection, and experiments were performed at the indicated times. The Flp-InTM 293 cells expressing TRPC4G503S or TRPC5G504S after induction in the presence of tetracycline (10 μg/ml) were grown on glass coverslips and used up to 48 h after plating or induction. The human Dicer cDNA (pIRES-neo2-Dicer) was a gift from Dr. Gunter Meister (Universität Regensburg, Regensburg, Germany). The calmodulin cDNA was subcloned in the pcAGGS-IRES-GFP vector. The Tarpb2Δ209–234 mutant was generated by PCR using the primer pair 5′-GGA AGT GCC ACT GCC AAT CTC AAT G-3′ and 5′-AAT GAG GCA GAG CCT GAT GAC GAT C-3′ and the plasmid p3xFLAG-Tarbp2 as template.
Antibodies, Coimmunoprecipitation, and Western Blot Analysis
In-house-generated, affinity-purified antibodies for TRPC4 and TRPC5 (25) were used, as well as antibodies against FLAG M2 (Sigma-Aldrich), GST (GE Healthcare Biosciences), calmodulin (Zymed Laboratories Inc.), Dicer, anti-Dicer-1133 (catalog no. D38E7, Cell Signaling Technology), and anti-Dicer-Ct (an epitope corresponding to amino acids 1701–1912, mapping at the C terminus of human Dicer according to the manufacturer; catalog no. sc30226, Santa Cruz Biotechnology). The cells transfected with the respective cDNAs were lysed in buffer containing 50 mm HEPES (pH 7.5), 150 mm NaCl, 1 mm CaCl2, 1.25% (w/v) n-dodecyl β-d-maltoside, and a mixture of protease inhibitors. After centrifugation at 100,000 × g, the antibodies for FLAG (4 μl), TRPC4 (10 μg), or TRPC5 (10 μg) were added to the supernatant and incubated at 4 °C for 2 h. As a control for the precipitating mouse and rabbit antibodies, precipitations were performed in parallel using nonspecific mouse and rabbit IgGs (10 μg each/precipitation). After equilibration in lysis buffer, protein A- and protein G-Sepharose (30 μl) were added, and the resulting suspension was incubated at 4 °C for at least 2 h under continuous shaking. After centrifugation (800 × g at 4 °C for 3 min), the Sepharose beads were washed four times in washing buffer (50 mm HEPES (pH 7.5), 150 mm NaCl, 1 mm CaCl2, and 0.5% n-dodecyl β-d-maltoside). Bound proteins were eluted by 2× concentrated denaturing sample buffer (60 μl), followed by incubation at 65 °C for 20 min. SDS-PAGE and Western blot analysis were performed as described previously (25). Proteins were detected with horseradish peroxidase-coupled secondary antibodies and Western Lightning Chemiluminescence Reagent Plus (PerkinElmer Life Sciences). Original scans were saved as TIFF files from LAS 3000 (Fujifilm), which were further processed in Adobe Photoshop. Stain intensities were analyzed by AIDA Image Analyzer software. Images were cropped, resized proportionally, and brought to the resolution required for publication.
Ca2+ Imaging
Intracellular live cell Ca2+ imaging experiments were performed essentially as described (25) using a Polychrome V and camera-based imaging system (ANDOR iXon 885, ANDOR Technology) from TILL Photonics (Martinsried, Germany) at a Zeiss Axiovert 200 m fluorescence microscope equipped with a Zeiss Fluar ×20/0.75 objective. Data acquisition was accomplished with the imaging software TILLvisION (TILL Photonics). Prior to the experiments, cells were incubated in medium supplemented with 4 μm of the Ca2+-sensitive fluorescent dye Fura-2/AM for 30 min in the dark at room temperature and washed four times with nominally Ca2+-free external solution (140 mm NaCl, 5 mm KCl, 1 mm MgCl2, 10 mm HEPES, and 10 mm glucose (pH 7.2)) to remove excess Fura-2/AM. The Fura-2-loaded cells grown on 2.5-cm glass coverslips were transferred to a bath chamber containing nominally Ca2+-free solution, and Fura-2 fluorescence emission was monitored at >510 nm after excitation at 340 and 380 nm for 10 ms each at a rate of 1 Hz for up to 1700 s. Cells were marked, and the ratio of the background-corrected Fura-2 fluorescence at 340 and 380 nm (F340/F380) was plotted versus time. During the measurement, cells were incubated in the presence of 1 μm thapsigargin (no Ca2+ in the bath solution), 2 mm CaCl2, and 100 μm carbachol. For the tetracycline-induced Flp-InTM-293 cells expressing the TRPC4G503S or TRPC5G504S mutants, 2 mm CaCl2 was added to the bath solution after reaching a stable F340/F380 base line, and changes of cytosolic Ca2+ were monitored as described previously (25).
To get an estimate of the intracellular Ca2+ concentrations, a calibration of the Fura-2 ratio to Ca2+ concentration was performed. According to Equation 1, values for the minimal ratio (Rmin - Ca2+-free Fura-2), maximal ratio (Rmax - Ca2+-bound Fura-2), the fluorescence at 380 nm excitation of Ca2+-free (F380low) and Ca2+-bound (F380high) Fura-2, and the apparent dissociation constant (KD) of Fura-2 for Ca2+ were determined experimentally. Therefore, Fura-2-loaded HEK293 cells were incubated in external solution (see above) with Ca2+ buffered to 0 by adding 10 mm EGTA or 10 mm 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid (BAPTA) and permeabilized by the Ca2+ ionophores ionomycin (10 μm) or 4Br-A23187 (20 μm). After reaching a stable ratio (Rmin, F380low), 10 mm Ca2+ was added (Rmax, F380high). To obtain the apparent KD of Fura-2 in vivo, Fura-2-loaded cells were incubated in external solution containing 10 μm ionomycin or 20 μm 4Br-A23187, and free Ca2+ was set to 256 nm using 10 mm EGTA plus 6 mm CaCl2, respectively, calculated with WebMaxC. The calibration experiments revealed an apparent KD, Rmax, Rmin, and b (F380low/F380high) of 462 nm, 3.29, 0.25, and 7.58, respectively.
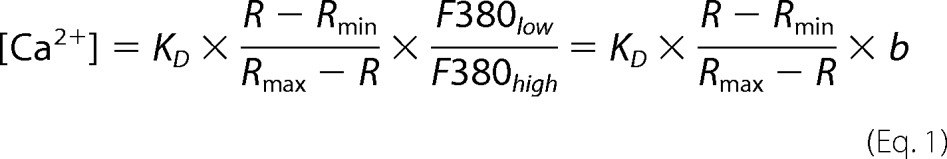 |
Electrophysiological Recordings and Solutions
Membrane currents were recorded in the tight seal whole-cell patch clamp configuration using an EPC-9 amplifier (HEKA Electronics, Lambrecht, Germany). Patch pipettes were pulled from glass capillaries (catalog no. GB150T-8P, Science Products, Hofheim, Germany) at a vertical puller (catalog no. PC-10, Narishige, Tokyo, Japan) and had resistances between 2 and 4 mΩ when filled with standard internal solution (120 mm cesium glutamate, 8 mm NaCl, 1 mm MgCl2, 10 mm HEPES, 10 mm 1,2-bis(2-aminophenoxy)ethane-N,N,N',N'-tetraacetic acid tetracesium salt, and 3.1 mm CaCl2 (100 nm free Ca2+, calculated with WebMaxC), pH adjusted to 7.2 with CsOH). Standard external solution contained 140 mm NaCl, 2 mm MgCl2, 1 mm CaCl2, 10 mm HEPES, and 10 mm glucose, pH adjusted to 7.2 with NaOH. The osmolarity of all solutions ranged between 285 and 315 mosm. Voltage ramps of 400 ms duration spanning a voltage range from −100 to 100 mV were applied at 0.5 Hz from a holding potential of 0 mV over a period of 300 s using PatchMaster software (HEKA). All voltages were corrected for a 10-mV liquid junction potential. Currents were filtered at 2.9 kHz and digitized at 100-μs intervals. Capacitive currents and series resistance were determined and corrected before each voltage ramp using the automatic capacitance compensation of the EPC-9. Inward and outward currents were extracted from each individual ramp current recording by measuring the current amplitudes at −80 and 80 mV, respectively, normalized to the cell capacitance to get the current density in picoampere/picofarad, and plotted versus time. Current-voltage (IV) relationships were extracted at the indicated time points.
GST-TRPC4 Pulldown Experiments
To characterize the TRPC4 binding domain for Tarbp2, the GST-TRPC4 fusion proteins shown in Fig. 3A were expressed in Escherichia coli and purified with GSH-Sepharose beads (12). Lysates of HEK293 cells expressing the FLAG-Tarbp2-cDNA were prepared as described above and incubated in the presence of 12 μg of GSH-bound GST-TRPC4 fragments at 4 °C for 2 h. The beads where washed four times with washing buffer. Bound proteins were eluted by 2× concentrated denaturing sample buffer (50 μl), followed by incubation at 65 °C for 20 min and subjected to SDS-PAGE followed by Western blot analysis. For calmodulin/Tarbp2 competition the FLAG-Tarbp2 cell lysates (containing 1 mm CaCl2) were incubated in the presence of GSH-bound GST-TRPC4 fragment LB5/6b (12 μg) and in the absence and presence of increasing amounts of calmodulin (0.595 μg, 2.97 μg, 5.95 μg, and 11.9 μg). After incubation, beads were washed four times with washing buffer, and proteins were processed for Western blot analysis as described above.
FIGURE 3.
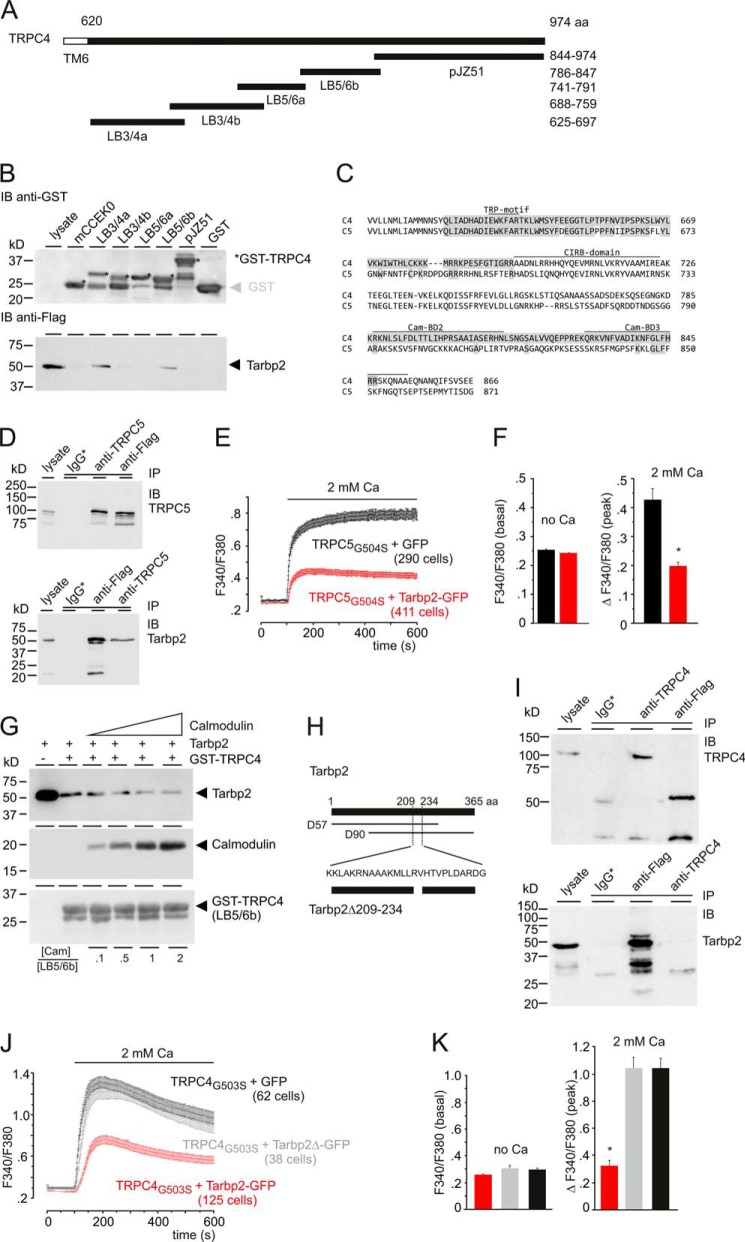
Identification of the aa sequences involved in Tarbp2-TRPC4 binding. A, TRPC4-GST fusion proteins covering the TRPC4 C terminus (aa 625–974). Numbers indicate aa residues. TM6, predicted transmembrane segment 6. B, pulldown of the Tarbp2 protein from HEK cell lysates by the recombinant TRPC4-GST fusion proteins (12 μg each) and GST (control) bound to GSH-Sepharose. Shown is an immunoblot (IB) using antibody for GST (top panel) and for FLAG-tagged Tarbp2 (bottom panel), respectively. The asterisk indicates GST-TRPC4 fusion proteins of expected Mr. C, alignment of parts of the C termini of TRPC4 and TRPC5 covering the sequences of the two Tarbp2-binding domains LB3/4a and LB5/6b (shaded in gray) identified in B. The positions of the TRP-motif, the CIRB-domain, and calmodulin-binding domains (Cam-BD) 2 and 3 are indicated. D, Western blot analyses showing reciprocal immunoprecipitation (IP) after coexpressing the FLAG-tagged Tarbp2 and TRPC5 cDNAs in HEK293 cells using the anti-TRPC5 and anti-FLAG antibodies. Immunoblots were incubated with antibodies for TRPC5 (top panel) and FLAG (bottom panel), respectively. IgG*, nonspecific IgGs used for control precipitations. E, HEK293 cells expressing TRPC5G504S (19 h after induction) and coexpressing the IRES-GFP (black trace) as control or the Tarbp2-IRES-GFP cDNA (red trace) were loaded (48 h after transfection) with Fura-2-AM and kept in nominally Ca2+-free bath solution. Ca2+ influx was challenged by adding 2 mm Ca2+ to the bath solution, and the cytosolic [Ca2+], represented by the Fura-2 fluorescence ratio (F340/F380) was measured versus time. Note that no agonist is present. F, summary. Data are mean ± S.E. *, p < 0001. G, calmodulin competes with Tarbp2 for TRPC4 binding. Shown is a pulldown of the Tarbp2 protein from HEK cell lysates by the recombinant TRPC4-GST fusion protein LB5/6b (12 μg) in the absence and presence of increasing amounts of calmodulin (0.59, 2.97, 5.95, and 11.9 μg). Immunoblots with antibodies for Tarbp2-FLAG (top), calmodulin (center) and GST (bottom) are shown. The molar ratio [Cam]/[LB5/6b] is as indicated. H–K, amino acid sequence of Tarbp2 required for binding of Tarbp2 to TRPC4. H, positions of aa residues are indicated by numbers, D57 and D90 indicate cDNA clones identified and isolated in the screen. The aa residues deleted in Tarbp2Δ209–234, common to both D90 and D57, are indicated. I, Western blot analyses. Both proteins, TRPC4 and Tarbp2Δ209–234, are detectable in cell lysate. Shown is immunoprecipitation after coexpressing the FLAG-tagged Tarbp2Δ209–234 and TRPC4 cDNAs in HEK293 cells using the anti-TRPC4 and anti-FLAG antibodies. Immunoblots were incubated with antibodies for TRPC4 (top panel) and FLAG (bottom panel), respectively. IgG*, nonspecific mouse (top panel) and rabbit IgGs (bottom panel) used for control precipitations. J, HEK-293 cells expressing TRPC4G503S (18 h after induction) and coexpressing IRES-GFP (black) as control, Tarbp2-IRES-GFP cDNA (red), or Tarbp2Δ209–234-IRES-GFP cDNA (gray) were loaded with Fura-2/AM and kept in nominally Ca2+-free bath solution. Ca2+ influx was challenged by adding 2 mm Ca2+ to the bath solution, and the cytosolic [Ca2+], represented by the Fura-2 fluorescence ratio (F340/F380), was measured versus time. Note that no agonist is present. K, summary of J. Data are mean ± S.E. *, p < 0.001.
miRNA Microarray Experiments
RNA was isolated from HEK293 cells and stably M2R/TRPC4 expressing HEK293 cells using a miRNeasy mini kit (Qiagen) according to the instructions of the manufacturer. RNA integrity was analyzed using Bioanalyzer 2100 (Agilent), yielding RNA integrity numbers between 9.8 and 10. RNA concentration and purity were measured using NanoDrop 2000 (Thermo Scientific), yielding A260/A230 ratios between 1.8 and 1.98.
A microarray analysis was performed according to the instructions of the manufacturer using SurePrint 8 × 60K human v16 miRNA microarrays (Agilent, catalog no. G4870A) that contain 40 replicates of each of the 1205 miRNAs of miRBase v. 16 with each RNA sample in duplicates. In brief, a total of 100 ng of total RNA was processed using the miRNA Complete Labeling and Hyb kit to generate fluorescently labeled miRNA. This method involves the ligation of one cyanine 3-pCp molecule to the 3′ end of a RNA molecule with greater than 90% efficiency. First, the RNA was dephosphorylated using calf intestinal alkaline phosphatase. After the dephosphorylation step, dimethyl sulfoxide, which is an effective RNA denaturant, was added to the samples, and the RNA was heat-denatured to minimize the effect of structure and sequence differences among miRNAs. Using T4 RNA ligase and 3′,5′-cytidine bisphosphate, labeled by a cyanine dye at its 3′ phosphate (pCp-Cy3), miRNA molecules with an additional 3′-cytidine and exactly one cyanine dye on their 3′end were produced. After the labeling reaction, the mixture was dried in a vacuum centrifuge and resuspended in hybridization mixture containing hybridization buffer and blocking reagent. Then the microarrays were loaded and incubated for 20 h at 55 °C and 20 rpm. To check whether the labeling and hybridization were successful, labeling and hybridization spike-in controls were added in the appropriate steps. After several washing steps, microarrays were scanned with an Agilent microarray scanner at 3 μm in double path mode. Microarray scan data were further processed using Feature Extraction software. Expression data were normalized using standard quantile normalization.
Miscellaneous Methods
The recombinant Dicer assay was carried out according to the protocol of the manufacturer (Recombinant Dicer enzyme kit, Genlantis, San Diego, CA) using 0.5 units of Dicer. As a substrate, the double-stranded RNA (dsRNA) 37ab (27) was generated as follows. The single strands (37a, 5′-UGA GGU AGU AGG UUG UAU AGU UUG AAA GUU CAC GAU U-3′; 37b, 5′-UCG UGA ACU UUC AAA CUA UAC AAC CUA CUA CCU CAU U-3′) were synthesized, and 50 pmol of 37a was end-labeled by T4 polynucleotide kinase (20 units) in the presence of 50 pMol [γ-32P]ATP. After running on a G25 MicroSpin column, the 5′-labeled 37a was annealed with the 37b, and the resulting dsRNA was diluted to 600 cpm/μl. After stopping the Dicer reaction by addition of 1.2 volumes of loading buffer (95% formamide, 18 mm EDTA, 0.025% SDS, 0.1% xylene cyanol, and 0.1% bromphenol blue), samples were heated for at 75 °C for 10 min, put on ice, and run on a 15% polyacrylamide/7 M urea gel in buffer containing 44.5 mm Tris base, 44.5 boric acid, and 1 mm EDTA. The activity of Dicer in HEK cell lysates was essentially carried out as described. Lysates of M2R TRPC4 HEK cells were prepared as described above, except that the lysis buffer contained 50 mm HEPES (pH 7.5), 150 mm NaCl, 10 mm BAPTA, and 1.25% n-dodecyl β-d-maltoside. The lysate (1 μl) was preincubated in a final volume of 5 μl (13.2 mm Hepes/40 mm Tris, 80 mm NaCl, 3.32 mm MgCl2, 2% (v/v) glycerin, 0.25% n-dodecyl β-d-maltoside, 0.32 mm dithiothreitol, 2 mm BAPTA, and RNase inhibitor) in the absence or presence of 6 mm CaCl2 at 21 °C for 30 min. Thereafter, 5 μl of dsRNA 37ab (600 cpm/μl) was added, and incubation continued for the indicated times at 37 °C. Reactions were stopped by addition of 1.2 volumes of loading buffer, incubated at 75 °C for 10 min, and put on ice. Thereafter, samples were run on polyacrylamide/urea gels as described above. The gels were exposed to PhosphorImager screens, and densitometric analysis was carried out with AIDA software.
RESULTS
Identification of Tarbp2 as a Potential Binding Partner of TRPC4
To define components and associated proteins of TRPC4 channels in the brain, we screened a mouse brain cDNA library with the N terminus (amino acid (aa) residues 1–329) and the C terminus (aa 625–974) of TRPC4 in yeast. Both the N terminus and C terminus are localized in the cytosol (Fig. 1A), and, accordingly, we assayed for protein-protein interactions outside of the nucleus, relying on a modified yeast two-hybrid system on the basis of the restoration of the Ras signal transduction pathway. The target cDNAs were fused to the cDNA of a myristoylation factor, and the translated fusion proteins were anchored to the cell membrane so that the screen for interactions occurs in the cytoplasm. The average size of 22 randomly chosen cDNA clones was ∼1.8 kb (Fig. 1B). In this screen, several proteins were identified as putative TRPC4 binding partners. Sequence analysis revealed that one of the potential TRPC4-interacting proteins was the human immunodeficiency virus transactivating response RNA-binding protein 2 (Tarbp2, synonym TRBP2), which is also an integral component of a Dicer-containing complex. Two independent Tarbp2 cDNA clones, D57 (nt 1–823) and D90 (nt 277–1096) were identified, which cover the complete protein coding Tarbp2 cDNA (Fig. 1C). Their overlapping DNA encodes aa 93–274 and, thus, about 50% of the Tarbp2 protein, including the double-stranded RNA-binding domain (dsRBD) 2, followed by a sequence that has been implicated in protein-protein interaction (19). Clones from colonies that exhibited galactose-dependent growth at 37 °C were verified by retransformation (Fig. 1D). False positive clones like Ras grf1, the mouse ortholog of hSos and CDC25, are able to grow independently from the bait protein on galactose medium at 37 °C. Only clones that exhibit bait-dependent growth on galactose medium at 37 °C were considered true interaction partners. This was the case for Tarbp2 and the C terminus of TRPC4 as bait (Fig. 1D).
The Tarbp2-TRPC4 Interaction Modulates TRPC4 Function
To confirm binding of Tarbp2 to TRPC4, we cotransfected HEK293 cells with the TRPC4 cDNA and the full-length Tarbp2 cDNA fused to the cDNA of the FLAG tag. From cell lysates, coimmunoprecipitations were performed using an anti-TRPC4 antibody and the FLAG antibody for the tagged Tarbp2. Fig. 2A shows that TRPC4 is precipitated by the anti-TRPC4 antibody but is also present among the proteins associated with Tarbp2 precipitated by the anti-FLAG antibody. Fig. 2B shows that Tarbp2 (calculated Mr including the FLAG tag, 42,755) is precipitated by the anti-FLAG antibody but is also present among the proteins associated with TRPC4 precipitated by the anti-TRPC4 antibody.
FIGURE 2.
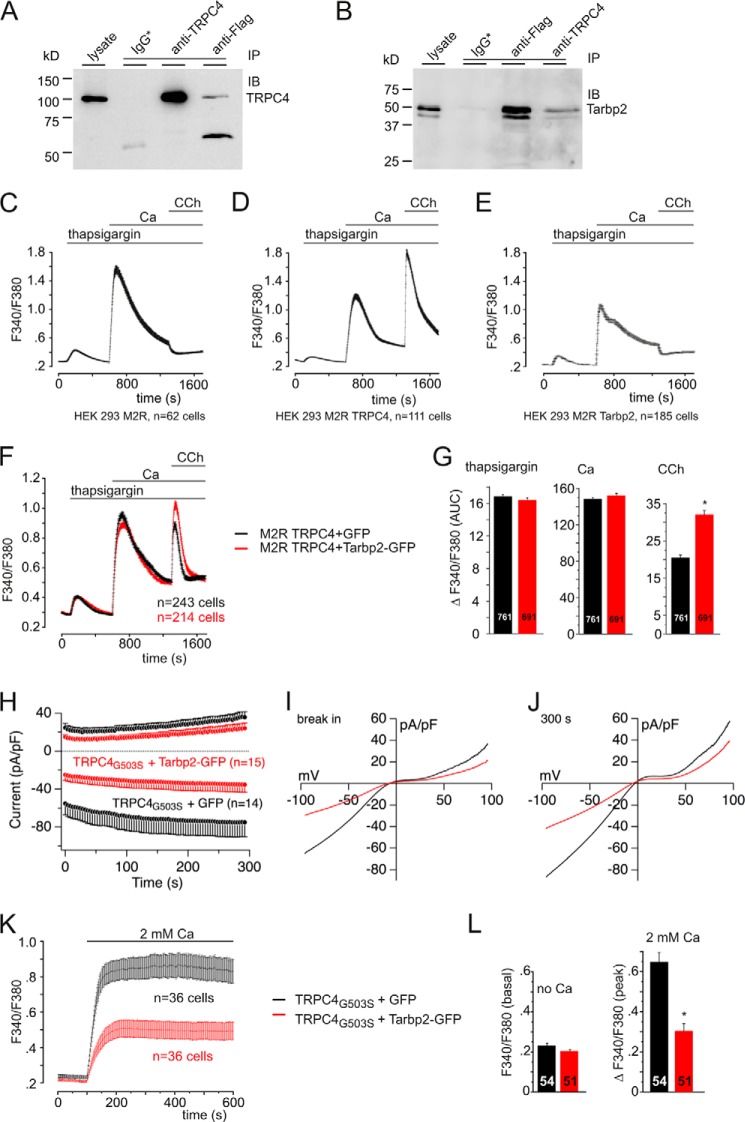
Tarbp2 interacts with TRPC4 and modulates TRPC4 activity. A and B, Western blot analysis showing reciprocal immunoprecipitation (IP) after coexpressing the FLAG-tagged Tarbp2 and TRPC4 cDNAs in HEK293 cells using the anti-TRPC4 and anti-FLAG antibodies. Immunoblots (IB) were incubated with antibodies for TRPC4 (A) and FLAG (B), respectively. IgG*, nonspecific mouse (A) or rabbit IgGs (B) used for control precipitations. C–E, changes of cytosolic [Ca2+] in Fura-2-loaded HEK cells stably expressing the cDNAs of the metabotropic M2R (C), of the metabotropic M2R and TRPC4 (M2R TRPC4, D), and in Fura-2-loaded M2R cells coexpressing the Tarbp2 cDNA (E). CCh, carbachol. F and G, Tarbp2 increases carbachol-induced (100 μm), TRPC4-dependent Ca2+ entry (red trace) without affecting Ca2+ release in the presence of thapsigargin and in nominally Ca2+-free bath solution or Ca2+ entry after readdition of extracellular Ca2+ (F). The Tarbp2-IRES-GFP vector (red trace) and the IRES-GFP vector (black trace) as control were coexpressed in M2R TRPC4α cells. G, summary of three experiments like the one in F. Data are mean ± S.E. with the number of cells indicated. *, p < 0.001. AUC, area under the curve. H–J, Tarbp2 (red traces) reduces the fully activated current of TRPC4G503S mutant channels. H, inward and outward currents at −80 and 80 mV, respectively, from HEK293 cells expressing TRPC4G503S 24 h after induction plotted versus time in the absence (black traces) and in the presence of coexpressed Tarbp2-IRES-GFP cDNA (red trace). pA, picoampere; pF, picofarad. Shown are corresponding current-voltage relationships immediately after break-in (I) and after 300 s (J). n, number of cells. K and L, HEK293 cells expressing TRPC4G503S (18 h after induction) and coexpressing IRES-GFP (black trace) as control or the Tarbp2-IRES-GFP cDNA (red trace) were loaded with Fura-2-AM and kept in nominally Ca2+-free bath solution. Ca2+ influx was challenged by adding 2 mm Ca2+ to the bath solution, and the cytosolic [Ca2+], represented by the Fura-2 fluorescence ratio (F340/F380), was measured versus time (K). Note that no agonist is present. L, summary of two experiments. Data are mean ± S.E. The number of cells is indicated. *, p < 0.001.
Next, the effects of Tarbp2 on TRPC4-dependent Ca2+ entry was studied by coexpressing the TRPC4 and Tarbp2 cDNAs. To distinguish changes of cytosolic [Ca2+] resulting from depletion of intracellular stores, store-operated Ca2+ entry and agonist-induced, TRPC4-dependent Ca2+ entry, the cells were first treated with thapsigargin (in nominal Ca2+-free bath solution), followed by readdition of 2 mm Ca2+ and by addition of the muscarinic receptor agonist carbachol to the bath solution (Figs. 2, C–E). Carbachol-induced, TRPC4-dependent Ca2+ entry is readily detectable in HEK293 cells stably expressing the muscarinic receptor type 2 (M2R) and the TRPC4α cDNA (Fig. 2D), whereas no carbachol-induced Ca2+ entry is observed in M2R cells (Fig. 2C) or M2R cells coexpressing the Tarbp2 cDNA (Fig. 2E). The carbachol-induced, TRPC4-dependent Ca2+ entry was increased significantly in M2R TRPC4 cells coexpressing the Tarbp2 cDNA compared with cells coexpressing the GFP cDNA only (Fig. 2F). Fig. 2G summarizes the results of three independent experiments like the one shown in Fig. 2F. The carbachol-induced, TRPC4-dependent Ca2+ entry calculated as area under the curve was increased significantly by 55.8% in the presence of Tarbp2 (Fig. 2G), whereas the thapsigargin- and Ca2+-induced changes are not altered (Fig. 2, F and G).
In the open conformation of the TRPC4 channel, the TRPC4 C terminus might be in a more favorable configuration to facilitate interaction with TRPC4-binding partners, and Tarbp2 may preferentially bind to the open rather than to the closed configuration of the TRPC4 channel. Therefore, we made use of the TRPC4G503S mutant, which renders the channel constitutively active (25). The measurable phenotype, in this case ensemble conductance in the absence of any agonist, comprises the typical, doubly rectifying TRPC4 current (Fig. 2, I and J). The Tarbp2 cDNA was transfected into a tetracycline-inducible HEK293 cell line expressing this TRPC4G503S mutant. Upon establishing the whole-cell configuration (Fig. 2H) or upon addition of extracellular Ca2+ (Fig. 2K), constitutive current activity (Fig. 2, H–J) and Ca2+ entry (Fig. 2, K and L) were detectable, which were inhibited by >53% (inward current and F340/F380) in the presence of Tarbp2 (Fig. 2, H–L). Apparently, Tarpb2 binding by TRPC4 is facilitated by the activated TRPC4 channel configuration.
Mapping of the Amino Acid Sequences Involved in Tarbp2-TRPC4 Binding
To narrow down the TRPC4-Tarbp2 interaction, GST-TRPC4 fusion proteins LB3/4a, LB3/4b, LB5/6a, LB5/6b, and pJZ51 were generated in E. coli, covering the complete TRPC4 C terminus (Fig. 3A). The fusion proteins were used to pull down the Tarbp2 protein present in HEK293 cells. As shown in Fig. 3B, Tarbp2 is readily detectable in cell lysate and is retained by two of the GST-TRPC4 fusion proteins, LB3/4a and LB5/6b, but not by GST alone. LB3/4a covers aa 625–697 of the TRPC4 C terminus just adjacent to the predicted transmembrane helix S6 and the N-terminal of the calmodulin- and inositol 1,4,5-trisphosphate receptor-binding (CIRB) domains of the TRPC4 protein (11) (Fig. 3C). It includes the so-called TRP motif EWKFAR, and its sequence is very similar to the corresponding aa sequence present in TRPC5 (Fig. 3C). The fusion protein LB5/6B covers aa 786–847, which are present in TRPC4α but not TRPC4β, and comprises two additional calmodulin-binding domains, Cam-BD2 (11) and Cam-BD3 (12) (Fig. 3C). There are no obvious amino acid sequence similarities of the two TRPC4 Tarbp2-binding domains (14.5% sequence identity).
Because of the sequence similarity of TRPC4 binding domain 1 for Tarpb2 (aa 625–697) with TRPC5 (Fig. 3C), we hypothesized that Tarbp2 might also interact with TRPC5. Fig. 3D shows the Western blot analysis of the coimmunoprecipitation of TRPC5 and Tarbp2. The TRPC5 protein is immunoprecipitated by the anti-TRPC5 antibody, and it is among the proteins associated with Tarbp2, which was precipitated by the anti-FLAG antibody (Fig. 3D, top panel). Reciprocal antibody immunoprecipitation demonstrates that the Tarbp2 protein is present among the proteins associated with TRPC5 (Fig. 3D, bottom panel). These results show that Tarbp2 also binds to TRPC5. We transfected the Tarbp2 cDNA into a tetracycline-inducible HEK293 cell line expressing TRPC5G504S (25) that, like TRPC4G503S, forms a constitutively active channel. Upon inducing TRPC5G504S expression and addition of extracellular Ca2+ (Fig. 3E), Ca2+ entry was detectable and was reduced by 54% in the presence of Tarbp2 (Fig. 3, E and F).
The sequence of TRPC4 binding domain 2 for Tarbp2 comprises two domains that have been shown to bind Ca2+-dependently calmodulin in vitro (11, 12). To study the ability of calmodulin to compete with Tarbp2 for binding to TRPC4, the Tarbp2 protein was pulled down from cell lysates by TRPC4-GST fusion protein LB5/6b in the absence of calmodulin and in the presence of increasing amounts of calmodulin. Fig. 3G shows that increasing the calmodulin/GST-TRPC4 molar ratio from 0.1 to 2.0 decreases the amount of Tarbp2 retained by TRPC4. Apparently, Tarbp2 competes with calmodulin for TRPC4 binding.
The Tarbp2 protein comprises two double-stranded RNA-binding domains, dsRBD1 (aa 32–96) and dsRBD2 (aa 153–225), both containing a KR-helix motif (aa 80–96 and aa 209–225) and a C-terminal region called Medipal (aa 298–365) that mediates protein-protein interactions (19). The dsRBD2, but not dsRBD1 and the Medipal, is part of the identified Tarbp2 cDNA clones isolated from the initial yeast two-hybrid screen (Figs. 1D and 3H). Therefore, a part of the dsRBD2 present in both cDNA clones, D57 and D90, including the complete KR-helix motif, was removed. The resulting Tarbp2Δ209–234-encoding cDNA and the TRPC4 cDNA were coexpressed in HEK cells. The Tarbp2Δ209–234 protein was detectable in the cell lysate (Fig. 3I, bottom panel), indicating that the deletion does not interfere with protein expression. From cell lysates, coimmunoprecipitations were performed using the anti-TRPC4 antibody and the FLAG antibody for the tagged Tarbp2Δ209–234. Fig. 3I shows that TRPC4 is precipitated by the anti TRPC4 antibody (Fig. 3I, top panel) but is not present among the proteins associated with Tarbp2 precipitated by the anti-FLAG antibody (Fig. 3I, top panel). The Tarbp2Δ209–234 protein is precipitated by the anti-FLAG antibody (Fig. 3I, bottom panel) but is not present among the proteins associated with TRPC4 precipitated by the anti-TRPC4 antibody (Fig. 3I, bottom panel). Coexpression of the Tarbp2 but not of the Tarbp2Δ209–234 cDNA does interfere with TRPC4G503S-dependent Ca2+ entry (Fig. 3, J and K). Taken together, these data confirm the TRPC4-Tarbp2 interaction and indicate that TRPC4-Tarbp2 interaction depends on the presence of amino acid residues 209–234 of Tarbp2.
Dicer-Tarbp2 Interaction and Ca2+-dependent Modulation of Dicer Activity
Tarbp2 also binds Dicer, and both proteins together with Argonaute represent the minimal RISC-loading complex and carry out RNA-induced silencing complex (RISC) activity by processing precursor miRNA (20, 28, 29). Therefore, we wondered whether the TRPC4 protein might interfere with Tarbp2 binding to Dicer and cotransfected HEK cells with the cDNAs of all three proteins. The expressed proteins, TRPC4, Dicer, and Tarbp2, are readily detectable in Western blot analyses of cell lysate (Fig. 4A), and they are readily precipitated by their respective antibodies. The anti-FLAG antibody precipitates the FLAG-tagged Tarbp2 (Fig. 4A, right panel, center), and among the proteins associated with the precipitated Tarbp2 is Dicer (Fig. 4A, right panel, bottom), whereas the Tarbp2 and Dicer and Tarbp2 and TRPC4 proteins are hardly detectable as associated proteins of the precipitated TRPC4 and Dicer proteins, respectively (Fig. 4A).
FIGURE 4.
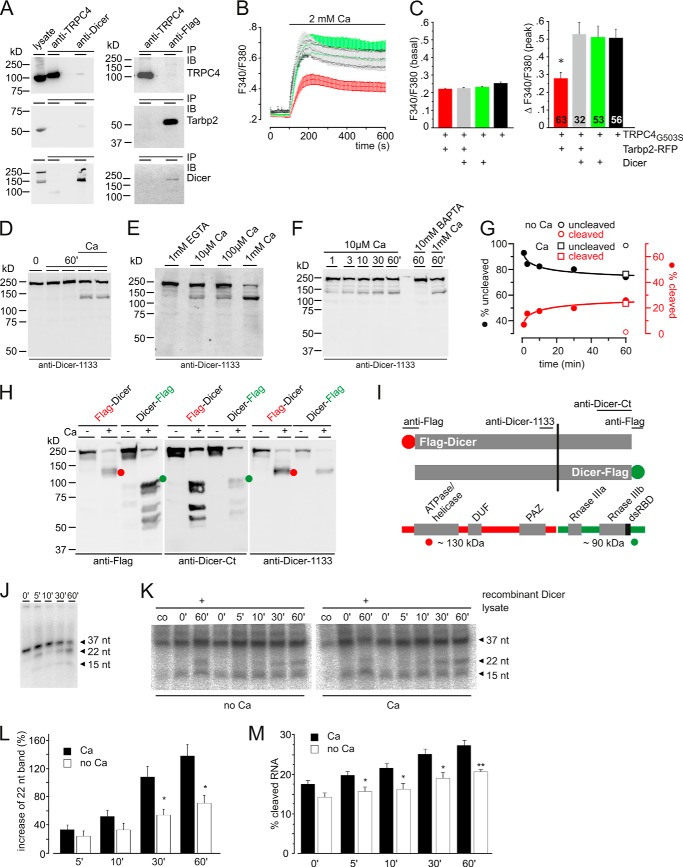
Interplay of Tarbp2, TRPC4, and Dicer. A, Western blot analyses. All proteins, TRPC4, Tarbp2, and Dicer, are detectable in cell lysate. Immunoprecipitation (IP) after coexpressing the cDNAs of FLAG-tagged Tarbp2, TRPC4, and Dicer in HEK293 cells using the anti-TRPC4, anti-Dicer, and anti-FLAG antibodies is shown. Immunoblots (IB) were incubated with antibodies for TRPC4 (top), Tarbp2-FLAG (center), and Dicer (bottom), respectively. B, HEK293 cells expressing TRPC4G503S (19 h after induction) and coexpressing the cDNAs of IRES-GFP plus TagRFP-T (black) as control, Tarbp2-TagRFP-T plus Dicer-GFP (gray), Dicer-GFP plus TagRFP-T (green), or Tarbp2-IRES-GFP plus TagRFP-T cDNA (red) were loaded (48 h after transfection) with Fura-2/AM and kept in nominally Ca2+-free bath solution. Ca2+ influx was challenged by adding 2 mm Ca2+ to the bath solution, and the cytosolic [Ca2+], represented by the fura-2 fluorescence ratio (F340/F380), was measured versus time. C, summary. Data are mean (peak after addition of Ca2+) ± S.E. *, p < 0.001. D, Ca2+-dependent cleavage of endogenous Dicer in HEK293/M2R/TRPC4 cells using Ca2+-free lysate buffer. Lysates (in duplicate) were incubated in the absence and presence of 2 mm Ca2+ for 60 min. Proteins were run on SDS-PAGE and blotted to a filter membrane, which was incubated with antibody for Dicer. E, Dicer cleavage in the absence and in the presence of increasing [Ca2+]. F and G, time course of cleavage in the presence of 10 μm Ca2+. F, Western blot analysis. G, densitometric analyses of band intensities. ○, values in the presence of 10 mm BAPTA (no Ca), 60 min; □, values in the presence of 1 mm Ca (Ca), 60 min. H, mapping of the Ca2+-dependent cleavage site using N-terminal (red) and C-terminal (green) FLAG-tagged human Dicer (expressed in HEK/M2R/TRPC4 cells) and antibodies for FLAG (anti-FLAG) and Dicer (anti-Dicer-Ct and anti-Dicer-1133). Cell lysates were prepared in the absence (−, 1 mm EGTA) and presence (+) of 1 mm Ca2+, and proteins were run on SDS-PAGE, blotted, and incubated with the indicated antibodies. Red, N-terminal tagged Dicer fragment; green, C-terminal tagged Dicer fragment. The red and green circles indicate the proteolytic fragment comprising the N-terminal (red) or C-terminal tag (green). I, summary of mapping experiments. Ca2+-dependent cleavage yields ∼130 (red) and ∼90 kDa (green) fragments that are visible in addition to the uncleaved full-length protein of ∼220 kDa (in H). The cleavage site maps near serine 1150 of human Dicer. J, time dependence of recombinant Dicer cleavage of the double-stranded, 37-bp RNA template 5′-phosphorylated at a single strand. Samples were run on polyacrylamide/urea gels and exposed to a PhosphorImager screen. K, Dicer activity at the time points indicated in HEK293 M2R TRPC4 lysates in the absence and presence of 2 mm Ca2+ using the double-stranded 37ab RNA as a template (3000 cpm/tube). As controls, the double-stranded 37-bp RNA template was applied without lysate (co), and recombinant Dicer (0.5 units) was spiked. Samples were run on polyacrylamide/urea gels and exposed to a PhosphorImager screen. L and M, densitometric analysis of Dicer-dependent cleavage of the double-stranded 37ab RNA. L, intensity of the 22-nt product in K normalized to the intensity of the 22-nt product obtained by recombinant Dicer in the absence of Ca2+. Data are the mean of four independent experiments ± S.E. *, p < 0.05. M, the percentage of cleaved dsRNA 37ab in K as calculated by the arbitrary units obtained by densitometric analysis of the 37- (a), 22- (b), and 15-nt (c) bands (percent of dsRNA 37ab cleavage = ((b + c) × 100)/(a + b + c)). Data are the mean of four independent experiments ± S.E. *, p < 0.05; **, p < 0.01.
Tarpb2 binding preferentially interferes with the activated TRPC4 channel configuration (Fig. 2, H–L). Therefore, we transfected the cDNAs for Tarbp2, Tarbp2 plus Dicer, and Dicer alone into the tetracycline-inducible HEK293 cell line expressing the constitutively active TRPC4G503S mutant (25). Upon addition of extracellular Ca2+ (Fig. 4B), Ca2+ entry was not significantly different in cells expressing TRPC4G503S (Fig. 4, B and C, black), TRPC4G503S plus Dicer (Fig. 4, B and C, green), or TRPC4G503S plus Dicer plus Tarbp2 (Fig. 4, B and C, gray) but was reduced by 45% in cells coexpressing TRPC4G503S plus Tarbp2 (Fig. 4, B and C, red) as shown in Figs. 2, K and L, and 3, J and K. Apparently, Tarbp2 binds TRPC4 and modulates TRPC4 function but is scavenged by the heterologously expressed Dicer, which by itself does not interact with TRPC4.
Because Tarbp2 binds TRPC4 and, thereby, modulates Ca2+ entry, the Dicer protein might be affected by changes of the cytosolic Ca2+ concentration. It is known that endogenous Dicer present in mammalian cells is stimulated by proteolysis (30). Therefore, we analyzed cell lysates for Ca2+-dependent proteolysis of endogenous Dicer. Fig. 4D shows the Western blot obtained after running cell lysates on SDS-PAGE and incubation with an anti-Dicer antibody. An additional, ∼130-kDa Dicer fragment in addition to the full-length, ∼220-kDa Dicer protein is detectable in the presence, but not in the absence, of Ca2+. Apparently, the increase in [Ca2+] promotes proteolysis of the Dicer protein. Proteolysis is already apparent at 10 μm [Ca2+] (Fig. 4E) and after 1 min (Fig. 4F). Fig. 4G summarizes the time-dependent cleavage of Dicer in the presence of 10 μm [Ca2+]. Using N- and C-terminal FLAG-tagged Dicer proteins, anti-FLAG, and two anti-Dicer antibodies that recognize a sequence of dicer around amino acid 1133 and its C terminus, respectively, the Ca2+-dependent cleavage site within Dicer was mapped (Fig. 4, H and I) to a region C-terminal of alanine1133, apparently near position 1150 (serine 1150). The N-terminal Dicer fragment contains the ATPase/hel, domain of unknown function (DUF) 283, and Piwi Argonaut and Zwille (PAZ) domains, and the other contains the two tandem RNase III domains and the C-terminal dsRBD. A very similar proteolytic processing of the endogenous Dicer protein has been identified in Caenorhabditis elegans (31), with a proteolytic cleavage near position 1200 that corresponds to position 1155 in the human enzyme. In addition, very similar, ∼130- and ∼90-kDa fragments were obtained by proteolysis of full-length recombinant human Dicer by endoproteinase Glu-C (32). The C-terminal fragment lacking regulatory domains (helicase, DUF, and PAZ) has been shown to be catalytically active (31, 32), although miRNA-specific (31).
Fig. 4J shows the time-dependent cleavage of a double-stranded, 37-bp RNA template 5′-phosphorylated at one strand by recombinant Dicer into 22-bp and 15-bp fragments. In the next experiment, the Dicer activity of HEK M2R TRPC4 cells coexpressing the Dicer cDNA was assayed in the absence and presence of Ca2+ using the same double-stranded, 37-nt RNA template. After densitometric analysis of the intensity of the 37-, 22-, and 15-bp fragments in the resulting autoradiographs (Fig. 4K), the time-dependent increase of the 22-bp fragment (Fig. 4L) and of cleaved RNA (Fig. 4M) is apparent. In addition, cleavage is increased in the presence of Ca2+.
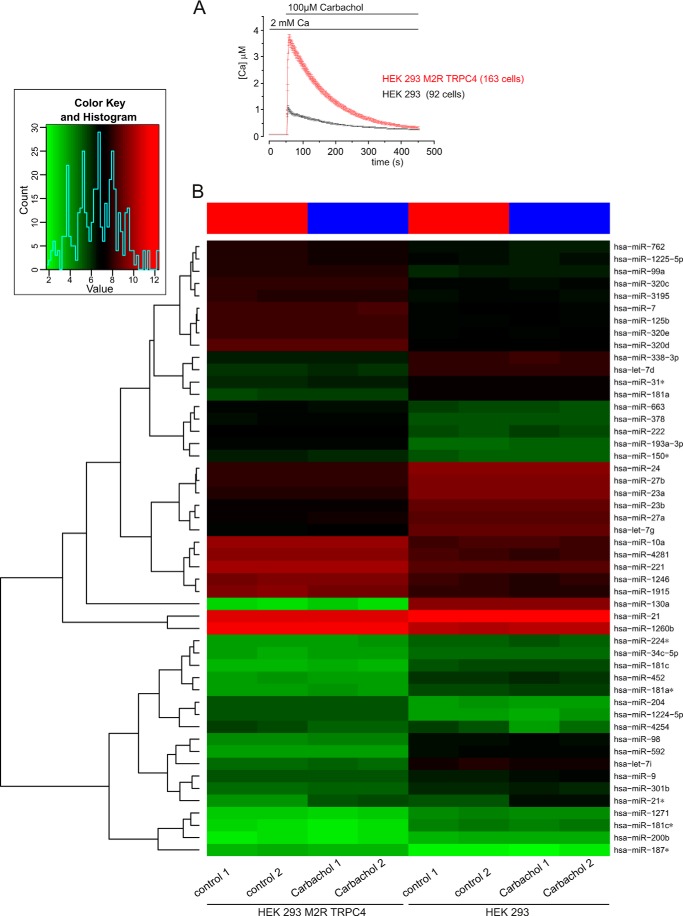
Next we incubated HEK293/M2R/TRPC4 cells in the presence of 100 μm carbachol for 15 min and performed miRNA analyses thereafter (Fig. 5). Under these conditions, cytosolic [Ca2+] increases to ∼4 μm (Fig. 5A), although, with its high affinity, the Fura-2 was very likely saturated and the real intracellular [Ca2+] might even be higher (33). Overall, about 297–306 miRNAs were detected in the analyzed samples. In the HEK293/M2R/TRPC4 group, we found 10 miRNAs (hsa-miR-1972, hsa-miR-425*, hsa-miR-124, hsa-miR-933, hsa-miR-142–3p, hsa-let-7b*, hsa-miR-455–5p, hsa-miR-431*, hsa-miR-548a-5p, and hsa-miR-3200–3p) that were detected in the controls but not after carbachol treatment of the cells. However, no miRNA was found that was induced because of carbachol treatment but not detected in the control cells. A total of 289 miRNAs were detected in all HEK293/M2R/TRPC4 samples. In the HEK293 control, we found one miRNA (hsa-miR-630) that was detected in the controls but not in the carbachol-treated cells, and three miRNAs (hsa-miR-1238, hsa-miR-93*, and hsa-miR-191*) were detected after carbachol treatment but not in the control cells. A total of 319 miRNAs were detected in all HEK293 samples.
FIGURE 5.
miRNA expression in TRPC4-expressing cells. A, monitoring cytosolic [Ca2+] in the presence of carbachol. B, heat map of miRNAs derived from cells that were grown in the absence or presence of carbachol. HEK293/M2R/TRPC4 cells and HEK 293 cells were incubated in the presence of 100 μm carbachol for 15 min. Controls were performed without carbachol treatment for both HEK293/M2R/TRPC4 cells and HEK 293 cells. Each experiment was done in duplicate (carbachol 1/2 and control 1/2). miRNA expression was analyzed using SurePrint 8 × 60K human v16 miRNA microarrays (Agilent) that contained 40 replicates of 1205 miRNAs. A heat map was generated using the 50 miRNAs with highest expression variance over all samples. The miRNA expression values are visualized by the red-green color code, where green means low expression and red means high expression. The samples (with/without carbachol) are arranged in columns and the 50 selected miRNAs in rows. The heat map shows that HEK293/M2R/TRPC4 cells and HEK 293 cells cluster separately, indicating that each cell line has a specific miRNA expression pattern. For each cell line there is a clear differentiation between the carbachol treatments and controls, as indicated by the red (controls) and blue (carbachol) bars at the top of the heat map. The duplicates cluster closely together, indicating the high reproducibility of the data. The dendrogram on the left side of the heat map shows miRNAs clusters on the basis of the similarity of their expression levels. Inset, summary of the overall distribution of all miRNA counts according to their expression level, with the lowest expression level in green and the highest expression in red.
Next we checked whether the expression levels of certain miRNAs were affected by carbachol treatment. Here we found three miRNAs (hsa-miR-4254, hsa-miR-574–3p, and hsa-miR-630) in the HEK293 carbachol-treated cells that were down-regulated more than 1.5-fold compared with controls and two miRNAs (hsa-miR-24–2* and hsa-miR-21*) that were up-regulated because of carbachol treatment. In HEK293/M2R/TRPC4 cells, three miRNAs (hsa-miR-1238, hsa-miR-2276, and hsa-miR-574–3p) were down-regulated more than 1.5-fold because of carbachol treatment, whereas two miRNAs (hsa-miR-24–2* and hsa-miR-21*) were up-regulated. Two miRNAs (hsa-miR-24–2* and hsa-miR-21*) were up-regulated in both cell lines, HEK293 and HEK293/M2R/TRPC4, because of carbachol treatment. One miRNA, namely hsa-miR-574–3p, was down-regulated in both cell lines because of carbachol treatment.
A heat map analysis was performed using the 50 miRNAs with the highest variance above all analyzed samples. Cells of the same cell line clustered together, independent of carbachol treatment. In each cluster we found a subclustering into carbachol-treated cells and controls. This indicates that each cell line has its specific miRNA expression pattern and that this pattern is affected by carbachol treatment (Fig. 5B).
DISCUSSION
In this study, we identified Tarbp2 as a novel TRPC4- and TRPC5-interacting protein. Tarbp2, or trans-activation response (TAR) RNA-binding protein (TRBP) 2, is one isoform of the cellular protein TRBP encoded by the tarbp2 gene, which was isolated by its ability to bind the human immunodeficiency virus, type 1 TAR RNA (18) and is characterized for its stimulation of the expression of the HIV long terminal repeat in human and murine cells (18). Tarbp2 is a double-stranded RNA-binding protein comprising two dsRBDs, aa 32–96 and aa 153–225), that associates with Dicer and is required for RNAi function mediated by both siRNAs and miRNAs (21, 22). The third domain of Tarbp2 (aa 298–366) has some structural homology to the dsRBDs, but this domain, also called C4, does not bind RNA but mediates protein-protein interaction and is sufficient for binding of Dicer and protein kinase R activator (19). The amino acid sequence of Tarbp2 involved in TRPC4 binding comprises a part of the dsRBD2 of Tarbp2, including the complete KR-helix motif, a stretch of 16, mainly basic amino acid residues with an α-helical structure (Fig. 3, H–K).
The binding site on TRPC4 was mapped to two regions within the C terminus of TRPC4, covering aa 625–697 and aa 786–847. The first region is just distal of the predicted transmembrane segment 6, includes the highly conserved EWKFAR motif of the “TRP box” found in several TRP subfamilies (4) and is almost identical to the corresponding sequence of TRPC5. Just C-terminal of this domain resides the sequence referred to as the CIRB domain (11), involved in TRPC4 interaction with calmodulin, the calmodulin binding domain (CBD 1), and inositol 1,4,5-trisphosphate receptor binding and conserved in all TRPC proteins. The aa sequence of the second region is not conserved in other TRPCs, including TRPC5. Two binding domains for calmodulin have been identified within this second region (11, 12), and calmodulin competes with Tarbp2 for TRPC4 binding (Fig. 3G).
The region encompassing the TRP box (4) has been suggested to be a determinant of TRPV1 subunit multimerization (34) and as a transduction domain for TRPV1 channel gating (35). In TRPM channels, this region appears to be required for phosphoinositide-mediated channel gating (36, 37) as well as for TRPM8 activation with menthol (38). In the Drosophila TRP and TRP1 proteins, the corresponding domains are required for efficient light response (39).
The C termini of most TRPCs bind directly to phosphoinositides (40), and phosphatidylinositol 4,5-bisphosphate has been shown to bind to the C terminus of TRPC4 and to selectively inhibit TRPC4α function (41). An essential region required for binding and for modulating TRPC4 activity is the KR-helix motif of Tarbp2. This region may not be directly involved in TRPC4 binding but could be responsible to stabilize a Tarbp2 confirmation suitable for TRPC4 binding. Tarbp2 contains several stretches of basic residues in addition to the two KR-helix motifs in dsRBD1 and dsRBD2, and it is conceivable that the positively charged side chains of these basic residues of TRPC4-bound Tarbp2 might interact with the negatively charged head groups of phosphatidylinositol 4,5-bisphosphate. Therefore, Tarbp2 might interfere with phosphatidylinositol 4,5-bisphosphate/TRPC4 interaction and increase agonist-dependent TRPC4 activity.
An interaction between the S4-S5 linker and a site in or C-terminal of the S6 transmembrane segment may stabilize the closed or gateable state of TRPC4 (and TRPC5) (25), very similar to the state-dependent interaction between the S4-S5 linker and a site in the S6 transmembrane segment suggested to stabilize the closed state of the depolarization-activated Kv7.1 (KCNQ1) channel (42, 43) and the open state of hyperpolarization-activated channels like KAT1 (44) and hyperpolarization-activated cyclic nucleotide-gated (HCN) channels (45). This interaction is disturbed when a mutation is introduced into the S4-S5 linker of TRPC4 and TRPC5, yielding fully active channels in the absence of any agonist (25). Under these conditions, binding of Tarbp2 to the C terminus of TRPC4 (and TRPC5) by itself might reduce the fraction of open channels and, thereby, reduce current and Ca2+ entry.
Binding sites for a couple of proteins have been identified on the C terminus of TRPC4, and, apparently, the available data show that these protein-protein interactions serve specific functions, including modulation of Ca2+ entry and appropriate assembly within signaling complexes. A proline-rich region C-terminal of the EWKFAR motif represents such a site of protein-protein interaction and has been implicated in the binding of protein 4.1 (15) and of the immunophilin FKBP52 (46) to TRPC4. Binding sites for calmodulin (47), the inositol 1,4,5-trisphosphate receptor (11), and SESTD1 (17) have been mapped to the CIRB domain, and it is assumed that both calmodulin and inositol 1,4,5-trisphosphate receptors play roles in gating TRPC channels, whereas SESTD1 may couple TRPC4 and TRPC5 channel function to lipid signaling. The binding site for αII-spectrin has been mapped C-terminally of the CIRB domain (14) and may influence TRPC4 surface expression. The PDZ-binding domain located at the distal C terminus of TRPC4 and TRPC5 (16) binds to the Na+/H+ exchange regulatory factor protein, and, in addition to coupling TRPC4 and TRPC5 to other members of the signaling cascade, such as phospholipase Cβ (16) and G protein-coupled receptors (48), Na+/H+ exchange regulatory factor protein may indirectly link TRPC4 to the cytoskeleton.
Tarbp2 is predominantly present in the cytoplasm, whereas Dicer can be observed in the nucleus and in the cytoplasm (49). The Dicer binding site in Tarpb2 is the 69 aa C4 domain, located at the C-terminal end of Tarbp2 (49). Tarbp2 mutants lacking the C4 domain are partially associated to the cell membrane (49). Considering the plasma membrane localization of TRPC4, it appears feasible that Tarbp2 colocalizes with and binds to TRPC4, especially under conditions of low Dicer concentrations. Dicer is endogenously expressed in HEK cells, but after overexpression of the Dicer cDNA in these cells, Tarbp2 is no longer associated with TRPC4 (Fig. 4A) nor is the Tarbp2-mediated modulation of TRPC4-induced Ca2+ entry detectable (Fig. 4B).
Endogenous Dicer is strongly stimulated by proteolysis (30, 31, 50), and TRPC4-dependent Ca2+ entry could promote activation of Ca2+-activated proteases to fine-tune Dicer activity. As shown in Fig. 4, D–I, Dicer is cleaved in the presence of Ca2+, yielding an N-terminal, ∼130-kDa fragment containing the ATPase/hel, DUF283, and PAZ domains and a C-terminal fragment containing the two tandem RNase III domains and the C-terminal dsRBD. At the same time, Dicer/ribonuclease III activity increases in the presence of Ca2+ in crude cell lysates (Fig. 4, K–M), and the miRNA expression pattern of HEK293/M2R/TRPC4 cells is affected (Fig. 5).
These results correspond with a model that Tarbp2, upon binding to TRPC4, modulates Ca2+ entry, which then promotes proteolytic activation of Dicer. Recently, Ca2+ influx through TRPC1 channels has been shown to activate a calpain protease that reduces axon outgrowth through proteolytic cleavage of the integrin-binding protein talin (51). In synaptosomes, activity-dependent calpain cleavage stimulates Dicer activity, which could result in enhanced miRNA production at activated synapses (50). Accordingly, TRPC-dependent Ca2+ entry can be expected to activate Ca2+-sensitive proteases, which have been localized to spines and postsynaptic densities (50, 52). MicroRNAs, a class of noncoding RNAs, are capable of regulating mRNA translation by various mechanisms (28) and, thereby, are also elements for the regulation of plasticity-related mRNAs by neurons (53). By the mechanism suggested above, Dicer activity could be dynamically regulated in neurons that do express TRPC4 and TRPC5 and induce profound changes in neuronal plasticity given the slow turnover rates of Dicer-dependent noncoding RNAs in these cells (54).
Acknowledgments
We thank Karin Wolske, Heidi Löhr, Stephanie Buchholz, and Martin Simon-Thomas for technical assistance.
This work was supported by the Deutsche Forschungsgemeinschaft (to A. B., P. L., and V. F.), by the Forschungsausschuss der Medizinischen Fakultät “HOMFOR,” and by the Forschungsausschuss der Universität des Saarlandes (to P. L., E. M., C. F.-T., and V. F.).
- TRP
- transient receptor potential
- TRPC
- transient receptor potential canonical
- BAPTA
- 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid
- aa
- amino acid(s)
- dsRBD
- double-stranded RNA-binding domain
- M2R
- muscarinic receptor type 2
- CIRB
- calmodulin- and inositol 1,4,5-trisphosphate receptor-binding
- miRNA
- microRNA
- TAR
- trans-activation response
- TRBP
- trans-activation response RNA-binding protein
- nt
- nucleotide(s).
REFERENCES
- 1. Philipp S., Cavalié A., Freichel M., Wissenbach U., Zimmer S., Trost C., Marquart A., Murakami M., Flockerzi V. (1996) A mammalian capacitative calcium entry channel homologous to Drosophila TRP and TRPL. EMBO J. 15, 6166–6171 [PMC free article] [PubMed] [Google Scholar]
- 2. Gees M., Colsoul B., Nilius B. (2010) The role of transient receptor potential cation channels in Ca2+ signaling. Cold Spring Harb. Perspect. Biol. 2, a003962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wu L. J., Sweet T. B., Clapham D. E. (2010) International Union of Basic and Clinical Pharmacology. LXXVI. Current progress in the mammalian TRP ion channel family. Pharmacol. Rev. 62, 381–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Flockerzi V. (2007) An introduction on TRP channels. Handb. Exp. Pharmacol. 179, 1–19 [DOI] [PubMed] [Google Scholar]
- 5. Munsch T., Freichel M., Flockerzi V., Pape H. C. (2003) Contribution of transient receptor potential channels to the control of GABA release from dendrites. Proc. Natl. Acad. Sci. U.S.A. 100, 16065–16070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Phelan K. D., Mock M. M., Kretz O., Shwe U. T., Kozhemyakin M., Greenfield L. J., Dietrich A., Birnbaumer L., Freichel M., Flockerzi V., Zheng F. (2012) Heteromeric canonical transient receptor potential 1 and 4 channels play a critical role in epileptiform burst firing and seizure-induced neurodegeneration. Mol. Pharmacol. 81, 384–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Freichel M., Suh S. H., Pfeifer A., Schweig U., Trost C., Weissgerber P., Biel M., Philipp S., Freise D., Droogmans G., Hofmann F., Flockerzi V., Nilius B. (2001) Lack of an endothelial store-operated Ca2+ current impairs agonist-dependent vasorelaxation in TRP4−/− mice. Nat. Cell Biol. 3, 121–127 [DOI] [PubMed] [Google Scholar]
- 8. Tiruppathi C., Freichel M., Vogel S. M., Paria B. C., Mehta D., Flockerzi V., Malik A. B. (2002) Impairment of store-operated Ca2+ entry in TRPC4−/− mice interferes with increase in lung microvascular permeability. Circ. Res. 91, 70–76 [DOI] [PubMed] [Google Scholar]
- 9. Tsvilovskyy V. V., Zholos A. V., Aberle T., Philipp S. E., Dietrich A., Zhu M. X., Birnbaumer L., Freichel M., Flockerzi V. (2009) Deletion of TRPC4 and TRPC6 in mice impairs smooth muscle contraction and intestinal motility in vivo. Gastroenterology 137, 1415–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Strübing C., Krapivinsky G., Krapivinsky L., Clapham D. E. (2001) TRPC1 and TRPC5 form a novel cation channel in mammalian brain. Neuron 29, 645–655 [DOI] [PubMed] [Google Scholar]
- 11. Tang J., Lin Y., Zhang Z., Tikunova S., Birnbaumer L., Zhu M. X. (2001) Identification of common binding sites for calmodulin and inositol 1,4,5-trisphosphate receptors on the carboxyl termini of trp channels. J. Biol. Chem. 276, 21303–21310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Trost C., Bergs C., Himmerkus N., Flockerzi V. (2001) The transient receptor potential, TRP4, cation channel is a novel member of the family of calmodulin binding proteins. Biochem. J. 355, 663–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yuan J. P., Zeng W., Huang G. N., Worley P. F., Muallem S. (2007) STIM1 heteromultimerizes TRPC channels to determine their function as store-operated channels. Nat. Cell Biol. 9, 636–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Odell A. F., Van Helden D. F., Scott J. L. (2008) The spectrin cytoskeleton influences the surface expression and activation of human transient receptor potential channel 4 channels. J. Biol. Chem. 283, 4395–4407 [DOI] [PubMed] [Google Scholar]
- 15. Cioffi D. L., Wu S., Alexeyev M., Goodman S. R., Zhu M. X., Stevens T. (2005) Activation of the endothelial store-operated ISOC Ca2+ channel requires interaction of protein 4.1 with TRPC4. Circ. Res. 97, 1164–1172 [DOI] [PubMed] [Google Scholar]
- 16. Tang Y., Tang J., Chen Z., Trost C., Flockerzi V., Li M., Ramesh V., Zhu M. X. (2000) Association of mammalian trp4 and phospholipase C isozymes with a PDZ domain-containing protein, NHERF. J. Biol. Chem. 275, 37559–37564 [DOI] [PubMed] [Google Scholar]
- 17. Miehe S., Bieberstein A., Arnould I., Ihdene O., Rütten H., Strübing C. (2010) The phospholipid-binding protein SESTD1 is a novel regulator of the transient receptor potential channels TRPC4 and TRPC5. J. Biol. Chem. 285, 12426–12434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gatignol A., Buckler C., Jeang K. T. (1993) Relatedness of an RNA-binding motif in human immunodeficiency virus type 1 TAR RNA-binding protein TRBP to human P1/dsI kinase and Drosophila Staufen. Mol. Cell. Biol. 13, 2193–2202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Daniels S. M., Gatignol A. (2012) The multiple functions of TRBP, at the hub of cell responses to viruses, stress, and cancer. Microbiol. Mol. Biol. Rev. 76, 652–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rossi J. J. (2005) Mammalian Dicer finds a partner. EMBO Rep. 6, 927–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chendrimada T. P., Gregory R. I., Kumaraswamy E., Norman J., Cooch N., Nishikura K., Shiekhattar R. (2005) TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 436, 740–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Haase A. D., Jaskiewicz L., Zhang H., Lainé S., Sack R., Gatignol A., Filipowicz W. (2005) TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 6, 961–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Meister G. (2013) Argonaute proteins. Functional insights and emerging roles. Nat. Rev. Genet. 14, 447–459 [DOI] [PubMed] [Google Scholar]
- 24. Shaner N. C., Lin M. Z., McKeown M. R., Steinbach P. A., Hazelwood K. L., Davidson M. W., Tsien R. Y. (2008) Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat. Meth. 5, 545–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Beck A., Speicher T., Stoerger C., Sell T., Dettmer V., Jusoh S. A., Abdulmughni A., Cavalié A., Philipp S. E., Zhu M. X., Helms V., Wissenbach U., Flockerzi V. (2013) Conserved gating elements in TRPC4 and TRPC5 channels. J. Biol. Chem. 288, 19471–19483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Miller M., Shi J., Zhu Y., Kustov M., Tian J. B., Stevens A., Wu M., Xu J., Long S., Yang P., Zholos A. V., Salovich J. M., Weaver C. D., Hopkins C. R., Lindsley C. W., McManus O., Li M., Zhu M. X. (2011) Identification of ML204, a novel potent antagonist that selectively modulates native TRPC4/C5 ion channels. J. Biol. Chem. 286, 33436–33446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ma E., MacRae I. J., Kirsch J. F., Doudna J. A. (2008) Autoinhibition of human dicer by its internal helicase domain. J. Mol. Biol. 380, 237–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Filipowicz W., Bhattacharyya S. N., Sonenberg N. (2008) Mechanisms of post-transcriptional regulation by microRNAs. Are the answers in sight? Nat. Rev. Genet. 9, 102–114 [DOI] [PubMed] [Google Scholar]
- 29. MacRae I. J., Ma E., Zhou M., Robinson C. V., Doudna J. A. (2008) In vitro reconstitution of the human RISC-loading complex. Proc. Natl. Acad. Sci. U.S.A. 105, 512–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang H., Kolb F. A., Brondani V., Billy E., Filipowicz W. (2002) Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J. 21, 5875–5885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sawh A. N., Duchaine T. F. (2013) A truncated form of dicer tilts the balance of RNA interference pathways. Cell Rep. 4, 454–463 [DOI] [PubMed] [Google Scholar]
- 32. Ma E., Zhou K., Kidwell M. A., Doudna J. A. (2012) Coordinated activities of human dicer domains in regulatory RNA processing. J. Mol. Biol. 422, 466–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Blair N. T., Kaczmarek J. S., Clapham D. E. (2009) Intracellular calcium strongly potentiates agonist-activated TRPC5 channels. J. Gen. Physiol. 133, 525–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Valente P., García-Sanz N., Gomis A., Fernández-Carvajal A., Fernández-Ballester G., Viana F., Belmonte C., Ferrer-Montiel A. (2008) Identification of molecular determinants of channel gating in the transient receptor potential box of vanilloid receptor I. FASEB J. 22, 3298–3309 [DOI] [PubMed] [Google Scholar]
- 35. García-Sanz N., Valente P., Gomis A., Fernández-Carvajal A., Fernández-Ballester G., Viana F., Belmonte C., Ferrer-Montiel A. (2007) A role of the transient receptor potential domain of vanilloid receptor I in channel gating. J. Neurosci. 27, 11641–11650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Voets T., Nilius B. (2007) Modulation of TRPs by PIPs. J. Physiol. 582, 939–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rohács T., Lopes C. M., Michailidis I., Logothetis D. E. (2005) PI(4,5)P2 regulates the activation and desensitization of TRPM8 channels through the TRP domain. Nat. Neurosci. 8, 626–634 [DOI] [PubMed] [Google Scholar]
- 38. Bandell M., Dubin A. E., Petrus M. J., Orth A., Mathur J., Hwang S. W., Patapoutian A. (2006) High-throughput random mutagenesis screen reveals TRPM8 residues specifically required for activation by menthol. Nat. Neurosci. 9, 493–500 [DOI] [PubMed] [Google Scholar]
- 39. Wang T., Jiao Y., Montell C. (2005) Dissecting independent channel and scaffolding roles of the Drosophila transient receptor potential channel. J. Cell Biol. 171, 685–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kwon Y., Hofmann T., Montell C. (2007) Integration of phosphoinositide- and calmodulin-mediated regulation of TRPC6. Mol. Cell 25, 491–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Otsuguro K., Tang J., Tang Y., Xiao R., Freichel M., Tsvilovskyy V., Ito S., Flockerzi V., Zhu M. X., Zholos A. V. (2008) Isoform-specific inhibition of TRPC4 channel by phosphatidylinositol 4,5-bisphosphate. J. Biol. Chem. 283, 10026–10036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Labro A. J., Raes A. L., Grottesi A., Van Hoorick D., Sansom M. S., Snyders D. J. (2008) Kv channel gating requires a compatible S4-S5 linker and bottom part of S6, constrained by non-interacting residues. J. Gen. Physiol. 132, 667–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Labro A. J., Boulet I. R., Choveau F. S., Mayeur E., Bruyns T., Loussouarn G., Raes A. L., Snyders D. J. (2011) The S4-S5 linker of KCNQ1 channels forms a structural scaffold with the S6 segment controlling gate closure. J. Biol. Chem. 286, 717–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Grabe M., Lai H. C., Jain M., Jan Y. N., Jan L. Y. (2007) Structure prediction for the down state of a potassium channel voltage sensor. Nature 445, 550–553 [DOI] [PubMed] [Google Scholar]
- 45. Prole D. L., Yellen G. (2006) Reversal of HCN channel voltage dependence via bridging of the S4-S5 linker and Post-S6. J. Gen. Physiol. 128, 273–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sinkins W. G., Goel M., Estacion M., Schilling W. P. (2004) Association of immunophilins with mammalian TRPC channels. J. Biol. Chem. 279, 34521–34529 [DOI] [PubMed] [Google Scholar]
- 47. Zhu M. X. (2005) Multiple roles of calmodulin and other Ca2+-binding proteins in the functional regulation of TRP channels. Pfluegers Arch. Eur. J. Physiol. 451, 105–115 [DOI] [PubMed] [Google Scholar]
- 48. Hall R. A., Premont R. T., Chow C. W., Blitzer J. T., Pitcher J. A., Claing A., Stoffel R. H., Barak L. S., Shenolikar S., Weinman E. J., Grinstein S., Lefkowitz R. J. (1998) The β2-adrenergic receptor interacts with the Na+/H+-exchanger regulatory factor to control Na+/H+ exchange. Nature 392, 626–630 [DOI] [PubMed] [Google Scholar]
- 49. Daniels S. M., Melendez-Peña C. E., Scarborough R. J., Daher A., Christensen H. S., El Far M., Purcell D. F., Lainé S., Gatignol A. (2009) Characterization of the TRBP domain required for dicer interaction and function in RNA interference. BMC Mol. Biol. 10, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lugli G., Larson J., Martone M. E., Jones Y., Smalheiser N. R. (2005) Dicer and eIF2c are enriched at postsynaptic densities in adult mouse brain and are modified by neuronal activity in a calpain-dependent manner. J. Neurochem. 94, 896–905 [DOI] [PubMed] [Google Scholar]
- 51. Kerstein P. C., Jacques-Fricke B. T., Rengifo J., Mogen B. J., Williams J. C., Gottlieb P. A., Sachs F., Gomez T. M. (2013) Mechanosensitive TRPC1 channels promote calpain proteolysis of talin to regulate spinal axon outgrowth. J. Neurosci. 33, 273–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kennedy M. B. (2000) Signal-processing machines at the postsynaptic density. Science 290, 750–754 [DOI] [PubMed] [Google Scholar]
- 53. Schratt G. (2009) microRNAs at the synapse. Nat. Rev. Neurosci. 10, 842–849 [DOI] [PubMed] [Google Scholar]
- 54. Konopka W., Kiryk A., Novak M., Herwerth M., Parkitna J. R., Wawrzyniak M., Kowarsch A., Michaluk P., Dzwonek J., Arnsperger T., Wilczynski G., Merkenschlager M., Theis F. J., Köhr G., Kaczmarek L., Schütz G. (2010) MicroRNA loss enhances learning and memory in mice. J. Neurosci. 30, 14835–14842 [DOI] [PMC free article] [PubMed] [Google Scholar]