Background: Mia40 accelerates Cox19 folding through the specific recognition of the third Cys in the second CX9C motif.
Results: The chaperone catalysis renders a native-like intermediate that oxidizes in a slow uncatalyzed reaction into native Cox19.
Conclusion: The role of Mia40 is funnelling an already sequentially encoded, but rough, substrate folding landscape.
Significance: These results provide a rationale for the substrate promiscuity of the chaperone Mia40.
Keywords: Biophysics, Disulfide, Mitochondria, Oxidation-Reduction, Protein Folding, Protein Translocation, Cox19, Mia40, Protein Oxidative Folding
Abstract
Mia40-catalyzed disulfide formation drives the import of many proteins into the mitochondria. Here we characterize the oxidative folding of Cox19, a twin CX9C Mia40 substrate. Cox19 oxidation is extremely slow, explaining the persistence of import-competent reduced species in the cytosol. Mia40 accelerates Cox19 folding through the specific recognition of the third Cys in the second helical CX9C motif and the subsequent oxidation of the inner disulfide bond. This renders a native-like intermediate that oxidizes in a slow uncatalyzed reaction into native Cox19. The same intermediate dominates the pathway in the absence of Mia40, and chemical induction of an α-helical structure by trifluoroethanol suffices to accelerate productive folding and mimic the Mia40 folding template mechanism. The Mia40 role is to funnel a rough folding landscape, skipping the accumulation of kinetic traps, providing a rationale for the promiscuity of Mia40.
Introduction
A growing number of cysteine-containing proteins synthesized on cytosolic ribosomes are found to be targeted to the intermembrane space (IMS)3 of mitochondria as reduced species and to oxidize and fold into their functional forms only after entering this organelle compartment (1–6). The IMS oxidoreductase Mia40 (7), introduces disulfide bonds into these target proteins, facilitating their import and folding (4, 5, 8, 9). This tight folding control by disulfide bond cross-linking drives the vectorial translocation of proteins across the outer membrane to the IMS without an additional input of energy.
Mia40 substrates are usually small proteins containing twin CX3C or twin CX9C motifs. Twin CX9C proteins typically contain two disulfide bonds stabilizing a coiled-coil helix coiled-coil helix domain. The best characterized of these proteins are Cox17 and Mia40. Cox19, is a member of the CX9C family and an important accessory protein in the assembly of cytochrome c oxidase (CcO) in yeast. Cells lacking Cox19 are respiratory deficient and have reduced CcO activity (10). Cox19 was recently proposed to be involved also in copper efflux to mitochondria (11). So far, little structural information exists for this protein, except that it contains two disulfides bonds between its external (Cys30-Cys62) and internal (Cys40-Cys52) Cys in the native state (10, 12), a topological linkage thought to be conserved in the CX9C family (13).
Despite the observation that several Mia40 targets as well as covalent Mia40/substrate complexes have been structurally characterized, oxidative folding pathways followed by these substrates remain essentially unknown, especially in kinetic terms (4, 5). This information is relevant because reversible changes in the redox state of Cys residues regulate protein import, fold, and function in mitochondria (1, 5). Disulfide bond formation is error-prone, particularly at the early stages of packing, often resulting in the accumulation of mispaired intermediates. In these cases, reshuffling is required to attain the native disulfide connectivity. Compared with other small disulfide-rich proteins (14), Mia40 canonical substrates display less Cys and thus a lower number of potential intermediates that may trap their folding reactions. Despite this, Bien et al. demonstrated the presence of long-lived intermediates in the Cox19 Mia40-catalyzed pathway, whose accumulation could be proofread by glutathione (GSH) (12). Unfortunately, the methodology they used did not allow discrimination between intermediates and thus a detailed description of the pathway. Here, we report the first characterization of the oxidative folding reaction of a Mia40 substrate. By dissecting and monitoring the elementary steps of folding with chromatographic, disulfide, and conformational analysis of the trapped intermediates, we were able to determine that reduced Cox19 folds through a sequential oxidation of its cysteine residues involving the population of both natively bonded and non-native intermediates. The influence of Mia40 on Cox19 oxidative folding is also described. NMR data reveal a multiexponential kinetics, allowing a structural characterization of the major intermediate in the folding pathway. Overall, our results are consistent with a folding template mechanism, where by inducing the formation of a secondary structural element, Mia40 restricts the conformational space to be explored by Cox19 to attain its native conformation, thus accelerating the folding reaction. Our data indicate, however, that Mia40 acts by just funneling a folding code that is already imprinted in the substrate primary structure.
EXPERIMENTAL PROCEDURES
Cloning and Expression
Cox19 cDNA was cloned in a pET-21a(+) vector for polyhistidine purification using the following primers: forward 5′-ATACATATGTCAGGGAACCCAGGAAGCTCG-3′ and reverse 5′-GTGCTCGAGTTTATTATCGGTGGCGTCTTT-3′ and pGEX-6P-1 Cox19 as template. The primers contained restriction sites for NdeI and Xho1, respectively (underline). Cloning was confirmed by sequencing, and the plasmid was used for the transformation of BL21(DE3) cells. Protein purification was performed under denaturing conditions using 8 m urea. Following Ni2+ purification, the fractions containing Cox19 were dialyzed against Tris 50 mm, pH 8.4, NaCl 100 mm to promote oxidation and refolding. The dialyzed protein was concentrated and loaded into a Superdex 16/60 for polishing. Cox19 was homogeneous, and its molecular weight was confirmed by MALDI-TOF carried out in the Proteomics and Bioinformatics facility at UAB, a member of the ProteoRed-ISCIII network. Full oxidation of the protein was confirmed by RP-HPLC and folding by CD spectroscopy. Purified Cox19 contained at its C-terminal the additional LEHHHHHH amino acids that did not affect its folding. N15 and N15,C13-labeled Cox19 were prepared as described in Ref. 15. Mutagenesis of Cox19 was carried out using the Stratagene site-directed mutagenesis kit following the manufacturer's instructions.
Mass Spectrometry
MALDI-TOF and TOD/TOF analysis were performed on an Autoflex Speed mass spectrometer (Bruker, Bremen). The free cysteine residues of Peak I and Peak III folding intermediate species were alkylated by adding the samples, dissolved in 0.1% trifluoroacetic acid (30 μl), to an excess of 75 mm iodoacetamide in 50 mm ammonium bicarbonate buffer, pH 8, (70 μl), enough to increase the pH of the reaction mixture, to minimize the risk of disulfide reshuffling. The reaction was allowed to proceed for 30 min at room temperature. Samples before and after alkylation were then analyzed by MALDI-TOF mass spectrometry using sinapinic acid as matrix. A mixture of protein standards was used for internal calibration.
For fingerprint analysis, the alkylated proteins were first purified using ZipTip (Millipore) C18 micro columns, evaporated, redissolved in 1 m urea, 25 mm ammonium bicarbonate, subjected or not to reduction by treatment with 10 mm DTT for 1 h at room temperature, and digested with trypsin (3 h at 37 °C) or Glu-C (16 h at 37 °C). The digests were then purified using ZipTip C18 and analyzed by MALDI TOF/TOF mass spectrometry using HCCA as matrix.
Determination of Cox19 Redox Potential
Oxidized Cox19 was incubated overnight with DTT and OxDTT at various ratios at 25 °C. Reactions were acidified with TFA 1%, and the mixture analyzed by RP-HPLC. The redox potential was calculated as previously described (16).
AMS Mobility Assay
For reductive unfolding, DTT (20 mm) was added to fully oxidized Cox19, and at the indicated times, aliquots of the reaction mixture were quenched with TCA 20%. Following precipitation, the protein was recovered, incubated for at least 2 h with AMS 20 mm in SDS sample buffer, and loaded in a 16% SDS-PAGE Tricine gel. For oxidative folding, Cox19, previously reduced with 20 mm DTT was loaded in a PD mini trap G-25 column (GE) containing Sephadex G-25, and the protein free of reducing agent was recovered in Tris 50 mm, pH 8.4 NaCl 100 mm following the manufacturer's instructions. The protein was allowed to oxidize, and aliquots were removed at defined time points and quenched with 20 mm AMS in SDS sample buffer and loaded in a 16% SDS-PAGE Tricine gel.
NMR Spectroscopy
Experiments for the backbone assignment (HNCO, HNCA, HN(CO)CA, HNCB, HN(CA)CO, and HN(CO)CB) were acquired at 318 K on an 800 MHz Bruker Avance III spectrometer equipped with a cryoprobe. Spectra were processed with Topspin© (Bruker GmbH) and analyzed with Sparky (SPARKY 3, University of California, San Francisco). A total of 34 (37%) non-proline residues were successfully assigned. Cox19 refolding was monitored by measuring the signal decay/growth in a series of 1H,15N-HSQC experiments. A freshly reduced sample of Cox19 was immediately placed in the spectrometer with an approximate lag phase of 30 min. Measurements were performed in steps of 2 h for a total period of time of 7 days. Buffering conditions were 50 mm Tris at pH 7, 100 mm NaCl, and 0.5% of dodecyl phosphocholine-D38.
Fluorescence
Intrinsic tryptophan fluorescence was measured using Cox19 (50 μm). The spectra were measured in the 290–450-nm interval using a 280-nm excitation wavelength (5 nm excitation and emission slits, 0.1 s averaging time). ANS fluorescence emission spectra were measured in the 400–550-nm interval using 385 nm as the excitation wavelength for ANS (10 nm excitation and emission slits, 0.1 s averaging time).
CD Spectroscopy
Far UV CD spectra were acquired on a Jasco-710 spectropolarimeter continuously purged with nitrogen and thermostatted at 25 °C. For oxidative folding, reduced Cox19 was prepared as described above and allowed to air oxidize for 26 h. Scans were taken at 1-h intervals.
HPLC
Cox19 (0.2–0.5 mg/ml) was reduced in 50 mm Tris, pH 8.4, NaCl 100 mm, and 20 mm DTT for at least 2 h at 23 °C. To initiate folding, samples were passed through a PD-10 column (Sephadex-25, GE Healthcare) previously equilibrated with 50 mm Tris, pH 8.4, NaCl 100 mm. For some experiments selected concentrations of GndHCl, GSH, PDI, and Mia40 were added. To monitor the folding reaction, aliquots were removed at defined time points and quenched with 2% TFA and analyzed by RP-HPLC as follows. Samples were injected in a Waters 2690 HPLC coupled to a UV detector set to 280 nm. A linear gradient of 25–45% of 0.1% TFA in acetonitrile was applied for 60 min into a 250 × 4.6 (5 mm) C4 column (phenomenex) at a flow-rate of 0.75 ml/m.
Stop/Go folding experiments were executed as described in Ref. 17. Briefly, acid-trapped intermediates were freeze dried and allowed to resume folding by dissolving the sample in Tris 50 mm, pH 8.4, NaCl 100 mm. The samples were analyzed as described above.
RESULTS
Disulfide Bond Reduction Results in Cox19 Unfolding
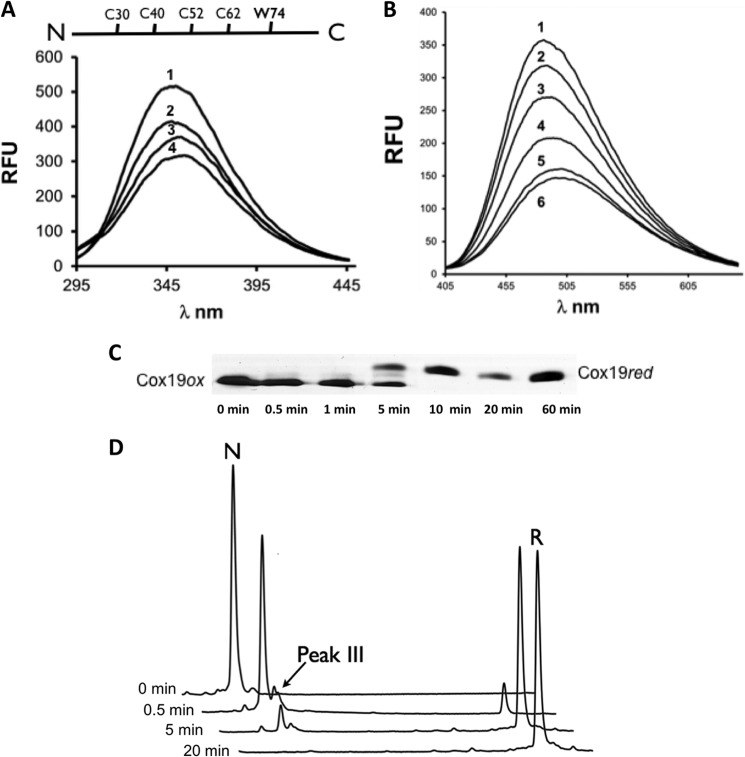
Mia40 substrates contain very stable disulfide bonds that require high concentration of reductants to be broken (16, 18, 19). We estimated the standard redox potential of Cox19 to be approximately −0.31 V (data not shown), very close to that reported for Cox17 (−0.34), Tim9 (−0.31), and Tim10 (−0.32), suggesting that Mia40 substrates have similar redox stability. According to this redox potential, 20 mm DTT was used to fully reduce Cox19. Changes in tryptophan (Fig. 1A) and ANS (Fig. 1B) fluorescence were consistent with Cox19 reduction promoting conformational changes leading to unfolding. Accordingly, the 1H-15N-HSQC NMR spectrum of reduced Cox19 displays poor dispersion in the proton dimension and an upfield shift of the e-TRP signal (data not shown). We confirmed that this changes resulted from Cys reduction by a thiol mobility assay using 4-acetomido-4′-maleimidylstilbene-2,2′-disulfonic acid (AMS), which reacts specifically with free thiols but not with disulfides (12) (Fig. 1C). Addition of DTT promoted the fast reduction of Cys residues. Apart from the fully reduced protein, an intermediate containing a single disulfide bond (2AMS) was detected during the reduction reaction. The presence of this intermediate was also evident from reductive unfolding experiments using RP-HPLC (Fig. 1D).
FIGURE 1.
Reductive unfolding of Cox19 involves single disulfide intermediates. A, Cox19 reduction and unfolding can be monitored by Cox19 W74 fluorescence. RFU, relative fluorescence units. DTT 20 mm was added to oxidized Cox19 and W74 emission spectrum recorded at time 0 (1), 30s (2), 2min (3), 10 min (4). B, Cox19 reduction and unfolding can be monitored by ANS fluorescence. DTT 20 mm was added to oxidized Cox19 in the presence of ANS (100 mm), and the probe emission spectra was recorded at time 0 (1), 30s (2), 1 min (3), 2 min (4), 10 min (5), and 25 min (6). C, reduction of fully oxidized Cox19 followed using AMS thiol mobility assay. In addition to the bands of the fully oxidized and fully reduced protein, an intermediate band was observed corresponding to a single disulfide intermediate. D, species involved in Cox19 reductive unfolding can be separated by RP-HPLC following reaction quenching with TFA, which blocks subsequent oxidation and reshuffling. Oxidized Cox19 (N), because of its compact structure compared with the reduced protein (R), has a shorter elution time. A small peak eluting close to N was identified at intermediate time points, probably corresponding to a single disulfide species (see below).
Slow Oxidation Leads to Cox19 Folding
Next, we analyzed the oxidative folding reaction of Cox19. For this purpose the protein was initially fully reduced in 20 mm DTT, and the reducing agent was further removed, allowing the protein to re-oxidize its free thiol groups into disulfides. This process should mimic protein folding after synthesis at the ribosomes (20, 21).
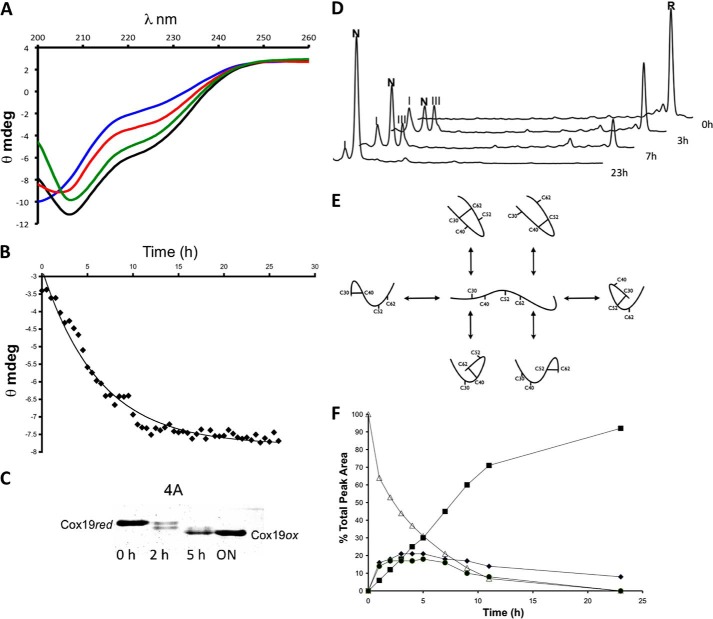
First we analyzed the acquisition of Cox19 secondary structure during oxidative folding using circular dichroism (CD). As expected, the reduced protein lacks any regular secondary structure signature; but as the reaction proceeds, the ellipticity of the protein recovers to yield a helical spectrum displaying the characteristic minima at ≈210 and ≈222 nm, highly similar to that previously reported for Cox17 (Fig. 2A). By plotting the changes in ellipticity at 215 nm versus time, it can be observed that in the absence of catalyzers Cox19 oxidative folding is extremely slow, taking around 24 h to complete (Fig. 2B). This slow folding rate was confirmed by monitoring the time dependence of ANS fluorescence recovery (see below) and by the AMS shift assay (Fig. 2C), with maximum fluorescence and protein oxidation occurring only after 24 h. This parsimonious folding suggested that, despite its small size, Cox19 does not fold through a “two-state” mechanism and accordingly that one or more transient intermediates should populate the pathway. In fact, the AMS shift assay already indicated the presence of such intermediate species (Fig. 2C). Acid-trapping and RP-HPLC analysis were used to confirm the presence of such intermediates. The results corroborate that oxidative folding proceeds very slowly, because also according to this technique, the conversion of fully reduced Cox19 into native Cox19 takes ≈24 h at pH 8.4 and 25 °C (Fig. 2D). The possible disulfide cross-linked intermediates for a protein containing four Cys comprise six single disulfide species (Fig. 2E) and two different fully scrambled forms. Thus, the detection of only two major peaks, peak I and III, both eluting close to the native protein, indicates that the folding of Cox19 proceeds through a funneled pathway. Using a C4 preparative column, we purified peaks I and III, and they were alkylated with iodoacetamide to evaluate their disulfide content by mass spectrometry. Both intermediates display two reduced Cys and therefore contain a single disulfide bond (supplemental Table S1). The quantitative analysis of disulfide species along the reaction (Fig. 2F) indicates that the presence of peaks I and III precede the formation of the native protein; thus showing that Cox19 oxidative folding proceeds via the sequential oxidation of single disulfide bonds.
FIGURE 2.
Slow oxidation of Cox19 results in folding. A, Cox19 was reduced in 20 mm DTT for at least 3 h. The reduced protein was loaded in a small PD10 column containing a G25 resin. The recovered protein, free of DTT, was allowed to air oxidized at 25 °C, and CD spectra were taken at 1-h intervals. Reduced Cox19 (blue, time 0) display features of an unfolded protein but slowly recovers features characteristic of a folded protein. Shown are the spectra at 4 (red), 8 (green), and 26 h (black). B, ellipticity at 215 nm, was plotted as a function of time. C, Cox19 air oxidation was followed using the AMS thiol shift assay. Full Cox19 oxidation takes about 20 h to occur, and the presence of single disulfide intermediates is clearly visible. D, time-dependent Cox19 oxidation was followed by RP-HPLC. Cox19 oxidation (R, fully reduced protein) displays slow kinetics and involves the formation of at least two intermediates eluting close to the native protein. Those peaks are identified as I (Cys40-Cys62) and III (Cys40-Cys52), N represents native protein. E, Cox19 contains 4 Cys residues arranged in two CX9C motifs that can form a maximum number of 6 different single disulfide species; only the Cys30-Cys62 and Cys40-Cys52 intermediates contain native disulfide bonds. F, peaks corresponding to Cox19 folding species were integrated, and their area plotted as a function of time, reduced Cox19 (▵), Native Cox19 (■), Peak I (♦), and Peak III (●). Peaks I and III display kinetics characteristics of folding intermediates as their formation precedes native protein accumulation.
Oxidative Folding of Cox19 Involves the Formation of Native and Non-native Single Disulfide Intermediates
We proceeded to determine the disulfide pairing of the two major detected intermediates in the Cox19 oxidative folding reaction. Peaks I and III were purified by RP-HPLC and alkylated with iodoacetamide. The derivatized intermediates were subsequently digested with trypsin or endoproteinase Glu-C, treated with or without DTT, and analyzed by mass spectrometry (supplemental Table S1). Mass fingerprint analysis of the resulting fragments indicates that the peaks III and peak I correspond to species containing a native and non-native disulfide bond, respectively. Peak III comprises the inner native disulfide bond Cys40-Cys52. The presence of a native disulfide bond on peak III was expected considering that its elution time coincides with that of the only intermediate identified during reductive unfolding. This points out that in Cox19, the inner disulfide is more protected against reduction than the external one. More surprising was to find out that the single disulfide in peak I species corresponds to the Cys40-Cys62 non-native linkage. This intermediate coexists with the native form even after 23 h of oxidation.
Peaks I and III Are Converted into Native Cox19
To further assess the kinetic role of the identified single-disulfide intermediates, we performed stop/go experiments on these species. Acid-trapped intermediates were isolated from oxidative folding mixtures, dried, and dissolved in standard buffer at pH 8.4 to allow them to resume folding. The formation of the native species from peak III is already evident at the beginning of the Stop/Go reaction with a subsequent conversion as it proceeds. In this case, formation of native Cox19 is straightforward, since only oxidation of the remaining free cysteines (Cys-30 and Cys-62) is required (Fig. 3A). Peak I also converts into native Cox19; however since it contains a non-native disulfide, reshuffling is needed. The formation of Peak III (Cys40-Cys52) at the beginning of the Stop/Go reaction and its persistence once Peak I (Cys40-Cys62) is exhausted, strongly suggest that disulfide rearrangement involves the formation of Peak III (Cys40-Cys52), which will be, thus, the only productive intermediate in the pathway (Fig. 3B).
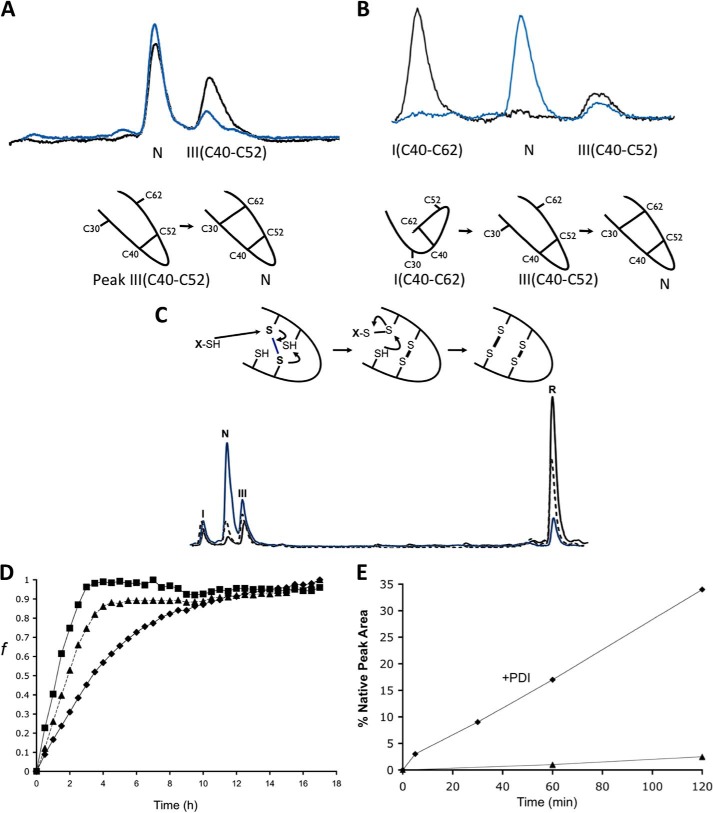
FIGURE 3.
Stop/Go folding of Cox19 intermediates. A, purified Peak III corresponding to Cys40-Cys52 was redissolved in Tris 50 mm NaCl, pH 8.4, and its evolution was followed by RP-HPLC. Sample corresponding to time 0 (black) and time 2 h (blue) are shown for comparison. B, purified Peak I, corresponding to Cys40-Cys62 was analyzed as above. Peak I converts to native protein probably after reshuffling to Cys40-Cys52. C, oxidative folding reactions in the presence of GSH (5 mm, blue) and GndHCl (300 mm, dashed) were quenched after 3 h and analyzed by RP-HPLC. Although displaying the same intermediates (peak I and III) both reactions were accelerated relative to the control (black). D, Cox19 folding can be monitored by fluorescence using ANS as a probe for the formation of a hydrophobic core. Reactions were performed in the presence of GSH (5 mm, ■) and GndHCl (300 mm, ▴), resulting in an increase in folding rate relative to control (♦). E, PDI (0.06 mg/ml) from bovine liver was added to reduced Cox19 (10 mm) to study its impact on oxidative folding (♦) relative to control (▴). Peak areas were integrated, and the % native protein (N) was plotted as a function of time in the reaction with and without PDI.
GSH and Protein Disulfide Isomerase Accelerate Cox19 Oxidative Folding
Bien et al. reported that GSH could accelerate the in vitro oxidative folding Cox19 and also promote its mitochondrial import (12). The physiological relevance of those results were recently confirmed in mammalian cells in vivo, where the GSH pool was shown fundamental for the efficient import of Cox19 and other Mia40 substrates (22). Low amounts of reducing agents are known to increase oxidative folding rates acting as thiol catalysts by counteracting the accumulation of partially oxidized non-native isomers (23). In Fig. 3C, we show the effect of 5 mm GSH on Cox19 oxidative folding as analyzed by acid-trapping and RP-HPLC. The formation of the fully oxidized Cox19 was clearly accelerated compared with the GSH-free reaction. The same result was obtained when we monitored folding using ANS fluorescence. GSH reduces the Cox19 folding time to about 4 h; thus increasing by 6-fold the oxidation rate (Fig. 3D). Despite the fact that the pathway involved the same intermediates observed in the control reaction, the ratio between peak III and peak I increased at any point in time, consistent with GSH assisting the reshuffling of the Cys40-Cys62 disulfide bond to form the Cys40-Cys52 native intermediate.
We also explored the effect of protein-disulfide isomerase (PDI), an enzyme that catalyzes both the formation and the isomerization of disulfide bonds in the eukaryotic endoplasmic reticulum, on Cox19 folding. Small PDI concentrations were selected to favor the isomerization reaction. The presence of PDI had dramatic effects on the rate of recovery of the native protein (Fig. 3E). In contrast to Mia40, PDI does not recognize any binding site in IMS substrates, and its effect together with that of GSH is a clear indication that increased reshuffling results in an overall increase in reaction rate. This implies that despite the fact that most cannot be detected because of their transient nature, the folding pathway involves the formation of non-natively bound intermediates whose conversion into on-pathway intermediates is catalyzed by both GSH and PDI.
Oxidative Folding Intermediates Display Native-like Secondary Structure
Although folding intermediates can be directly purified from oxidative folding reactions, they should be kept at acidic pH to prevent further oxidation and reshuffling, which precludes their conformational characterization under physiological conditions. To circumvent this problem, we constructed analogs in which the free Cys were replaced by Ser residues. Those mutants have the advantage of being chemically blocked; that is, they are frozen in a given conformation because of the lack of free Cys. As discussed above, for Cox19, there exist only six possible single disulfide intermediates, we constructed their analogs by mutating Cox19 Cys in pairs allowing for the presence of two free Cys in each mutant. The mutants were purified to homogeneity. They all displayed significantly lower RP-HLPC retention times than their respective reduced forms, indicating that despite most of them containing non-native disulfides, all were oxidized (data not shown).
Next, we analyzed the secondary structure of the oxidized mutants using CD. They displayed significant heterogeneity in their secondary structure content (Fig. 4A). The mutant containing the inner native disulfide bond presented an α-helical spectrum and exhibited the highest ellipticity, closely followed by the Cys40-Cys62 mutant. The rest of the mutants displayed clearly lower ellipticity and more random-coil spectra. Thus, the analogs exhibiting a native-like spectrum coincide precisely with the major intermediates detected in the Cox19 folding pathway. It is important to point out that the external native disulfide bond Cys30-Cys62 does not endorse Cox19 with significant secondary structure.
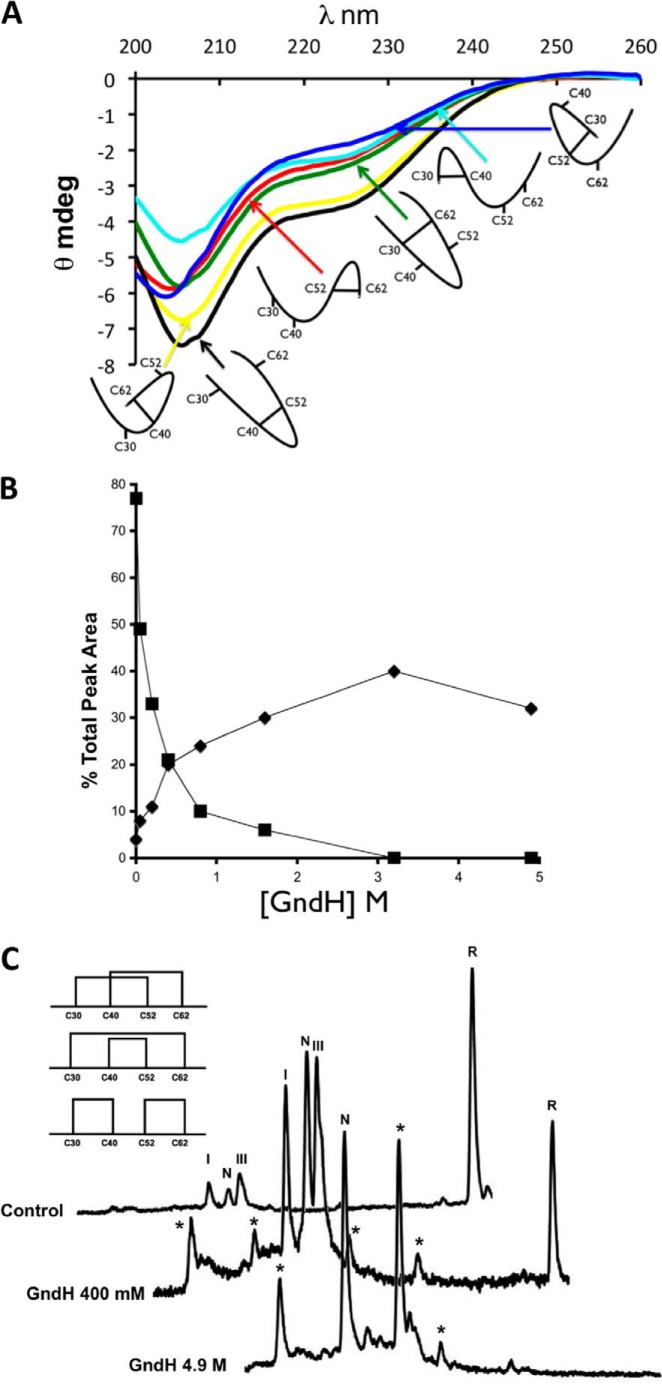
FIGURE 4.
Oxidative folding intermediates display secondary structural features. A, CD spectra of single disulfide mutants, Cys30-Cys40 (light blue), Cys30-Cys52 (dark blue), Cys30-Cys62 (green), Cys40-Cys52 (black), Cys40-Cys62 (yellow), and Cys52-Cys62 (red) were compared. B, Cox19 oxidative folding was performed in the presence of increasing concentrations of GndH. After 2 h, reaction samples were quenched and loaded in RP-HPLC for analysis. Peak areas were integrated in the chromatograms obtained, while GndH leads to an overall increase in protein oxidation rate shown by the decay in reduced protein peak area (■) that does not result in a concomitant increase in native protein (♦) formation as nonnative species are stabilized. C, indicated (*) are the species not detected in the control reaction. With the highest concentration of GndH tested (4.9 m), 3 main species were detected that probably correspond to the fully oxidized species of Cox19.
Intermediate III (Cys40-Cys52) and Intermediate I (Cys40-Cys62) Act as Kinetic Traps
GSH and PDI accelerate the Cox19 folding reaction but do not prevent the accumulation of Intermediate III (Cys40-Cys52) and Intermediate I (Cys40-Cys62), which suggests that they act as kinetic traps, preventing fast folding because of their native-like structure and metastability. In kinetic traps, the slow productive kinetics usually indicates that the rearrangement is somehow sterically hindered and thus that the transition to the native conformation involves overcoming relatively large barriers. Oxidative folding of Cox19 was performed in the presence of increasing concentrations of GdnHCl and analyzed by RP-HPLC to evaluate its impact on the trapping capability of the intermediates. At moderate concentrations, the denaturant is expected to destabilize the metastable kinetic traps without altering significantly the native stability. This effect would, in principle, lower the free energy barriers between the metastable states and the native conformation and accelerate folding. In agreement with this view, the rate of native Cox19 formation increases with the denaturant concentration, achieving a maximum increase (8-fold) at ≈3 m GdnHCl (Fig. 4B).
The III (Cys40-Cys52) and I (Cys40-Cys62) species are not expected to differ significantly in entropy from the other possible four single disulfide species, and according to CD data their kinetic stability responds to a higher number of non-covalent contacts. Disruption of these contacts by denaturants would decrease the energy barrier relative to the rest of intermediates and allow their accumulation in the pathway. Accordingly, in the presence of GdnHCl the population of four additional new peaks could be observed (Fig. 4C).
By adding enough denaturant to strongly destabilize III (Cys40-Cys52) and I (Cys40-Cys62) (4.9 m GdnHCl), the Cox19 pathway changed completely, and native Cox19 was no longer the only species at the end of the reaction; two additional peaks being observed. If we consider that there are only 3 possibilities to fully oxidize Cox19, those extra peaks likely correspond to scrambled species, which indicates that the formation of at least one of the detected intermediates is obligatory, and in its absence disulfide formation occurs randomly (Fig. 4C).
Structural Characterization of the Cox19 Main Intermediate by NMR Monitorization of the Oxidative Folding Process
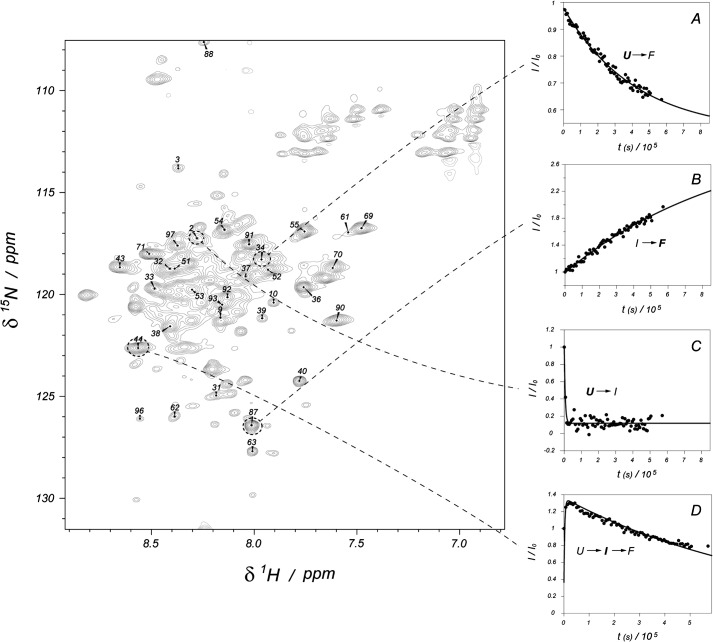
To collect structural evidence for the proposed Cox19 folding pathway, the reaction was monitored using NMR. 15N-labeled Cox19 was reduced, and its oxidation followed in real time. As previously stated, in the presence of a reducing agent, the 1H-15N-HSQC spectrum of Cox19 is consistent with the existence of a mostly unfolded species. However, after DTT removal, the protein spectrum acquires significant complexity with the appearance of a new set of signals leading to a total set of resonances exceeding the number of residues in the protein (Fig. 5). Therefore, the spectrum must report on more than one conformational state of the polypeptide. This hypothesis was confirmed by monitoring the evolution of the peak intensities over time. For this experiment, the pH was lowered to 7.0 to match the reaction kinetics with the NMR requirements. Under these conditions, the time evolution of the spectrum (between 0.5 h and ≈160 h) showed four types of peaks. A first group (group A, involving 20 peaks) presented a monotonic decay (fit to a single exponential) whereas a second set (group B, 23 peaks) showed a single exponential growth in the peak intensity (Fig. 5, A and B). These two groups of signals are likely associated to the slow transition between the unfolded (U) and the native (F) conformations, and the corresponding residues are largely insensitive to the population of any intermediate. A third group (group C, 15 peaks) followed a single exponential but with a very abrupt decay, losing 90% of the peak intensity in less than 30 min (Fig. 5C). This group is coupled to the fourth one (group D, 31 peaks), which shows a biexponential behavior, where a first increase in the intensity in about the same time (30 min) is followed by a slow decay of the acquired signal (Fig. 5D). Residues in group C disappear at a rate comparable to the first spectrum build-up, providing an explanation for the lower population of group C as compared with group D. We interpret the group C as the (fast) transition between U and a folding intermediate (I) and the residues belonging to the group D being sensitive to the pathway U→I→F.
FIGURE 5.
Assignment and kinetic curves for WT Cox19. Left figure shows the partial assignment of Cox19. The plots on the right show the time evolution of representative signals for each of the four kinetic processes detected (the monitored species is marked in bold): slow unfolded state decrease toward folded conformation (A), native folded state formation (B), fast disappearance of unfolded state toward intermediate formation (C), which is coupled to the appearance of the fast intermediate and its slow evolution to the native species (D). Points represent experimental intensities, whereas lines show the least minimum square fit to the appropriate exponential equations.
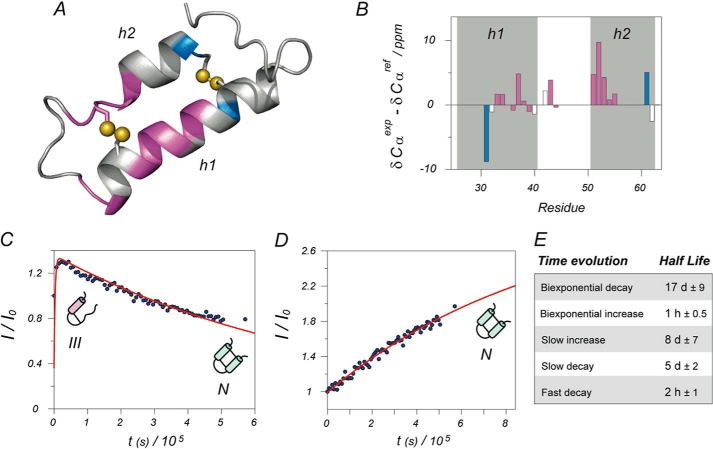
The conventional set of experiments for the NMR assignment (HNCO, HNCA, HNcoCA, HNCB, HNcoCB, and HNcaCO) were registered and subsequently analyzed for native Cox19. The pronounced chemical shift heterogeneity combined with the complex dynamic behavior of the peaks precluded full assignment of the protein, and only 37% of the peaks could be assigned unequivocally. This is unfortunate, but still very insightful, since assignment information could be obtained for most of the residues belonging to the structured regions of the protein. No three-dimensional structural information is available for Cox19; therefore, a structural homology model based on Cox17 was built (supplemental Fig. S1). It is shown in Fig. 6A where the residues with available chemical shift assignment are color-coded: blue for group B and pink for group D. Representative examples of the time evolution for peaks from the groups D and B are shown in Fig. 6, C and D, respectively. Cys-30 and Cys-60 belong to group B, and they are insensitive to the presence of intermediate species, consistent with the marginal role that the formation of Cys30-Cys60 plays in the oxidative folding of Cox19. On the other hand, most of the residues in the structured regions belong to group D, consistent with the intermediate formation affecting mostly to residues in these areas. Interestingly, the slow I to F transition indicate that the observed chemical shifts of the residues from group D belong to the detected intermediate, whose formation would trap the folding reaction. The Cα chemical shift is a good reporter of the secondary structure in a given conformation (24); accordingly the assigned chemical shifts were compared with the standard random coil values (Fig. 6B). Chemical shifts are consistent with Cys-52 being involved in a native disulfide bond in the intermediate, suggesting that it corresponds to I (Cys40-Cys52). Residues belonging to helix 1, according to the model, show chemical shifts consistent with a random coil conformation in the intermediate. Remarkably, residues from helix 2 have helical chemical shifts already in the intermediate structure. Thus, the NMR data are consistent with an intermediate containing the inner disulfide bond where helix 2 is already assembled while helix 1 is yet to be structured.
FIGURE 6.
Structural analysis and kinetic model. A, structural model of Cox19 based on homologous Cox17. Only the two structured helixes are shown; the rest of the protein is largely disordered. Assigned residues are color coded according to their kinetic evolution: purple for biexponential intermediate formation and blue for monoexponential increase of folded conformation. Most of the structured residues correspond to the intermediate state. Differences between alpha carbon chemical shifts minus reference random coil values are well known reporters of secondary structure (positive differences indicate helical structure). Structural analysis for Cox19 in the bar diagram B shows a higher helical content for helix 2 when compared with that of helix 1, suggesting that helix 2 is already formed in the intermediate. Shaded areas correspond to predicted helices according to the structural model, whereas the color code of the bars is the same as in A. Representative examples of kinetic curves for partially structured intermediate and native folded state are, respectively, shown in plots C and D, as well as schematic models for the proposed dominant species at each stage. Again, blue points represent experimental points and red lines the best least minimum square fit to the proper exponential functions. The average half-lives calculated from fits are shown in Table E.
Mia40 Catalyzes the Rapid Oxidation of Cox19 into Single Disulfide Intermediates
The fact that spontaneous oxidation of Cox19 occurs so slowly emphasizes the role played by Mia40 in the IMS. Yeast Mia40 is a membrane protein, but its C-terminal soluble part and constructs containing tags at the N termini are competent to oxidize substrates both in vitro and in vivo (25). We used a Mia40 construct with a GST-tag to test Mia40 activity by RP-HPLC, since this fusion, due to its size, does not elute in the same region as Cox19. Although Mia40 pathway consists of at least two more components, Erv1 and cytochrome c, Mia40 has been shown to fully catalyze substrate oxidation when added in excess (12, 25). Accordingly, purified GST-Mia40 was able to form intermolecular disulfide species with reduced Cox19 (data not shown) and to catalyze its complete oxidation.
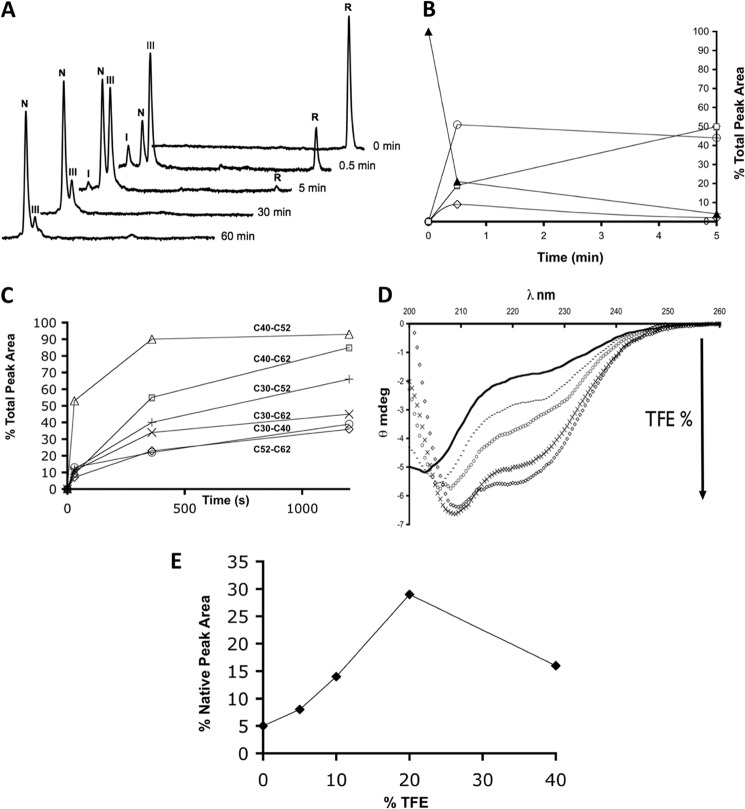
When reduced Cox19 (10 mm) was mixed with excess of oxidized Mia40 (100 mm), we observed the rapid conversion of the reduced Cox19 peak into the 3 major species detected previously: peak I, N and III (Fig. 7A, 30s). Mia40 promotes a fast and preferential oxidation of reduced Cox19 into I (Cys40-Cys52), as it is evident from the comparison of the kinetic profiles of catalyzed and uncatalyzed reactions (Fig. 7B). However, the presence of Mia40 cannot avoid the formation of a minor amount of I (Cys40-Cys62). The conversion of Cox19 into a single disulfide species takes about 5 min, and total protein oxidation takes less than an hour compared with the 24 h required for air oxidation. Considering that both intermediates display significant secondary structure, this would trap the substrate in the IMS, therefore providing a physiological mechanism for the import of Cox19. Our results also show that the reaction can be separated into two independent steps; that is, Mia40 does not release the fully oxidized substrate, in agreement with the observation that Mia40 only contains one redox center, and it can, in principle, just introduce one disulfide bond at a time (25–27).
FIGURE 7.
Mia40 catalyzes the rapid oxidation of Cox19 into a single disulfide species. A, Mia40 (100 μm) was added to reduced Cox19 (10 μm), and aliquots were removed at the indicated time points and analyzed by RP-HPLC. The retention times of the detected species coincide exactly with those detected in spontaneous Cox19 oxidative folding. B, different peaks were integrated, and their area plotted as a function of time; reduced Cox19 (▴), Native Cox19 (□), Peak I (♢), and Peak III (○). For clarity the first 5 min of reactions are shown, with a rapid conversion of reduced Cox19 into single disulfide species, mainly peak III that slowly converts into native protein. C, Mia40 (100 μm) was added to reduced Cox19 single disulfide mutants (10 μm); aliquots were removed at the indicated time points and analyzed. Peak areas were integrated and plotted as a function of time. At early stages, Mia40 is only able to catalyze formation of the Cys40-Cys52 disulfide. D, TFE was added to reduced Cox19 at different final percentages, and the protein CD spectrum analyzed: 0% (black), 5% (−), 10% (○), 20% (×), and 40% (♢). E, TFE was added to reduced Cox19, and 1 h folding mixtures were analyzed by RP-HPLC. The percentage of native protein formed was plotted as a function of TFE %.
Mia40 Only Catalyzes Cys40-Cys52 for Substrate Oxidation
In vivo and in vitro data support an oxidation mechanism involving the formation of an intermediate displaying a mixed disulfide between Mia40 CPC redox site and one of the substrate Cys (28). Mia40 recognizes a precise set of residues in the substrate, the ITS/MISS region, whose location on the polypeptide chain depends on the type of mitochondrial protein to be imported. In the TIM family binding occurs to the first Cys of the CX3C twin motif, whereas in Cox17 it involves the third Cys (28–30). In Cox19, the interaction is also expected to occur in the third Cys of the second twin CX9C motif, Cys-52 as it contains a conserved CXX[Hy][Hy]XX[Ar]XC ITS/MISS motif (31). The catalysis would then proceed with the formation of the inner disulfide bond as a consequence of the attack of Cys-40 on the intermolecular disulfide bond between Mia40 and Cys-52, a mechanism consistent with I (Cys40-Cys52) being the major product of Mia40 catalysis (50% of reduced Cox19 is converted into this species in 30s). However, the formation of I (Cys40-Cys62) in the presence of Mia40 is inconsistent with this catalytic model. We analyzed whether Mia40 could catalyze the direct formation of other disulfide bonds in Cox19 apart from Cys40-Cys52. We took advantage of the mobility shift in RP-HPLC to study the influence of Mia40 on the oxidation reaction of the six single disulfide analogs. Single-disulfide Cox19 mutants were reduced, mixed with Mia40, and its oxidation analyzed (Fig. 7C). As expected, Mia40 catalyzed the rapid formation of the inner disulfide bond Cys40-Cys52 and in 30s 50% of the mutant was already oxidized. In contrast, the ability to catalyze the reaction of the remaining mutants, including Cys40-Cys62 species during short time periods was very limited. Thus, Mia40 funnels Cox19 folding by promoting the selective and fast formation of the inner native disulfide bond. We can discard that the formation of the non-native off-pathway intermediate results from a specific interaction of Mia40 with Cys-40 or Cys-62. In addition, the fact that the formation of the Cys30-Cys62 disulfide bond cannot by catalyzed by Mia40 suggests that the formation of the second native disulfide on top of the Cys40-Cys52 intermediate will be mediated by oxygen and thus, as shown here, slowly, a view that is consistent with the data obtained for Cox17 in which the production of H2O2 as a co-product of that reaction was detected (25).
Low Concentrations of TFE Accelerate Cox19 Folding - Evidence for the Folding Template Mechanism
Banci et al. used an artificially arrested Mia40-substrate intermediate obtained with Cys-mutated forms of Cox17 and Mia40 to show by NMR that Mia40 binding and disulfide formation induced the selective folding of an initial α-helix stretch on the previously unfolded ITS/MISS region, with hydrophobic residues in the Mia40 substrate binding cleft acting as a scaffold (26). Our data argue that in Cox19, the equivalent disulfide and helix form spontaneously, although at a very low rate. We speculated that any factor favoring the formation of native helical structure should in principle accelerate the folding reaction. We investigated the effect of 2,2,2-trifluoroethanol (TFE) on the oxidative folding of Cox19. TFE is known to stabilize secondary elements, in particular α-helices, at low concentrations and it has been shown to promote and accelerate the folding of several proteins by stabilizing native helical structure in folding intermediates (32). When reduced Cox19 was titrated against TFE and monitored by far-UV CD, we observed that despite reduced Cox19 being essentially unfolded, in the presence of increasing amounts of TFE it gradually shows an increase in ellipticity consistent with the formation of α-helical structures (Fig. 7D).
Acid-trapping and RP-HPLC analysis show that the oxidative folding of Cox19 is highly accelerated by TFE. Interestingly, the accelerating effect is not linearly correlated with its concentration but passes through a maximum. The maximum increase in Cox19 oxidation rate, 6-fold faster than that under control conditions, was found at 20% (v/v) TFE. Further addition of TFE caused a deceleration of oxidative folding, consistent with the observation that over-stabilization of the secondary structure in folding intermediates results in the formation of non-native hydrogen bonds and causes a retardation of folding. In any case, we show that, as Mia40, by reducing the conformational search to attain the native secondary structure TFE accelerates the formation of the correctly disulfide-bonded Cox19 conformation (Fig. 7E).
DISCUSSION
In the present work, we show that Cox19 folds from the reduced and unfolded state to the native conformation through a sequential oxidation of cysteine residues. Although 8 different disulfide-bonded intermediates are possible, only two species populate significantly its oxidative folding pathway. Despite this apparent simplicity, the spontaneous oxidative folding of Cox19 is a slow process, taking around 24 h to complete. This parsimony is advantageous in the cell since oxidative folding is a strong kinetic competitor of mitochondrial import (16). The half time of Cox19 residence in the cytosol is around 8 min (22). In this time frame, the protein would still remain reduced and, according to our data, essentially unfolded, being thus in a conformational state in which it can diffuse to the IMS. The slow oxidative folding of Cox19 results from two different effects, the population of non-native isomers, identifiable in the presence of denaturant, and the presence of kinetic traps displaying significant helical structure.
For the first time, Mia40 influence on a substrate folding pathway could be determined with structural detail, as we were able to resolve the different intermediates populating the reaction. There has been considerable debate on whether Mia40 catalyzes formation of both disulfides in CX9C motifs; we demonstrate here that only the inner disulfide intermediate is rapidly catalyzed by Mia40. In fact, the Mia40 main action is to convert Cox19 into an inner disulfide cross-linked species, skipping the sampling of alternative intermediates. This species then oxidizes spontaneously into the native protein.
The Cox19 folding code is already imprinted in its primary structure; that is, with the proper difference in rates, Cox19 oxidative folding by air follows the same pathway as in the presence of Mia40. NMR data indicate that helix 2, which is expected to get “activated” upon Mia40-Cox19 interaction, already forms at the early stages of folding in the absence of the chaperone, and only then the remaining protein completes folding. The role of Mia40 is funnelling these intrinsic tendencies rather than changing or creating the pathway by activating a specific sequential element, as previously suggested (26). In fact, induction of α-helical structure by TFE suffices to mimic the Mia40 template mechanism. The Mia40 mechanism of action conforms to the central dogma in chaperone catalysis and explains how and why Mia40 is able to scope with such a dissimilar and numerous group of substrates. Apart from the CX9C motif, there are no clear sequential/structural relationships between Mia40 substrates of this family, implying that each folding process is perhaps unique, providing thus a large field of study, as 31 CX9C motif proteins have so far been described.
Acknowledgment
We thank Dr. Jan Riemer, TU Kaiserslautern, for pGEX-6p-1 Cox19 and pGEX-6p-1 scMia40 dam plasmids.
This work was supported by Grants BFU2010-14901 (Ministerio de Ciencia e Innovacion, Spain) and 2009-SGR-760 (Generalitat de Catalunya) grants.

This article contains supplemental Table S1 and Fig. S1.
- IMS
- intermembrane space
- AMS
- (4-acetamido-49-maleimidylstibene-2,29-disulfonic acid)
- ANS
- 8-anilino-1-naphthalenesulfonic acid
- Mia40
- mitochondrial intermembrane space import and assembly
- Erv1
- essential for respiration and vegetative grow
- TFA
- trifluoroacetic
- PDI
- protein-disulfide isomerase
- SDRs
- small disulfide-rich proteins
- TFE
- 2,2,2-trifluoroethanol.
REFERENCES
- 1. Riemer J., Bulleid N., Herrmann J. M. (2009) Disulfide formation in the ER and mitochondria: two solutions to a common process. Science 234, 1284–1287 [DOI] [PubMed] [Google Scholar]
- 2. Endo T., Yamano K. (2009) Multiple pathways for mitochondrial protein traffic. Biol. Chem. 390, 723–730 [DOI] [PubMed] [Google Scholar]
- 3. Deponte M., Hell K. (2009) Disulphide bond formation in the intermembrane space of mitochondria. J. Biochem. 146, 599–608 [DOI] [PubMed] [Google Scholar]
- 4. Riemer J., Fischer M., Herrmann J. M. (2011) Oxidation-driven protein import into mitochondria: Insights and blind spots. Biochim. Biophys. Acta 1808, 981–989 [DOI] [PubMed] [Google Scholar]
- 5. Koehler C. M., Beverly K. N., Leverich E. P. (2006) Redox Pathways of the Mitochondrion. Antioxid. Redox. Signal 8, 813–822 [DOI] [PubMed] [Google Scholar]
- 6. Hatahet F., Ruddock L. W., Ahn K., Benham A., Craik D., Ellgaard L., Ferrari D., Ventura S. (2009) Protein disulfide isomerase: a critical evaluation of its function in disulfide bond formation. Antioxid. Redox. Signal 11, 2807–2850 [DOI] [PubMed] [Google Scholar]
- 7. Chacinska A., Pfannschmidt S., Wiedemann N., Kozjak V., Sanjuán Szklarz L. K., Schulze-Specking A., Truscott K. N., Guiard B., Meisinger C., Pfanner N. (2004) Essential role of Mia40 in import and assembly of mitochondrial intermembrane space proteins. EMBO J. 23, 3735–3746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mesecke N., Terziyska N., Kozany C., Baumann F., Neupert W., Hell K., Herrmann J. M. (2005) A disulfide relay system in the intermembrane space of mitochondria that mediates protein import. Cell 121, 1059–1069 [DOI] [PubMed] [Google Scholar]
- 9. Hell K. (2008) The Erv1-Mia40 disulfide relay system in the intermembrane space of mitochondria. Biochim. Biophys. Acta 1783, 601–609 [DOI] [PubMed] [Google Scholar]
- 10. Rigby K., Zhang L., Cobine P. A., George G. N., Winge D. R. (2007) Characterization of the cytochrome c oxidase assembly factor Cox19 of Saccharomyces cerevisiae. J. Biol. Chem. 282, 10233–10242 [DOI] [PubMed] [Google Scholar]
- 11. Leary S. C., Cobine P. A., Nishimura T., Verdijk R. M., de Krijger R., de Coo R., Tarnopolsky M. A., Winge D. R., Shoubridge E. A. (2013) COX19 mediates the transduction of a mitochondrial redox signal from SCO1 that regulates ATP7A-mediated cellular copper efflux. Mol. Biol. Cell 24, 683–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bien M., Longen S., Wagener N., Chwalla I., Herrmann J. M., Riemer J. (2010) Mitochondrial disulfide bond formation is driven by intersubunit electron transfer in Erv1 and proofread by glutathione. Mol. Cell 37, 516–528 [DOI] [PubMed] [Google Scholar]
- 13. Longen S., Bien M., Bihlmaier K., Kloeppel C., Kauff F., Hammermeister M., Westermann B., Herrmann J. M., Riemer J. (2009) Systematic Analysis of the Twin Cx9C Protein Family. J. Mol. Biol. 393, 356–368 [DOI] [PubMed] [Google Scholar]
- 14. Ventura S., Chang J. Y. (2011) Folding of Disulfide Proteins, Berlin, Heidelberg & New York [Google Scholar]
- 15. Marley J., Lu M., Bracken C. (2001) A method for efficient isotopic labelling of recombinant proteins. J. Biomol. NMR 20, 71–75 [DOI] [PubMed] [Google Scholar]
- 16. Morgan B., Lu H. (2008) Oxidative folding competes with mitochondrial import of the small Tim proteins. Biochem. J. 411, 115–122 [DOI] [PubMed] [Google Scholar]
- 17. Arolas J. L., Bronsoms S., Ventura S., Aviles F. X., Calvete J. J. (2006) Characterizing the tick carboxypeptidase inhibitor: molecular basis for its two-domain nature. J. Biol. Chem. 281, 22906–22916 [DOI] [PubMed] [Google Scholar]
- 18. Voronova A., Meyer-Klaucke W., Meyer T., Rompel A., Krebs B., Kazantseva J., Sillard R., Palumaa P. (2007) Oxidative switches in functioning of mammalian copper chaperone Cox17. Biochem. J. 408, 139–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tienson H. L., Dabir D. V., Neal S. E., Loo R., Hasson S. A., Boontheung P., Kim S. K., Loo J. A., Koehler C. M. (2009) Reconstitution of the mia40-erv1 oxidative folding pathway for the small tim proteins. Mol. Biol. Cell 20, 3481–3490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Harrison P., Hegyi H., Tompa P. (2008) Intrinsically Disordered Proteins Display No Preference for Chaperone Binding In Vivo. PLoS Comput. Biol. 4, e1000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fersht A. R., Daggett V. (2002) Protein Folding and Unfolding at Atomic Resolution. Cell 108, 573–582 [DOI] [PubMed] [Google Scholar]
- 22. Fischer M., Horn S., Belkacemi A., Kojer K., Petrungaro C., Habich M., Ali M., Kuttner V., Bien M., Kauff F., Dengjel J., Herrmann J. M., Riemer J. (2013) Protein import and oxidative folding in the mitochondrial intermembrane space of intact mammalian cells. Mol. Biol. Cell 24, 2160–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Arolas J. L., Aviles F. X., Chang J.-Y., Ventura S. (2006) Folding of small disulfide-rich proteins: clarifying the puzzle. Trends Biochem. Sci. 31, 292–301 [DOI] [PubMed] [Google Scholar]
- 24. Wishart D. S., Sykes B. D., Richards F. M. (1992) The chemical shift index: a fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. Biochemistry 31, 1647–1651 [DOI] [PubMed] [Google Scholar]
- 25. Banci L., Bertini I., Cefaro C., Ciofi-Baffoni S., Gallo A., Martinelli M., Sideris D. P., Katrakili N., Tokatlidis K. (2009) MIA40 is an oxidoreductase that catalyzes oxidative protein folding in mitochondria. Nat. Struct. Mol. Biol. 16, 198–206 [DOI] [PubMed] [Google Scholar]
- 26. Banci L., Bertini I., Cefaro C., Cenacchi L., Ciofi-Baffoni S., Felli I. C., Gallo A., Gonnelli L., Luchinat E., Sideris D., Tokatlidis K. (2010) Molecular chaperone function of Mia40 triggers consecutive induced folding steps of the substrate in mitochondrial protein import. Proc. Natl. Acad. Sci. U.S.A. 107, 20190–20195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bourens M., Dabir D. V., Tienson H. L., Sorokina I., Koehler C. M., Barrientos A. (2012) Role of Twin Cys-Xaa9-Cys Motif Cysteines in Mitochondrial Import of the Cytochrome c Oxidase Biogenesis Factor Cmc1. J. Biol. Chem. 287, 31258–31269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sideris D. P., Tokatlidis K. (2007) Oxidative folding of small Tims is mediated by site-specific docking onto Mia40 in the mitochondrial intermembrane space. Mol. Microbiol. 65, 1360–1373 [DOI] [PubMed] [Google Scholar]
- 29. Milenkovic D., Ramming T., Müller J. M., Wenz L. S., Gebert N., Schulze-Specking A., Stojanovski D., Rospert S., Chacinska A. (2009) Identification of the Signal Directing Tim9 and Tim10 into the Intermembrane Space of Mitochondria. Mol. Biol. Cell 20, 2530–2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Milenkovic D., Gabriel K., Guiard B., Schulze-Specking A., Pfanner N., Chacinska A. (2007) Biogenesis of the Essential Tim9 Tim10 Chaperone Complex of Mitochondria: site-specific recognition of cysteines residues by the intermembrane space receptor Mia40. J. Biol. Chem. 282, 22472–22480 [DOI] [PubMed] [Google Scholar]
- 31. Sideris D. P., Petrakis N., Katrakili N., Mikropoulou D., Gallo A., Ciofi-Baffoni S., Banci L., Bertini I., Tokatlidis K. (2009) A novel intermembrane space-targeting signal docks cysteines onto Mia40 during mitochondrial oxidative folding. J. Cell Biol. 187, 1007–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chiti F., Taddei N., Webster P., Hamada D., Fiaschi T., Ramponi G., Dobson C. M. (1999) Acceleration of the folding of acylphosphatase by stabilization of local secondary structure. Nat. Struct. Mol. Biol. 6, 380–387 [DOI] [PubMed] [Google Scholar]