Background: Many lantibiotic peptides induce their own biosynthesis through histidine kinase receptors.
Results: Bovicin HJ50 and BovK form a signaling complex; substitutions of key amino acids in each protein result in disrupted signal transduction.
Conclusion: Bovicin HJ50 activates BovK through hydrophobic and electrostatic interaction to start signal transduction.
Significance: A novel peptide activating multitransmembrane histidine kinase mechanism was identified.
Keywords: Antimicrobial Peptides, Histidine Kinases, Membrane Proteins, Protein-Protein Interactions, Signal Transduction, BovK, Bovicin HJ50, Lantibiotics
Abstract
Lantibiotic bovicin HJ50 is produced by Streptococcus bovis HJ50 and acts as the extracellular signal to autoregulate its own biosynthesis through BovK/R two-component system. Bovicin HJ50 shows a linear N-terminal and glubolar C-terminal structure, and the sensor histidine kinase BovK contains eight transmembrane segments lacking any extensive surface-exposed sensory domain. The signal recognition mechanism between bovicin HJ50 and BovK is still unknown. We performed saturated alanine scanning mutagenesis and other amino acid substitutions on bovicin HJ50 using a semi-in vitro biosynthesis. Results of the mutants inducing activities indicated that several charged and hydrophobic amino acids in ring B of bovicin HJ50, as well as two glycines were key residues to recognize BovK. Circular dichroism analyses indicated that both glycines contributed to bovicin HJ50 structural changes in the membrane. Biotin-labeled bovicin HJ50 could interact with the N-terminal sensor of BovK, and several charged residues and a conserved hydrophobic region in the N-terminal portion of BovK sensor domain were important for interacting with the signal bovicin HJ50. By combining the results, we suggested a mechanism of bovicin HJ50 recognizing and activating BovK mainly through electrostatic and hydrophobic interactions.
Introduction
Two-component signal transduction systems containing a membrane-bound histidine kinase (HK)3 and a response regulator (RR) are used primarily to sense and respond to environmental stimuli in bacteria. Spanning all species of bacteria, there are thousands of HK/RR systems in which HK identifies varied specific signal molecules that triggers its autophosphorylation, and RR receives the phosphoryl group transferred from HK to activate or repress target gene transcription (1). The typical histidine kinase is a transmembrane protein comprising an N-terminal sensor domain that identifies and senses signals and a conserved C-terminal cytoplasmic kinase transmitter domain. Based on the architecture of the N-terminal sensor domain, HKs are classified into three major groups (2): Group 1 is the prototypical periplasmic-sensing HKs, which is characterized by the presence of an extracellular sensory domain that detects external signals. This is the largest group of HKs. Group 2 HKs contains 2 to 20 transmembrane segments and lacks an apparent sensory domain. Their signal sensing regions is believed to be either membrane associated or occur directly within the membrane interface. Group 3 is cytoplasmic-sensing HKs, which contain cytoplasmic input domains.
Now thousands of HKs have been identified in bacteria, fungi, archaea, yeast, and some plants, however, because many HKs have complicated membrane-spanning domains that have made structural analyses particularly difficult, and only a handful of ligands have been defined for HKs, analyses of ligand-receptor interactions are still limited to a few HKs with well defined periplasmic sensing domains and known ligands (3–12). Depending on structural analysis, histidine kinase periplasmic sensor domains may be mixed α-β-folds, all α-folds, or sensor domains similar to HK29s (13). Group 2 HKs that contain multiple transmembrane segments often lack obvious extracellular sensory domains and show little conservation, thus, little is known about how the ligand-sensor interacts with each other and ligand binding affects signaling activity in this important class of histidine kinases.
Lantibiotics are usually lactic acid bacteria or actinobacteria produced and ribosomally synthesized antimicrobial peptides containing unusual amino acids, such as dehydrated and lanthionine residues that are post-translationally modified (14). The biosynthesis of some lantibiotics including nisin, subtilin, salivaricin, SA-FF22, mersacidine, bovicin HJ50 and so on are regulated by a two-component system LanK/R. LanKs are involved in identifying extracellular signals to initiate phosphate transfer cascade to activate correlated downstream gene expression, such as the lantibiotic biosynthesis structural gene lanA, the immunity gene lanIFEG, or the regulatory gene lanRK. Some lantibiotics were also reported to function as the inducer of their own biosynthesis through the LanK/R system (15–21), whereas there is no report on how LanK identifies its ligand.
Bovicin HJ50 is a type AII lantibiotic consisting of 33 amino acids produced by Streptococcus bovis (S. bovis) HJ50. It shows a typical linear N terminus and globular C terminus structure of lantibiotics in lacticin group 481, which is the largest group and contains a specific disulfide bridge rarely seen in other lantibiotics (22). Bovicin HJ50 was initially synthesized as BovA, which consists of an N-terminal leader peptide and a C-terminal propeptide. The propeptide moiety was then post-translationally modified by BovM, and two threonine residues were dehydrated to be (Z)-2,3-didehydrobutyrines and cyclized with cysteine thiols resulting in two 3-methyllanthionines (Abu-S-Ala, in which Abu is an aminobutyric acid) to form thioether rings A and B. Ring C, which is a disulfide ring, is also formed. The mature bovicin HJ50 is produced by the removal of leader peptide from modified prepeptide by the specific protease BovT and translocated across the cytoplasmic membranes.
Our previous study (20) revealed that bovicin HJ50 acts as the extracellular inducer to autoregulate the transcription of its own structural gene bovA through a unique BovK/R two-component system. However, it remains unclear how bovicin HJ50 binds to BovK and activates BovK/R to start the signal transduction. BovK is a group 2 histidine kinase with eight transmembrane segments in its N terminus and no apparent extracytoplasmic sensory domain. Here we labeled bovicin HJ50 with biotin and observed that bovicin HJ50 could interact with BovK N-terminal sensor domain. We performed a saturated alanine scanning mutagenesis of bovicin HJ50 by a semi-in vitro biosynthesis (23), as well as different substitutions of key amino acids, and examined their inducing abilities. Meanwhile, several charged amino acids and a conserved hydrophobic region in the sixth membrane segment of BovK were observed and found to be involved in identifying bovicin HJ50 to carry on the lantibiotic signal transduction. We proposed a model for the interaction of bovicin HJ50 and BovK.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions
Strains and plasmids used in this study are listed in supplemental Table S1. S. bovis HJ50 and derivatives were routinely cultured in M17 medium supplemented with 0.5% (w/v) glucose (GM17 medium) at 37 °C. Escherichia coli DH5α, BL21, and MC1061 used as hosts for cloning procedures were maintained in Luria-Bertani (LB) broth at 37 °C. Micrococcus flavus NCIB8166 was maintained in SI medium (0.8% tryptone (w/v), 0.5% yeast extract (w/v), 0.5% glucose (w/v), 0.2% disodium hydrogen phosphate (w/v), 0.5% sodium chloride (w/v), 0.1% Tween 20 (v/v)) at 30 °C. Where appropriate, antibiotics were added as follows: kanamycin, 50 μg/ml (E. coli); chloramphenicol, 10 (E. coli) and 5 μg/ml (S. bovis); or erythromycin, 100 (E. coli) and 5 μg/ml (S. bovis).
Plasmid Constructions
The main plasmids and primers used in this study are listed in supplemental Tables S1 and S2, respectively. pET-bovAM co-expression plasmid and site-directed mutagenesis were constructed as described (23). To disrupt bovKR, two fragments flanking bovKR genes were amplified with primers KR-LF/KR-LR and KR-RF/KR-RR and then subcloned into pSET5s to yield pDKR. An erythromycin resistance cassette digested by BamHI from the temperature-sensitive plasmid pDX (24) was isolated and cloned into pDKR to create plasmid pDEKR. BovKR disruption mutant DKR strain was produced by transformation of pDEKR to S. bovis HJ50 and obtained as described (20). To complement bovKR expression in the DKR strain, the 205-bp P32 promoter and the downstream bovKR DNA fragments were amplified and cloned into pSET1 to construct the pSET1-bovKR plasmid. Site-directed mutagenesis of bovK was performed according to the protocols reported (25), generating a series of pSET1-bovKR derivatives.
Bovicin HJ50 and Mutant Biosynthesis
Bovicin HJ50 and its mutants were produced by semi-in vitro biosynthesis as described (23). Briefly, E. coli BL21(DE3) cells containing pET-bovAM were incubated at 37 °C and protein expressions were induced by 0.5 mm isopropyl 1-thio-β-d-galactopyranoside when A600 reached 0.6. The culture was grown at 18 °C for 20 h. The cells were sonicated and the His6-tagged modified prepeptide BovAm was purified by nickel-nitrilotriacetic acid chelating resin. Using immobilized metal affinity chromatography, BovAm was enriched and purified. The protease BovT150 was expressed and purified by the similar procedures. Purified His6-BovAm and BovT150 were co-incubated at 25 °C overnight to remove the leader peptide of BovAm. The mature peptide bovicin HJ50 was separated by HPLC and the molecular weight was identified using MALDI-TOF MS.
RNA Extraction, RT-PCR, and qRT-PCR Analysis
Overnight cultures of the S. bovis HJ50 DM strain were diluted 1:30 in fresh GM17. When A600 of the cells reached ∼0.4, bovicin HJ50 and the mutants purified as described previously were added to the culture (100 ng/ml), if required. The cells were further incubated for about 4 h and harvested by centrifugation. Total RNAs were isolated and quantitated as described (26).
For quantitative RT-PCR (qRT-PCR), each RNA sample was treated by DNase I (ThermoFisher) for 30 min at 37 °C to exclude contamination by DNA. Primers specific for bovA and 16 S rRNA of S. bovis HJ50 were designed. qRT-PCR was performed with SYBR Premix ExTag (TaKaRa) by Rotor-Gene Q (Qiagen) and all samples were analyzed in triplicate. Data were collected using Rotor-Gene Q Pure Detection software, normalized against endogenous 16 S rRNA gene levels, and calculated using the comparative cycle threshold method as described (27). Analyzed data were presented as fold-changes over WT levels. Student's unpaired t test was used to determine statistical significance. A p value of ≤0.05 was considered significant.
Bovicin HJ50 Labeling with Biotin
2 mg of bovicin HJ50 was produced by semi-in vitro biosynthesis and labeled with biotin (Beijing Biosynthesis Biotechnology Co., Ltd.), the product that bovicin HJ50 labeled with one biotin molecule was purified by HPLC and identified by MALDI-TOF MS and named b-bovicin HJ50.
Proteins Extraction and Gel Blot Assays
500 ml of S. bovis HJ50 cells with late-exponential phase were harvested and resuspended in ice-cold PBS buffer (NaCl, 137 mm; KCl, 2.7 mm; Na2HPO4, 10 mm; KH2PO4, 2 mm, pH 7.5) with 1 mm PMSF. After lysing by sonication, cells were centrifuged at 6,000 × g for 1 h to remove intact cells. The supernatant was ultracentrifuged at 120,000 × g for 2 h to separate soluble (cytoplasmic fraction) and insoluble (cell membrane fraction) proteins. The cytoplasmic portion of the BovK protein (cBovK, negative control) was purified as described (20). Far Western blot assays were performed using a modified protocol as described by Wu et al. (28). Briefly, the purified proteins were separated on a 12% SDS-PAGE gel and transferred to a PVDF membrane on which the proteins were denatured and renatured. The membrane was blocked with 5% (w/v) milk in PBS and probed with b-bovicin HJ50, and then incubated with an antibody conjugated to streptavidin-horseradish peroxidase (HRP). For Western blotting, a replicate membrane was probed with anti-cBovK primary antibodies (20) and then incubated with secondary antibodies conjugated to HRP. Interacting proteins or control proteins were detected by adding enhanced chemiluminescence (ECL) reagents and visualized by x-ray film exposure.
Analysis of Bovicin HJ50 Antimicrobial Activity
S. bovis HJ50 DKR and the complementary strains were cultured at 37 °C for 12 h, diluted by 1:30 in fresh GM17. When A600 of the cells reached ∼1.0, the culture supernatants were used to measure the bacteriocin activity against the indicator strain M. flavus NCIB8166 as described (29). The plate in which each well was filled with 20 μl of culture supernatant was incubated at 30 °C overnight, and the inhibition zones were examined.
Cross-linking Reactions and Analysis
500 ml of S. bovis HJ50 cells with late-exponential phase were harvested, washed with PBS buffer (pH 7.4), and resuspended in 20 ml of PBS buffer prior to cross-linking. A CuPhen (copper(II)-1,10-phenanthroline) stock solution was prepared by dissolving Cu(II)-SO4 and 1,10-phenanthroline (Acros Organics) in a 4:1 water/ethanol solution to concentrations of 150 and 500 mm, respectively. 20 μl (1:1000) of CuPhen stock was added to the cells and incubated with agitation at 20 °C for 20 min. Cells were harvested and lysed by sonication. Membrane proteins were isolated as described above, separated on a 10% SDS-PAGE gel, and transferred to a PVDF membrane. The proteins were treated with anti-cBovK primary antibodies and the secondary antibodies were conjugated to HRP. Proteins were detected by adding enhanced chemiluminescence (ECL) reagents and visualized by x-ray film exposure.
Circular Dichroism
The mean residue molar ellipticities of bovicin HJ50 and mutants were determined in H2O and a helix-inducing 3% SDS solvent, respectively, at 25 °C by circular dichroism (CD) spectroscopy with a Chirascan plus circular dichroism detector (Applied Photophysics). 200 μl of peptide analogs (100 μg/ml) were loaded into a 1-mm Quartz SUPPASIL precision cell, and ellipticities were scanned from 200 to 260 nm. Analysis of the secondary structure was performed with CDNN software (version 2.1).
RESULTS
The N Terminus and Ring BC Are Necessary for Bovicin HJ50 Inducing Activity
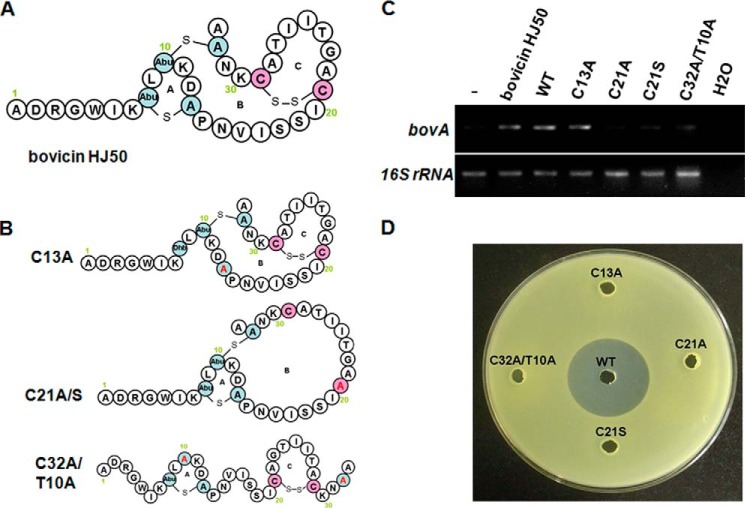
Bovicin HJ50 contains a linear N terminus and a globular C terminus formed by two thioether rings (ring A and ring B) and a disulfide bridge (forming ring C), which is uncommon in lantibiotics (Fig. 1A). To clarify the importance of these rings for its inducing activity, bovicin HJ50 ring A, ring B, and ring C disruption mutants C13A, C21A, and C32A/T10A (Fig. 1B) were produced by a semi-in vitro biosynthesis. BovA mutants tagged by His6 at its N terminus and BovM were co-expressed in E. coli, and the modified prepeptide was purified by a nickel-nitrilotriacetic acid chelating resin. After removing the leader peptide by the protease BovT150 in vitro, bovicin HJ50 mutants were separated by HPLC and their molecular weights were identified using MALDI-TOF MS (Table 1). The ring formation in bovicin HJ50 mutants were identified by their molecular mass compared with bovicin HJ50 wild type (molecular mass was 3428 Da). For example, the molecular mass of C13A was 3396 Da, 32 Da increased compared with bovicin HJ50 wild type, corresponding to the cysteine (121 Da) substitution of alanine (89 Da), and Abu (101 Da) positioned at 8 changing to (Z)-2,3-didehydrobutyrines (101 Da). Similarly, the molecular weight of C21A was 3398, corresponding to the broken disulfide bridge (two hydrogens were added to Cys-21 and Cys-29) and residue substitution at position 21 in ring B. The molecular weight of C32A/T10A was 3398, corresponding to residue substitutions and one molecular H2O (18 Da) was added at position 10 in ring C.
FIGURE 1.
Inducing and antimicrobial activity analyses of bovicin HJ50 structural mutants. A, the structure of bovicin HJ50. The Abu and A in the ring structures indicate Thr and Cys residues, respectively, which were post-translationally modified including dehydration and cyclization; B, the structure of bovicin HJ50 ring A, ring B, and ring C disruption mutants corresponding to C13A, C21A/C21S, and C32A/CT10A, respectively. Dhb indicates that (Z)-2, 3-didehydrobutyrines is a dehydrated form of Thr; C, RT-PCR analysis of bovA gene expression in the S. bovis HJ50 DM strain induced by 100 ng/ml of bovicin HJ50 purified from S. bovis HJ50, bovicin HJ50 was biosynthesized by semi-in vitro biosynthesis (WT), and ring A, ring B, and ring C disruption mutants; D, antimicrobial analysis of bovicin HJ50 structural mutants.
TABLE 1.
The molecular mass of bovicin HJ50 and its mutants
| Name | Molecular mass | Name | Molecular mass |
|---|---|---|---|
| Da | Da | ||
| Bovicin HJ50 | 3428 | N15A | 3385 |
| C13A | 3396 | V16A | 3400 |
| C21A | 3398 | I17A | 3386 |
| C21S | 3414 | S18A | 3412 |
| C32A/T10A | 3384 | S19A | 3412 |
| D2A | 3384 | I20A | 3386 |
| R3A | 3343 | G23A | 3442 |
| G4A | 3442 | δG23 | 3371 |
| W5A | 3313 | T24A | 3398 |
| I6A | 3386 | I25A | 3386 |
| K7A | 3371 | I26A | 3386 |
| L9A | 3386 | T27A | 3398 |
| K11A | 3371 | K30A | 3371 |
| K11D | 3415 | K30D | 3415 |
| K11E | 3429 | K30E | 3429 |
| K11H | 3437 | K30H | 3437 |
| D12A | 3384 | K30R | 3456 |
| D12K | 3441 | N31A | 3385 |
| D12E | 3442 | δADRGW | 2843 |
| P14A | 3402 |
Our previous study (20) demonstrated that there is no bovA (structural gene encoding bovicin HJ50) mRNA transcript detected in the S. bovis DM strain (bovM encoding the bovicin HJ50 post-translational modification enzyme was knocked out from S. bovis HJ50), because the lack of modification enzyme BovM resulted in disruption of mature bovicin HJ50 production. When induced by bovicin HJ50, which is an autoinducer of its own structural gene bovA expression, the BovK protein expressed in the DM strain could identify and transduce the lantibiotic signal, and then activate bovA transcription. To examine the critical structure of bovicin HJ50 in activating BovK protein, bovicin HJ50 and its structural mutants were added to DM strain cells, and their inducing abilities were compared by monitoring the level of the bovA transcript. As shown in Fig. 1C, the abilities of bovicin HJ50 C21A and C32A/T10A to activate the expression of bovA were attenuated obviously, whereas C13A shows a similar inducing ability with bovicin HJ50 either extracted from S. bovis HJ50 or biosynthesized with semi-in vitro biosynthesis. The mutant C21S, another ring B disruption mutant (Fig. 1B), was also constructed and showed no inducing activity. These results demonstrated that ring A in bovicin HJ50 is not necessary to activate BovK to induce bovA expression, whereas both ring B and ring C are important for its inducing activity. A truncated mutant ΔADRGW in which five amino acids on the N terminus of bovicin HJ50 were deleted was further constructed and its inducing ability was also attenuated vigorously (Fig. 2B). These results indicated that the N terminus of bovicin HJ50 was also necessary and only ring BC was not enough to carry on its inducing activity. So the linear N terminus of at least the first five amino acids and the integrity of ring BC were necessary for bovicin HJ50 to interact with BovK to start the signal transduction. Meanwhile, the antimicrobial activities of all the ring disruption mutants were lost (Fig. 1D).
FIGURE 2.
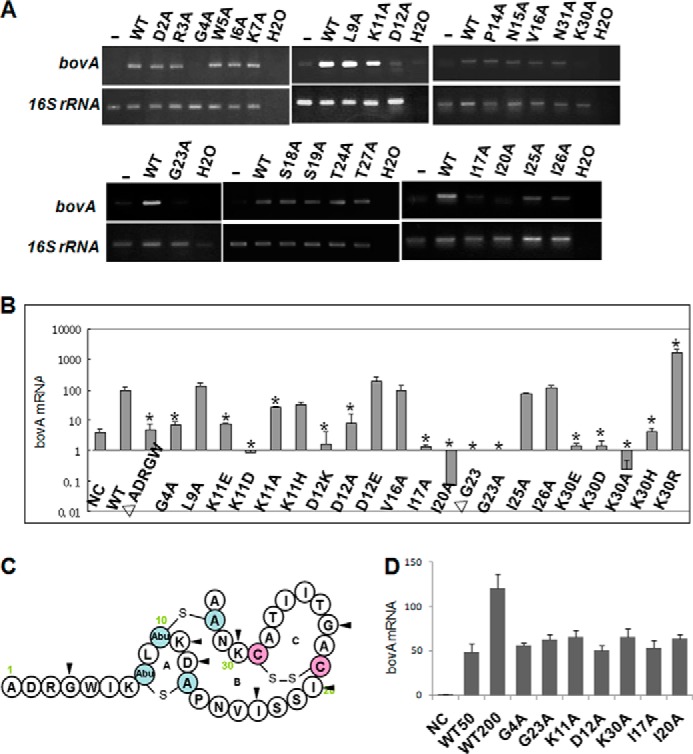
Inducing activity analyses of various mutants of S. bovis HJ50. A, RT-PCR analysis of bovA gene expression in the S. bovis DM strain. Transcript levels of bovA in S. bovis DM induced by bovicin HJ50 alanine scanning mutants (100 ng/ml) were measured by RT-PCR with 16 S rRNA as an internal control. B, determination of transcript levels of bovA in the S. bovis DM strain by qRT-PCR. Transcript levels of bovA in S. bovis DM induced by bovicin HJ50 mutants (100 ng/ml) measured by qRT-PCR with 16 S rRNA as an internal control. Values and standard deviations were calculated from at least two independent experiments performed in triplicate. C, key amino acids involved in bovicin HJ50 inducing activity are indicated with black triangles. D, qRT-PCR analyses of bovA expression induced by bovicin HJ50 competed with several analogs. Transcript levels of bovA in S. bovis DM induced by bovicin HJ50 (50 ng/ml) added with mutants (150 ng/ml) were measured by qRT-PCR with 16 S rRNA as an internal control. Values and standard deviation were calculated from at least two independent experiments performed in triplicate. WT50 and WT100 indicate S. bovis DM induced by 50 and 100 ng/ml of bovicin HJ50, respectively.
Critical Sites Affecting Bovicin HJ50 Inducing Activities
To further identify the critical sites of bovicin HJ50 on activating BovK, we performed a saturated alanine scanning mutagenesis on bovicin HJ50. The analogs of bovicin HJ50 were biosynthesized by semi-in vitro biosynthesis and their inducing abilities were tested according to bovA expression in the S. bovis DM strain after induction. The abilities of several bovicin HJ50 analogs including G4A, K11A, D12A, I17A, I20A, G23A, and K30A to activate the expression of bovA were attenuated vigorously (Fig. 2, A and B). We next determined whether these seven analogs could competitively inhibit wild type bovicin HJ50, and found that all these analogs failed to inhibit bovicin HJ50-induced bovA expression (Fig. 2D). This result demonstrated that each substitution disrupted the binding of bovicin HJ50 to the receptor BovK.
The substitution of each glycine residue at positions 4 and 23 in bovicin HJ50 could not induce its own biosynthesis, indicating that they were important to activate BovK to start the bovA expression. As glycine is the most flexible amino acid, we supposed that these two residues might contribute to conformation changing of bovicin HJ50 when interacted with the BovK protein. To make sure whether these two mutations affected bovicin HJ50 structural changes, circular dichroism (CD) spectra of G4A and G23A under H2O solvent and membrane conditions were recorded to assess their secondary structures. The results showed that in membrane conditions (3% SDS), the helical propensities of both mutants were increased (72.7–93.8 and 72.7–93.6%, respectively), whereas the anti-parallel, parallel, and random coils are obviously decreased (Table 2). Meanwhile there was no big difference between bovicin HJ50 and mutants in the H2O solvent (data not shown). These results indicated that Gly-4 and Gly-23 were important for bovicin HJ50 to change its conformation in the membrane to produce a matched structure for interacting with BovK.
TABLE 2.
Relative contents of secondary structures in bovicin HJ50 analogs
| Bovicin HJ50 | G4A | G23A | I17A | I20A | K11A | D12A | K30A | |
|---|---|---|---|---|---|---|---|---|
| Helix (%) | 72.7 | 93.8 | 93.6 | 83.3 | 82.6 | 67.5 | 74.5 | 78.4 |
| Antiparallel (%) | 2.6 | 0.7 | 0.7 | 1.7 | 1.7 | 3.1 | 2.6 | 2.0 |
| Parallel (%) | 2 | 0.5 | 0.5 | 1.2 | 1.2 | 2.5 | 1.8 | 1.6 |
| β-Turn (%) | 11.9 | 8 | 7.6 | 10.4 | 10.4 | 12.5 | 11.9 | 10.8 |
| Random coil (%) | 7.8 | 1.6 | 2.2 | 4.4 | 4.5 | 9.8 | 6.7 | 6.5 |
| Total sum (%) | 97 | 104.5 | 104.6 | 100.9 | 100.5 | 95.4 | 97.6 | 99.4 |
Substitutions of charged lysine and aspartic acid with the neutral alanine resulting in decreased bovA expression suggested that the positive charges of lysine at positions 11 and 30 and negative charge of aspartic acid at position 12 are important for bovicin HJ50 inducing activity. Results of CD spectra showed that no obvious structural differences happened in K11A, D12A, and K30A analogs (Table 2), indicating these three residues might not contribute to the structural conformation. To further determine the importance of these charges for bovicin HJ50 inducing activity, we examined the inducing abilities of an additional nine biosynthesized analogs K11H, K11D, K11E, D12K, D12E, K30H, K30R, K30D, and K30E. qRT-PCR analysis (Fig. 2B) showed that the expressions of bovA were obviously attenuated induced by K11D and K11E in which the positive lysine was substituted with the negative aspartic acid and glutamic acid, respectively. Meanwhile the analog K11H in which lysine is substituted with another positive amino acid histidine retained 31.5% of the wild type bovicin HJ50 activity to induce expression of bovA, confirming that the positive charge of lysine at position 11 is important for the activity of bovicin HJ50. BovA expression was obviously attenuated induced by D12K in which the negative aspartic acid was substituted with the positive lysine, compared with nearly 2-fold of increased bovA expression induced by D12E in which the negative aspartic acid is substituted with the negative glutamic acid. This result confirmed that the negative charge of aspartic acid at position 12 is also important for the activity of bovicin HJ50. K30D and K30E in which the positive lysine was substituted with the negative aspartic acid and glutamic acid showed little inducing activities, whereas K30R showed more than 16-fold of increased inducing activity, indicating that the positive charge of aspartic acid at position 30 is important for the inducing activity of bovicin HJ50. K30H in which the positive lysine was substituted with the positive histidine showed unexpectedly little inducing activity, and we supposed that the bigger imidazole side chain of histidine might make K30H difficult to interact with BovK. The above results demonstrated that the positive charges afforded by lysines at positions 11 and 30, together with the negative charge afforded by aspartic acid at position 12 in bovicin HJ50 might be involved in activating the BovK protein to start its own biosynthesis.
Another two important residues were isoleucines that exhibited strong hydrophobicity at positions 17 and 20 in ring B of bovicin HJ50. Substitution of each isoleucine with alanine resulted in vigorous attenuation of bovA expression, indicating that the hydrophobicity afforded by these two residues might be involved in interacting with BovK. Results of CD spectra showed that there was little structural changes in I17A and I20A analogs (Table 2), indicating these two residues contributed little to bovicin HJ50 structural changes when they interact with BovK.
Bovicin HJ50 Interacts with the BovK N-terminal Transmembrane Domain
BovK is a membrane protein with predicted eight transmembrane regions and was difficult to heterologously express or purify from E. coli. To identify the interaction between bovicin HJ50 and BovK, we labeled bovicin HJ50 with biotin, purified by HPLC and identified by MALDI-TOF MS (Fig. 3A). The peak at 3654 Da shows a 226-Da increase in molecular mass compared with bovicin HJ50 (molecular mass 3428 Da) corresponding to bovicin HJ50 labeled with one biotin molecule, and named b-bovicin HJ50. The antimicrobial activity and inducing ability of b-bovicin HJ50 were both comparable with bovicin HJ50 (Fig. 3, B and C). These results indicate that b-bovicin HJ50 was a valid substitution of bovicin HJ50 to study its interaction with BovK.
FIGURE 3.
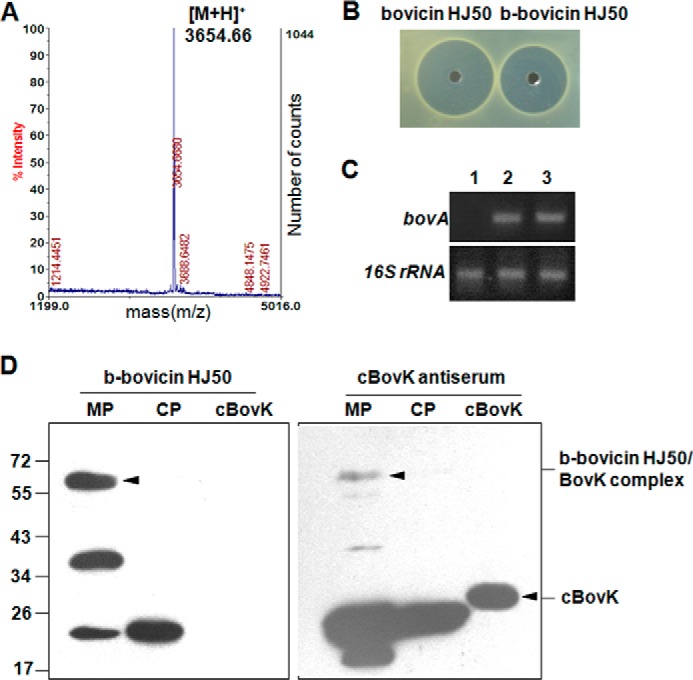
Bovicin HJ50 labeled by biotin could interact with the BovK N-terminal transmembrane region. A, molecular weight of bovicin HJ50 labeled by biotin (b-bovicin HJ50) was measured by MALDI-TOF MS. 3654.66 Da corresponds to the molecular mass of b-bovicin HJ50; B, antimicrobial analysis of bovicin HJ50 and b-bovicin HJ50; C, RT-PCR analysis of bovA gene expression in the S. bovis DM strain induced by bovicin HJ50 and b-bovicin HJ50. 1, 2, and 3 indicate no inducer, bovicin HJ50 as an inducer, and b-bovicin HJ50 as an inducer, respectively; D, far Western blot and Western blot analysis of b-bovicin HJ50 binding to BovK. Membrane proteins (MP) and cytoplasmic proteins (CP) from S. bovis HJ50 were probed with b-bovicin HJ50 and BovK antiserum with cBovK (cytoplasmic portion of BovK) protein as a negative control. The b-bovicin HJ50/BovK complex and cBovK protein are indicated with black triangles.
We studied the interaction between b-bovicin HJ50 with the membrane and cytoplasmic proteins extracted from S. bovis HJ50 by far Western blotting. The cytoplasmic portion of BovK (cBovK, 26 kDa) as mentioned previously (20) was expressed in E. coli and purified as a control. As shown in Fig. 3D, incubation of the above proteins with b-bovicin HJ50 labeled three membrane proteins and one cytoplasmic protein from S. bovis HJ50, whereas no interation was detected between b-bovicin HJ50 and cBovK. One of the 58-kDa membrane proteins was supposed to be the BovK (molecular mass of 58 kDa) protein that was labeled with b-bovicin HJ50. Western blot analysis utilizing polyclonal cBovK antiserum against the same proteins showed that BovK and cBovK were visualized at positions of 58 and 26 kDa, respectively, which indicated that b-bovicin HJ50 could identify the BovK whole protein but not the cytoplasmic portion of BovK. These results demonstrated that BovK could interact with b-bovicin HJ50 through its N-terminal transmembrane sensor domain. Two other membrane proteins and one cytoplasmic protein labeled by b-bovicin HJ50 were unknown and will be defined further.
Amino Acids Important for Ligand Detection and Signal Transduction in BovK
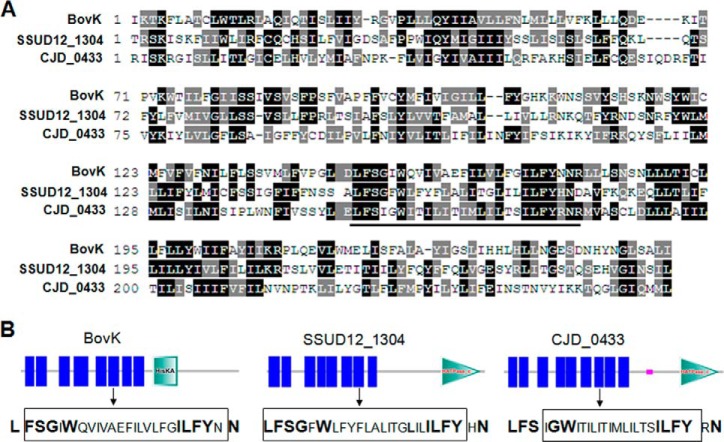
The N-terminal sensor domain of BovK shows little conservation with other lantibiotic related or not related histidine kinases. As several bovicin HJ50-like lanthipeptides had been found through BlastP searches (30), we analyzed the predicted biosynthesis gene cluster of each lanthipeptide and observed three relevant histidine kinases, SSUD12_1304 (from Streptococcus suis D12), SSUSC84_0850 (from S. suis SC84), and CJD_0439 (from Clostridium perfringens JGS1721), which may be involved in the regulation of bovicin HJ50-like lanthipeptides encoded by SSUD12_1310, SSUSC84_0866, and CJD_0433, respectively. As SSUD12_1304 and SSUSC84_0850 show 100% protein sequence similarity, we aligned the sequence of BovK with SSUD12_1304 and CJD_0433. As shown in Fig. 4A, a conserved region enriched in hydrophobic amino acids was identified as LFS(X)G(X)W … ILFYXN (position 201–225), which were mainly located in the sixth transmembrane segment of BovK (Fig. 4B).
FIGURE 4.
BovK sequence alignment and the histidine kinases domain architectures. A, BovK sequence alignment with SSUD12_1304 and CJD_0433 through ClustalW and shaded with BoxShade. The conserved LFS(X)G(X)W … ILFYXN region is indicated above a line. B, the domain architectures of BovK, SSUD12_1304, and CJD_0433 were produced using SMART. Vertical lines indicate the predicted transmembrane segments; HisKA in BovK indicates the His kinase A phosphoacceptor domain that is a dimerization and phosphoacceptor domain of histidine kinase; HATPase_c indicates the histidine kinase-like ATPase found in several ATP-binding proteins. Each sequence corresponding to the LFS(X)G(X)W … ILFYXN region is indicated and amino acids framed are in the transmembrane segments. Conserved amino acids are shown in bold.
To test whether the conserved region is involved in interacting with bovicin HJ50, we constructed a S. bovis DKR strain in which bovKR genes were destroyed and could not be transcribed in S. bovis HJ50 (Fig. 5A). The culture supernatant of DKR showed no antimicrobial activity against M. flavus NCIB8166 compared with that of S. bovis wild type (Fig. 5B). We constructed a pSET1-bovKR plasmid in which bovKR was controlled by the P32 promoter and transformed to S. bovis DKR producing the CKR strain in which disruption of bovKR was complemented. As shown in Fig. 5C, S. bovis CKR recovered the ability of producing bovicin HJ50 and the supernatant showed the antimicrobial activity. To examine the importance of the conserved LFS(X)G(X)W … ILFYXN region in BovK, each of the 10 conserved residues was substituted with alanine in pSET1-bovKR to construct a series of plasmids and transformed to DKR producing strains S. bovis L201A, F202A, S203A, G204A, W206A, I220A, L221A, F222A, Y223A, and N225A expressing relevant BovK mutants, respectively. The abilities of BovK mutants to identify bovicin HJ50 were monitored according to bovA expression and the antimicrobial activities of their culture supernatant. Besides L221A, which showed similar bovA expression, and I220A retaining about 25% bovA expression compared with CKR, eight other mutated S. bovis strains showed no or dramatically decreased bovA expressions (Fig. 5C). The antimicrobial activities of their culture supernatants showed similar results (Fig. 5D). These results indicated that most residues in the conserved hydrophobic region in the sixth transmembrane segment of BovK were necessary for identifying the bovicin HJ50 signal to synthesize its own molecule. Two non-conserved residues in the LFS(X)G(X)W … ILFYXN region in BovK were also substituted with alanine, and produced strains I205A and N224A. As shown in Fig. 5C, the culture supernatant of I205A showed similar antimicrobial activity compared with that of CKR, whereas N224A exhibited no antimicrobial activity. These results demonstrated that the non-conserved isoleucine at position 205 was not necessary to identify bovicin HJ50 and asparagine at position 224 as a non-conserved residue was also important to identify bovicin HJ50. We supposed that this asparagine might be involved in the specific identification of bovicin HJ50, which needs to be further defined.
FIGURE 5.
S. bovis DKR characterization and phenotypes of BovK mutant-complemented strains. A, RT-PCR analysis of bovK and bovR gene expressions in S. bovis HJ50 and DKR strains. B, antimicrobial analysis of culture supernatant of S. bovis HJ50 and DKR strains. C, qRT-PCR analyses of bovA expression levels in S. bovis DKR complemented with wild type BovKR (CKR) and mutated BovKR with site mutation of BovK. 16 S rRNA expression levels were measured as internal controls. D, antimicrobial analysis of the culture supernatant of S. bovis DKR complemented with wild type BovKR (CKR) and mutated BovKR with site mutation of BovK. E, far Western blot and Western blot analyses of b-bovicin HJ50 binding to BovK mutants. Membrane proteins from S. bovis DKR complemented with BovKR mutants were probed with b-bovicin HJ50 and BovK antiserum. F, key amino acids involved in BovK identifying bovicin HJ50 are indicated with white triangles. G, in vivo cross-linking analysis of BovK. The BovK monomer and dimer are indicated as 58 and 116 kDa, respectively, with arrows.
To clarify whether the important residues in BovK affected the interaction between bovicin HJ50 and BovK, the membrane proteins of DKR strains complemented with BovK mutants were extracted and the binding abilities of BovK mutants to b-bovicin HJ50 were detected by far Western blotting. As Fig. 5E shown, no binding abilities of most BovK mutants were detected, and the binding abilities of D200A and G204A were dramatically decreased. These results demonstrated that the residues in the conserved hydrophobic region in the sixth transmembrane segment of BovK were important to interact with bovicin HJ50.
BovK has eight transmembrane segments linked with four small extracellular loops (20), we supposed that some charged amino acids in the four extracellular loops might interact with the positive lysines at positions 11 and 30 and the negative aspartic acid at position 12, which were important for the bovicin HJ50 inducing activity as mentioned above. Predicted by SMART (31), there are 10 charged amino acids in BovK extracellular loops including Asp-31 and Glu-32 in the first loop, Asp-112, Glu-113, Lys-114, Lys-119 in the second loop, Asp-200 in the third loop, Lys-254, Arg-255, and Glu-259 in the forth loop. To test whether the above 10 charged residues are important for BovK to identify bovicin HJ50 signal molecules, each residue was substituted with alanine in pSET1-bovKR and transformed into the DKR producing strains S. bovis D31A, E32A, D112A, E113A, K114A, K119A, D200A, K254A, R255A, and E259A expressing relevant BovK mutants, respectively. As shown in Fig. 5C, bovA expressions of D31A, E32A, D112A, K114A, and D200A were obviously decreased, whereas substitution of Glu-113, Lys-119 in the second loop and all three residues Lys-254, Arg-255, and Glu-259 in the forth loop showed similar bovA expressions with wild type BovK. The antimicrobial activities of their culture supernatants showed similar results (Fig. 5D). Far Western blotting analyses also indicated that the binding abilities of D31A, E32A, D112A, and K114A to b-bovicin HJ50 were disrupted, and D200A showed dramatically decreased binding ability (Fig. 5E). These results demonstrated that the especially charged negative amino acids in the first three extracellular loops of BovK were important, whereas the forth loop may be not involved in BovK interacting with bovicin HJ50. Fig. 5F showed all of the necessary residues we detected in BovK to identify bovicin HJ50.
Determination of the Oligomeric State of BovK by Cross-linking Analysis
As many histidine kinases are known to function as dimers (1, 32), we checked the oligomeric state of BovK in vivo with covalent chemical cross-linking analysis, which has been shown to be a useful technique to check protein-protein interactions. The oxidative catalyst CuPhen is a small membrane permeable reagent that efficiently catalyzes disulfide bond formation in the membrane to cross-link cysteines residues in the transmembrane domain of proteins (33). There are three cysteines, Cys-145, Cys-178, and Cys-239 located in transmembrane helixs TM4, TM5, and TM7, respectively, in the N-terminal sensor domain of BovK. If the cysteine residues in each protein are close to each other (<12 Å between two Cα), they will be cross-linked and the protein complex could be identified with Western blotting. As shown in Fig. 5G, BovK in untreated S. bovis cells revealed a band of ∼58 kDa, corresponding to the apparent molar mass of the monomer. When treated with CuPhen, BovK produced an additional band of about 116 kDa, corresponding to the molar mass of the BovK dimer. Meanwhile the quantity of BovK monomer was reduced obviously. This result demonstrated that when BovK accepted the ligand bovicin HJ50, it might function as dimers to sense and transduce the signal.
DISCUSSION
Bacteria usually respond to diverse small molecule signals using a quorum sensing machinery. The Gram-positive bacteria recognize peptides as quorum sensing pheromones to activate a regulatory cascade (HK/RR two-component system) to control its own biosynthesis or other cellular processes. Most of the reported peptide signals are linear peptides, such as 17-residue competence-stimulating peptidess regulating competence, 10-residues comX, and a series of competence and sporulation factors regulating competence and sporulation, various linear peptides regulating virulence or plasmid-mediated conjugation, or a small peptide with a thiolactone ring, such as 8-residue autoinducing peptides controlling pathogenesis and biofilm formation (34, 35). Lantibiotics that contain 19–38 amino acids are relatively bigger molecules and have more complex ring structures. Now lantibiotics are paid more attention for their antimicrobial activities against competitive microorganisms, whereas little is known about how they function as peptide signals to regulate their own biosynthesis or other processes. Bovicin HJ50 not only shows antimicrobial activities to many Gram-positive bacteria but also acts as a signal to regulate its own biosynthesis through BovK/R. BovK belongs to the HPK7 subfamily as noted by Grebe and Stock (36), different with other peptide signal-inducible histidine kinases, which belong to the HPK10 subfamily classified by their C-terminal conservation (34). Here we clarified the important structures and amino acids of bovicin HJ50 and the sensor histidine kinase BovK, as well as their protein interactions. This study will extend our understanding of peptide signaling in Gram-positive bacteria.
Either ring B or ring C disruptions of bovicin HJ50 resulted in the lost of bovicin HJ50 inducing activity. This result meant that the thioether bridge forming ring B and the disulfide bridge forming ring C are both necessary for bovicin HJ50 to form an effective structure to identify and activate BovK. Although ring A is conserved in bovicin HJ50-like lanthipeptides (30) and the bovicin HJ50 ring A disruption mutant showed no antimicrobial activity, the disruption of ring A did not lose the ability of inducing bovA expression. These results demonstrated that C-terminal ring BC is a key structure for bovicin HJ50 to identify BovK. Ring B was especially important as most of the key amino acids including the charged lysine (Lys-11 and Lys-30) and aspartic acid (Asp-12), as well as the hydrophobic isoleucine (Ile-17 and Ile-20) were positioned in ring B. Ring B formed by 16 amino acids of bovicin HJ50 was bigger than that of other class AII lantibiotics, such as lacticin 481 (10 amino acids), which meant that ring B of bovicin HJ50 was more flexible. We supposed ring B was the main functional portion of bovicin HJ50 to interact with BovK.
The glycine at position 4 is well conserved in class AII lantibiotics (37), and was a key residue for the antimicrobial activity of nukacin ISK-1 (38) and bovicin HJ50,4 however, the detailed contribution of this residue to their antimicrobial activities is unknown. Here we found that this conserved residue was also involved in activating bovicin HJ50 inducing activity. CD spectra indicated that substitution of glycine at position 4 resulted in obvious secondary structural changes of bovicin HJ50 in membrane conditions. As glycine is the most flexible amino acid, we supposed that Gly-4 could help bovicin HJ50 to change its conformation in the cell membrane to identify BovK effectively. The glycine at position 23 in ring C is specific in class AII lantibiotics, and was also found to be important for bovicin HJ50 inducing activity. Gly-23 substitution also caused obvious secondary structural changes of bovicin HJ50 in membrane conditions, we supposed that this residue might provide the flexibility necessary for bovicin HJ50 active sites to bind to BovK protein.
Bovicin HJ50 is a cationic lantibiotic in which the charged Lys-11, Asp-12, and Lys-30 residues were all essential for its inducing activity. Substitutions of these residues by amino acids with opposite charges showed almost no ability to induce bovA expression, indicating the charges of these three residues played key roles in interacting with BovK. The solution structure of bovicin HJ50 was determined by NMR spectroscopy,4 and a relatively stable structure showed that Lys-11, Asp-12, and Lys-30 were positioned on the same surface. We speculated that these three residues might construct an important charged cluster to identify BovK. As four negative charged residues, Asp-31, Glu-32, Asp-112, Asp-200, and one positive charged residue, Lys-114, in BovK extracellular loops were important to identify bovicin HJ50, we suggested that bovicin HJ50 somewhat identified BovK through electrostatic attraction between the interfaces formed by Lys-11, Asp-12, and Lys-K30 of bovicin HJ50 and Asp-31, Glu-32, Asp-112, Asp-200, and Lys-114 of BovK as shown in Fig. 6. However, the detailed interaction between these amino acids is unknown because the purification and crystal structure analysis of the membrane protein BovK is not so easy.
FIGURE 6.

A model for bovicin HJ50 and BovK interaction. Bovicin HJ50 could bind to cell membrane through the N-terminal amphiphilic region. The conformation of bovicin HJ50 might be changed with the help of glycines at positions 4 and 23 in the membrane. When the concentration of bovicin HJ50 reached a suitable level, they could activate BovK function. Two BovK molecules dimerize and the charged residues Lys-11, Asp-12, and Lys-30 of bovicin HJ50 identified the BovK extracellular charged residues Asp-31, Glu-32, Asp-112, Asp-200, and Lys-114 through electrostatic attraction. The LFS(X)G(X)W … ILFYXN regions form a hydrophobic pocket that the C-terminal ring BC, especially residues I17 and I20 of bovicin HJ50 could insert into and produced a more stable complex to activate BovK to transduce the signal. The critical sites in bovicin HJ50 and BovK are indicated.
We have demonstrated that the hydrophobic LFS(X)G(X)W … ILFYXN region in the sixth transmembrane segment was conserved and might be involved in BovK interacting with bovicin HJ50. As BovK functioned as dimers (Fig. 5F), we speculated that the two LFS(X)G(X)W … LFYXN regions of the BovK dimer complex could form a hydrophobic pocket, which the C-terminal globular ring BC of bovicin HJ50 (two molecules were supposed to interact with BovK dimer) could form “wedge”-like structures (bovicin HJ50 NMR structure)4 and insert into it (Fig. 6). The isoleucines at positions 17 and 20 in ring B of bovicin HJ50 might interact with some residues in the pocket to form a relatively stable complex to activate BovK to transfer the signal, whereas the detailed interaction between the amino acids needs further research.
Here we suggested a model of the lantibiotic bovicin HJ50-activating BovK mechanism (Fig. 6). When the concentration of bovicin HJ50 reached a certain level (inducing concentration) and met BovK, the conformation of bovicin HJ50 was changed in the cell membrane with the help of glycines at positions 4 and 23. The bovicin HJ50 and BovK complex might be formed by interaction between key residues Lys-11, Asp-12, and Lys-30 of bovicin HJ50 and BovK extracellular charged residues Asp-31, Glu-32, Asp-112, Asp-200, and Lys-114 through electrostatic attraction. Meanwhile, the C-terminal ring BC of bovicin HJ50 might insert into the hydrophobic pocket formed by LFS(X)G(X)W … ILFYXN regions of BovK dimers through hydrophobic interactions and stabilize the ligand-receptor complex, then activate BovK. It is interesting that histidine kinases SUD12_1304 and CJD_0433 related to bovicin HJ50-like lanthipeptides as mentioned above also contained multitransmembrane regions, and the conserved hydrophobic LFS(X)G(X)W … ILFYXN region were mainly located in or near the seventh transmembrane segment of them (Fig. 4B). We supposed that a similar model might be used when bovicin HJ50-like lantibiotics activate the cognate histidine kinases. Our results indicated that the N-terminal first three extracellular loops and the sixth transmembrane segment were key positions for BovK to interact with bovicin HJ50. This finding was consistent with the phenomena found in some multimembrane histidine kinases in the HPK10 subfamily that the N-terminal halves of sensory domains could respond to different peptide ligands. For example, autoinducing peptide 1 activates AgrC-1 activity through binding to its first two extracellular loops, amino acids important for PlnB to be activated would be located on the first extracellular loop (39), and so on.
Research in the past decade tells us that lantibiotics are not limited to antimicrobial peptides. Besides their own biosynthesis induced by themselves, several other cellular events were coordinately regulated with lantibiotic production. For example, mutacin I production is coregulated with biofilm formation, acid tolerance, and cell competence (40, 41), Smb is regulated in concert with cell competence formation (42), cytolysin is biosynthesized only when target cells are present (43), however, the detailed complex pathways still remained unknown. It seems likely that various other cellular processes are also regulated with lantibiotic production, which has remained unrecognized so far.
Acknowledgments
We thank Dr. Yuanming Luo and Dr. Qian Wang with MALDI-TOF MS measurements.
This work was supported by grants from the National Natural Science Foundation of China (31070041 and 31200046) and the Knowledge Innovation Program of the Chinese Academy of Sciences (KSCX2-EW-Q-14, KSCX2-EW-G-14, and KSCX2-EW-J-6).

This article contains supplemental Tables S1 and S2.
J. Zhang, unpublished data.
- HK
- histidine kinase
- RR
- response regulator
- qRT
- quantitative RT
- CuPhen
- copper(II)-1,10-phenanthroline
- Abu
- aminobutyric acid.
REFERENCES
- 1. Heermann R., Altendorf K., Jung K. (1998) The turgor sensor KdpD of Escherichia coli is a homodimer. Biochim. Biophys. Acta 1415, 114–124 [DOI] [PubMed] [Google Scholar]
- 2. Mascher T., Helmann J. D., Unden G. (2006) Stimulus perception in bacterial signal-transducing histidine kinases. Microbiol. Mol. Biol. Rev. 70, 910–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cheung J., Hendrickson W. A. (2009) Structural analysis of ligand stimulation of the histidine kinase NarX. Structure 17, 190–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cheung J., Hendrickson W. A. (2008) Crystal structures of C4-dicarboxylate ligand complexes with sensor domains of histidine kinases DcuS and DctB. J. Biol. Chem. 283, 30256–30265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Neiditch M. B., Federle M. J., Miller S. T., Bassler B. L., Hughson F. M. (2005) Regulation of LuxPQ receptor activity by the quorum-sensing signal autoinducer-2. Mol. Cell 18, 507–518 [DOI] [PubMed] [Google Scholar]
- 6. Neiditch M. B., Federle M. J., Pompeani A. J., Kelly R. C., Swem D. L., Jeffrey P. D., Bassler B. L., Hughson F. M. (2006) Ligand-induced asymmetry in histidine sensor kinase complex regulates quorum sensing. Cell 126, 1095–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhou Y. F., Nan B., Nan J., Ma Q., Panjikar S., Liang Y. H., Wang Y., Su X. D. (2008) C4-dicarboxylates sensing mechanism revealed by the crystal structures of DctB sensor domain. J. Mol. Biol. 383, 49–61 [DOI] [PubMed] [Google Scholar]
- 8. Sevvana M., Vijayan V., Zweckstetter M., Reinelt S., Madden D. R., Herbst-Irmer R., Sheldrick G. M., Bott M., Griesinger C., Becker S. (2008) A ligand-induced switch in the periplasmic domain of sensor histidine kinase CitA. J. Mol. Biol. 377, 512–523 [DOI] [PubMed] [Google Scholar]
- 9. Krämer J., Fischer J. D., Zientz E., Vijayan V., Griesinger C., Lupas A., Unden G. (2007) Citrate sensing by the C4-dicarboxylate/citrate sensor kinase DcuS of Escherichia coli: binding site and conversion of DcuS to a C4-dicarboxylate- or citrate-specific sensor. J. Bacteriol. 189, 4290–4298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Reinelt S., Hofmann E., Gerharz T., Bott M., Madden D. R. (2003) The structure of the periplasmic ligand-binding domain of the sensor kinase CitA reveals the first extracellular PAS domain. J. Biol. Chem. 278, 39189–39196 [DOI] [PubMed] [Google Scholar]
- 11. Cho U. S., Bader M. W., Amaya M. F., Daley M. E., Klevit R. E., Miller S. I., Xu W. (2006) Metal bridges between the PhoQ sensor domain and the membrane regulate transmembrane signaling. J. Mol. Biol. 356, 1193–1206 [DOI] [PubMed] [Google Scholar]
- 12. Pappalardo L., Janausch I. G., Vijayan V., Zientz E., Junker J., Peti W., Zweckstetter M., Unden G., Griesinger C. (2003) The NMR structure of the sensory domain of the membranous two-component fumarate sensor (histidine protein kinase) DcuS of Escherichia coli. J. Biol. Chem. 278, 39185–39188 [DOI] [PubMed] [Google Scholar]
- 13. Cheung J., Hendrickson W. A. (2010) Sensor domains of two-component regulatory systems. Curr. Opin. Microbiol. 13, 116–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. McAuliffe O., Ross R. P., Hill C. (2001) Lantibiotics: structure, biosynthesis and mode of action. FEMS Microbiol. Rev. 25, 285–308 [DOI] [PubMed] [Google Scholar]
- 15. Wescombe P. A., Upton M., Dierksen K. P., Ragland N. L., Sivabalan S., Wirawan R. E., Inglis M. A., Moore C. J., Walker G. V., Chilcott C. N., Jenkinson H. F., Tagg J. R. (2006) Production of the lantibiotic salivaricin A and its variants by oral streptococci and use of a specific induction assay to detect their presence in human saliva. Appl. Environ. Microbiol. 72, 1459–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Upton M., Tagg J. R., Wescombe P., Jenkinson H. F. (2001) Intra- and interspecies signaling between Streptococcus salivarius and Streptococcus pyogenes mediated by SalA and SalA1 lantibiotic peptides. J. Bacteriol. 183, 3931–3938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schmitz S., Hoffmann A., Szekat C., Rudd B., Bierbaum G. (2006) The lantibiotic mersacidin is an autoinducing peptide. Appl. Environ. Microbiol. 72, 7270–7277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kuipers O. P., Beerthuyzen M. M., de Ruyter P. G., Luesink E. J., de Vos W. M. (1995) Autoregulation of nisin biosynthesis in Lactococcus lactis by signal transduction. J. Biol. Chem. 270, 27299–27304 [DOI] [PubMed] [Google Scholar]
- 19. Stein T., Borchert S., Kiesau P., Heinzmann S., Klöss S., Klein C., Helfrich M., Entian K. D. (2002) Dual control of subtilin biosynthesis and immunity in Bacillus subtilis. Mol. Microbiol. 44, 403–416 [DOI] [PubMed] [Google Scholar]
- 20. Ni J., Teng K., Liu G., Qiao C., Huan L., Zhong J. (2011) Autoregulation of lantibiotic bovicin HJ50 biosynthesis by the BovK-BovR two-component signal transduction system in Streptococcus bovis HJ50. Appl. Environ. Microbiol. 77, 407–415 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21. Wang J., Gao Y., Teng K., Zhang J., Sun S., Zhong J. (2014) Restoration of bioactive lantibiotic suicin from a remnant Lan locus of pathogenic Streptococcus suis serotype 2. Appl. Environ. Microbiol. 80, 1062–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xiao H., Chen X., Chen M., Tang S., Zhao X., Huan L. (2004) Bovicin HJ50, a novel lantibiotic produced by Streptococcus bovis HJ50. Microbiology 150, 103–108 [DOI] [PubMed] [Google Scholar]
- 23. Lin Y., Teng K., Huan L., Zhong J. (2011) Dissection of the bridging pattern of bovicin HJ50, a lantibiotic containing a characteristic disulfide bridge. Microbiol. Res. 166, 146–154 [DOI] [PubMed] [Google Scholar]
- 24. Liu G., Zhong J., Ni J., Chen M., Xiao H., Huan L. (2009) Characteristics of the bovicin HJ50 gene cluster in Streptococcus bovis HJ50. Microbiology 155, 584–593 [DOI] [PubMed] [Google Scholar]
- 25. Chiu J., Tillett D., Dawes I. W., March P. E. (2008) Site-directed, ligase-independent mutagenesis (SLIM) for highly efficient mutagenesis of plasmids greater than 8 kb. J. Microbiol. Methods 73, 195–198 [DOI] [PubMed] [Google Scholar]
- 26. Sambrook J., Fritsch E., Maniatis T. (1989) Molecular Cloning: a Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 27. Livak K. J., Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔ CT) method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 28. Wu Y., Li Q., Chen X. Z. (2007) Detecting protein-protein interactions by far Western blotting. Nat. Protoc. 2, 3278–3284 [DOI] [PubMed] [Google Scholar]
- 29. Cintas L. M., Rodriguez J. M., Fernandez M. F., Sletten K., Nes I. F., Hernandez P. E., Holo H. (1995) Isolation and characterization of pediocin L50, a new bacteriocin from Pediococcus acidilactici with a broad inhibitory spectrum. Appl. Environ. Microbiol. 61, 2643–2648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Georgalaki M., Papadimitriou K., Anastasiou R., Pot B., Van Driessche G., Devreese B., Tsakalidou E. (2013) Macedovicin, the second food-grade lantibiotic produced by Streptococcus macedonicus ACA-DC 198. Food Microbiol. 33, 124–130 [DOI] [PubMed] [Google Scholar]
- 31. Letunic I., Copley R. R., Pils B., Pinkert S., Schultz J., Bork P. (2006) SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 34, D257–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Moore J. O., Hendrickson W. A. (2009) Structural analysis of sensor domains from the TMAO-responsive histidine kinase receptor TorS. Structure 17, 1195–1204 [DOI] [PubMed] [Google Scholar]
- 33. Lemmin T., Soto C. S., Clinthorne G., DeGrado W. F., Dal Peraro M. (2013) Assembly of the transmembrane domain of E. coli PhoQ histidine kinase: implications for signal transduction from molecular simulations. PLoS Comput. Biol. 9, e1002878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Thoendel M., Kavanaugh J. S., Flack C. E., Horswill A. R. (2011) Peptide signaling in the staphylococci. Chem. Rev. 111, 117–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Miller M. B., Bassler B. L. (2001) Quorum sensing in bacteria. Annu. Rev. Microbiol. 55, 165–199 [DOI] [PubMed] [Google Scholar]
- 36. Grebe T. W., Stock J. B. (1999) The histidine protein kinase superfamily. Adv. Microb. Physiol. 41, 139–227 [DOI] [PubMed] [Google Scholar]
- 37. Dufour A., Hindré T., Haras D., Le Pennec J. P. (2007) The biology of lantibiotics from the lacticin 481 group is coming of age. FEMS Microbiol. Rev. 31, 134–167 [DOI] [PubMed] [Google Scholar]
- 38. Islam M. R., Shioya K., Nagao J., Nishie M., Jikuya H., Zendo T., Nakayama J., Sonomoto K. (2009) Evaluation of essential and variable residues of nukacin ISK-1 by NNK scanning. Mol. Microbiol. 72, 1438–1447 [DOI] [PubMed] [Google Scholar]
- 39. Johnsborg O., Diep D. B., Nes I. F. (2003) Structural analysis of the peptide pheromone receptor PlnB, a histidine protein kinase from Lactobacillus plantarum. J. Bacteriol. 185, 6913–6920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Merritt J., Kreth J., Shi W., Qi F. (2005) LuxS controls bacteriocin production in Streptococcus mutans through a novel regulatory component. Mol. Microbiol. 57, 960–969 [DOI] [PubMed] [Google Scholar]
- 41. Wen Z. T., Burne R. A. (2004) LuxS-mediated signaling in Streptococcus mutans is involved in regulation of acid and oxidative stress tolerance and biofilm formation. J. Bacteriol. 186, 2682–2691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yonezawa H., Kuramitsu H. K. (2005) Genetic analysis of a unique bacteriocin, Smb, produced by Streptococcus mutans GS5. Antimicrob. Agents Chemother. 49, 541–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Coburn P. S., Pillar C. M., Jett B. D., Haas W., Gilmore M. S. (2004) Enterococcus faecalis senses target cells and in response expresses cytolysin. Science 306, 2270–2272 [DOI] [PubMed] [Google Scholar]