Abstract
Multiple scientific disciplines require the isolation of specific subsets of blood cells from patient samples for gene expression analysis by microarray or RNA-sequencing, preserving disease- or treatment-related signatures. However, little is known with respect to the impact of different cell isolation methods on gene expression and the effects of positive selection, negative selection and fluorescence activated cell sorting (FACS) have not previously been assessed in parallel. To address this knowledge gap, CD4+ T cells, CD8+ T cells, B cells and monocytes were isolated from blood samples from 5 independent donors using positive immunomagnetic selection, negative immunomagnetic selection and FACS. We hypothesized that positive selection and FACS would yield higher purity but may have an impact on gene expression since both methods utilize antibodies that bind surface receptors of the cell type of interest. Moreover, FACS might upregulate stress response genes due to passage of the cells through the sorter. Microarray gene expression data was generated and subjected to unsupervised clustering and differential gene expression analysis. Surprisingly, these analyses revealed that gene expression signatures were more similar between cells isolated by negative selection and FACS compared to cells isolated by positive selection. Moreover, genes that are involved in the response to stress generally had the highest expression in cells isolated by negative or positive selection and not FACS. Thus, FACS is the recommended method for isolation of leukocyte subsets for gene expression studies since this method results in the purest subset populations and does not appear to induce a stress response.
Keywords: negative immunomagnetic selection, positive immunomagnetic selection, fluorescent activated cell sorting, CD4+ T cell, CD8+ T cell, B cell, monocyte, gene expression, microarray
INTRODUCTION
With the advance of microarray and sequencing technologies, multiple disciplines profile gene expression in the entire transcriptome of patient samples in order to investigate the mechanism of disease pathogenesis (1-3). Ideally, cells from patients should undergo minimal handling prior to analysis to preserve gene expression signatures pertinent to a disease or a therapeutic intervention and not to incur gene expression perturbations related to cell isolation. Thus, peripheral blood mononuclear cells (PBMCs) have been used in many studies aimed to build diagnostic or prognostic classifiers, due to the relative ease of sampling (4,5). However, PBMCs are not suitable for specific pathogenesis analyses (5). Studying diseases that affect cells in the blood compartment (e.g., viral infections, autoimmune diseases and stress) by PBMC gene expression profiling poses problems associated with the signal of gene expression being derived from a mixture of different blood cell types (6,7). For example, HIV primarily infects CD4+ T cells and it would be desirable to profile gene expression in this subset as opposed to PBMCs when investigating viral-induced pathogenesis. Furthermore, although monocytes may also be infected by HIV (8-10), the course of infection may be quite different compared to CD4+ T cells (11), and combined analysis of CD4+ T cells and monocytes in the PBMC compartment may obscure important distinctions between these two cell subsets. Isolation of cell subsets is also highly desirable when proportion of a particular cell subset is changed due to the disease, and thus gene expression signature would reflect mainly cell composition. For example, in systemic lupus erythematosus PBMCs contain elevated numbers of immature granulocytes, and a previous gene expression profiling study simply reflected the increased amount of these cells in patient samples compared to controls (12). Moreover, studies of disease pathogenesis often need to isolate individual cell types from a limited amount of blood, and therefore it is relevant to optimize the generation of high-quality gene expression data for these samples.
One potential problem posed by isolation of individual cell types is the effect of isolation method on gene expression. Two commonly used methods for cell isolation are positive and negative selection, which differ greatly in terms of the approach used to isolate the target cell type. Positive selection utilizes cell receptor antibodies to target the specific cell type of interest and may potentially turn on activation cascades through these receptors (13-16) or cause receptor blockade and inhibit the downstream functions of the isolated cells (17,18). Negative selection methods were developed to avoid these issues by using antibody cocktails to remove non-target cells, which may result in target cell enrichment rather than purification. To address the impact of positive and negative selection on gene expression, Lyons and colleagues (19) previously analyzed gene expression in leukocyte subsets (i.e., CD4+ T cells, CD8+ T cells and CD14+ monocytes) that were isolated by these two approaches. Magnetic cell sorting with Miltenyi microbeads (20) was used for positive and negative selection, and they concluded that gene expression differences between the two methods for the same cell subset was the result of differences in sample purity. Specifically, they found that CD4+, CD8+ T cells and CD14+ monocytes were contaminated with other cells after negative, not positive selection, and that genes that were identified as differentially expressed reflected this contamination.
Fluorescence activated cell sorting (FACS) (21) is an additional method of cell subset isolation which results in high cell purity and facilitates the isolation of multiple cell types in a single run (22). However, it is currently unknown whether the stresses associated with passing cells through the flow cytometer result in changes in gene expression. Moreover, FACS uses antibodies to stain cells for detection during sorting, thus the same concerns with respect to activation cascades or receptor inhibition may be relevant as discussed above for positive selection. Therefore, the aim of the current study was to assess the effects of FACS, positive selection and negative selection on gene expression when isolating four different cell subsets (i.e., CD4+ and CD8+ T cells, B cells and monocytes) from a total of 5 donor samples. We focused on immediate rather than long-term effects of cell isolation on gene expression to mimic the scenario when gene expression signatures from patients with a disease or under therapeutic treatment are of interest where the time of cell handling needs to be minimized. Furthermore, since the majority of sample repositories consist of viably cryopreserved PBMCs, the effect of these isolation methods on gene expression was evaluated using previously frozen cells rather than cells isolated from fresh blood.
MATERIALS AND METHODS
Sample collection
Blood samples from 5 healthy male donors (a mean age of 25) were collected by venipuncture following protocols approved by the Human Research Protections Program at the University of California San Diego. PBMCs were isolated using Ficoll–Hypaque (Histopaque-1077, Sigma Chemical Company, St. Louis, MO) and stored in 10×106 cell aliquots in liquid nitrogen until use. Please refer to the Supplemental Methods for details on PBMC purification, cryopreservation and thawing procedures.
Immunomagnetic positive and negative selection of leukocyte subsets
For positive selection, an aliquot of 10×106 frozen PBMCs was used for sequential isolations of B cells followed by CD8+ T cells and another 10x106 frozen PBMC aliquot was used for sequential isolation of monocytes followed by CD4+ T cells. All isolations were performed using the following kits from StemCell Technologies, Inc. (Vancouver, Canada): human CD19 positive selection kit (B cells), human CD8 positive selection kit (CD8+ T cells), human CD33 positive selection kit (monocytes), human CD4 positive selection kit (CD4+ T cells) according to manufacturer's instructions. For negative selection, independent aliquots of 10×106 frozen PBMCs were used to isolate CD4+ and CD8+ T cells, B cells and monocytes. EasySep enrichment kits (human CD4+ T cell enrichment kit, human CD8+ T cell enrichment kit, human B cell enrichment kit, human monocyte enrichment kit) from StemCell Technologies, Inc. (Vancouver, Canada) were used according to manufacturer's instructions. Purity was assessed for all cells isolated by positive and negative selection. The following antibodies from BD Biosciences, Inc. (San Diego, CA) were used: CD4-FITC (Multiclone Leu3a+3b), CD14-PE (MoP9), CD8-PE-Cy7 (SK1), CD19-APC (HIB19), CD3-PerCP-Cy5.5 (SK7). For further isolation and purity assessment details, please refer to the Supplemental Methods. Cell suspensions were immediately centrifuged and pellets were resuspended in RLT buffer from RNeasy Mini kit (Qiagen, Inc., Valencia, CA) supplemented with β-mercaptoethanol and stored at −80°C until RNA extraction.
Fluorescence activated cell sorting of leukocyte subsets
An aliquot of 10×106 frozen PBMCs was used to isolate CD4+ and CD8+ T cells, B cells and monocytes using CD4-FITC (Multiclone Leu3a+3b), CD8-PE-Cy7 (SK1), CD19-APC (HIB19) and CD33-PE (P67.6) antibodies, respectively, and the CD45-APC-H7 (2D1) antibody was used to discriminate PBMCs. All antibodies were supplied by BD Biosciences, Inc. (San Diego, CA) and used at the minimal saturating concentration, determined by titration prior to the experiment. PBMCs were stained with the antibody combination for 20 minutes at room temperature, washed twice, and resuspended in PBS supplemented with 1% FCS. Leukocytes subsets were sorted on a MoFlo cell-sorter (Beckman Coulter Instruments, Brea, CA) and kept at 4°C throughout sorting. Live PBMCs were gated based on forward versus scatter profiles, followed by CD45+ based gating for leukocyte identification. Monocytes were identified and selected based on their double positivity for CD45 and CD33. The CD33 negative population was subdivided by CD8/CD4 expression and used for the identification of CD4+ (CD4+CD8–) and CD8+ (CD4–CD8+) T cells; double negative cells (CD4–CD8–) were further selected by CD19 positivity for the isolation of B cells (Fig. S1). Purity was determined for subsets isolated by FACS for CD4+ and CD8+ T cells and one sample of B cells. The remaining B cell samples and monocyte samples were not assessed for purity due to low yields. Sorted cells were immediately centrifuged and pellets were resuspended in RLT buffer from RNeasy Mini kit (Qiagen Inc., Valencia, CA), supplemented with β-mercaptoethanol, and stored at −80°C until RNA extraction.
RNA extraction and hybridization to microarrays
RNA was extracted using Qiagen RNeasy micro kit (Qiagen Inc., Valencia, CA)according to manufacturer's instructions. RNA concentration was determined using a NanoDrop 1000 (Thermo Fisher Scientific, Wilmington, DE). Out of the 60 samples (5 donors, 3 isolation methods, and 4 cell types) sufficient RNA yield for microarray analysis was obtained for 50 samples. RNA quality was spot checked in the majority of samples (74%) by measuring RNA integrity number (RIN) using Agilent 2100 Bioanalyzer (Agilent Technologies Inc., Santa Clara, CA). RINs were all above 6.0 (average 7.94, standard deviation ±1.35), and deemed of sufficient quality for microarray analysis. cRNA preparations and hybridizations to Illumina (San Diego, CA) HT12 version 4 beadchips (47,324 probes) were performed by Expression Analysis, Inc. (Durham, NC). One hundred nanograms of total RNA was used as starting material for all cell types except B cells, for which 50 ng was used. Gene expression data are available at the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE50008.
Microarray data analysis
Microarray data quality was assessed using MA-plots in the affyPLM package (23), implemented in the R statistical computing environment version 2.8.0, followed by construction of median vs interquartile range (IQR) plots to identify outlier arrays (defined as falling outside of two standard deviations by median and/or IQR). The lumi package (24) in Bioconductor was used to transform (i.e., variance-stabilizing) and normalize (i.e., robust-spline) the data and the genefilter package (25) was used to filter genes based on an IQR cut-off of 0.7.
To assess the similarities between samples based on gene expression, two unsupervised approaches in R version 2.14.0 were used: bootstrapped clustergrams using the pvclust package (26) and principal component analysis (PCA) implemented in the ClassDiscoverypackage in OOMPA (http://bioinformatics.mdanderson.org/Software/OOMPA/Current/oompa-cd.pdf). For these analyses, expression data was normalized using z-score normalization. The approximately unbiased p-value calculated by pvclust was used to determine the significance of sample clustering such that a percentage of 95% corresponds to p<0.05). For further details about statistics that are used by pvclust and PCA please refer to Supplemental Methods.
To identify differentially expressed genes (DEGs) between the 3 isolation methods for CD4+ and CD8+ T cell subsets, a repeated measures (RM) ANOVA was implemented with a post hoc Tukey test using R. Correction for the false discovery rate (FDR) associated with multiple testing was performed using Benjamini-Hochberg method (27). The RM ANOVA code implemented in R is available in the Supplemental Methods. Genes with FDR-corrected p-values <0.05 were considered significant. Venn diagrams were constructed using Venny (http://bioinfogp.cnb.csic.es/tools/venny/index.html). Since samples from only two groups (isolated by positive and negative selection and not by FACS) were available for B cells and monocytes, DEGs were identified using univariate paired t-test with FDR correction implemented in BRB-Array Tools (28), rather than using a RM ANOVA.
Gene ontology (GO) terms related to biological processes that were significantly over-represented for differentially expressed genes were identified using the Biological Networks Gene Ontology (BiNGO) plugin version 2.42 in Cytoscape version 2.7.0 (29). GO terms with p-values <0.05 derived from a hypergeometric test and subsequent FDR-correction were considered significant. The list of all the probes present on the Illumina HT12 v4 platform was selected as background when performing hypergeometric tests to identify significant GO terms.
Validation of gene expression by droplet digital PCR (ddPCR)
Absolute quantification of target RNA molecules was facilitated by ddPCR using the QX100 system (Bio-Rad, Hercules, CA), which requires only 5 ng or less total RNA per reaction. TaqMan Gene Expression Assays (Applied Biosystems [ABI], Foster City) were used to confirm microarray results for the following targets: the spleen focus forming virus proviral integration oncogene (SPI1, Hs00231368_m1), the CD14 molecule (CD14, Hs02621496_s1), and the serum deprivation response (SDPR, Hs00190538_m1) gene. The TaqMan Gene Expression Assay (Hs03044961_g1) for the ribosomal protein L27 (RPL27) gene was selected for a normalizer (30). The data was expressed as copies of target mRNA molecules per million copies of RPL27 mRNA molecules and then log2 transformed. RM ANOVAs with post hoc Tukey tests were performed to compare expression of SPI1 and CD14 in CD4+ T cells and monocytes isolated by positive and negative selection, and to compare expression of SDPR between all three isolation methods in monocytes. Genes differentially expressed with Tukey corrected p-values <0.05 were considered significant. For further details on ddPCR procedure please refer to Supplemental Methods.
RESULTS
The effect of isolation method on target cell yield and purity
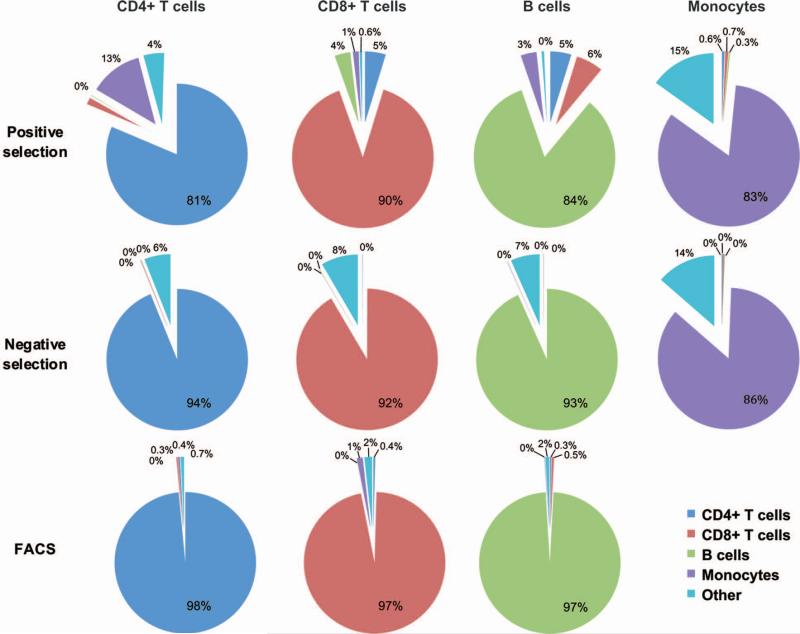
CD4+ and CD8+ T cells, B cells and monocytes were isolated from the PBMC samples taken from 5 donors using 3 different isolation methods: immunomagnetic positive or negative selection, or FACS (N = 60 samples). Positive and negative immunomagnetic selection resulted in similar yields of each cell subtype, with CD4+ and CD8+ T cells having the highest yields and B cells the lowest (Fig. S2). FACS resulted in lower yields for all the cell types, particularly monocytes. In the majority of cases, percentages of all contaminating cell subsets were significantly higher in the positively isolated samples compared to samples isolated by negative selection or FACS, with FACS having the least contamination (Fig. 1 and Table S1). Monocytes were the exception having similar contamination with each cell subset for positive and negative selection. All samples isolated by negative selection had low levels of contamination with CD4+ T cells, CD8+ T cells and B cells compared to positive selection, but had higher level of contamination with unidentified cells denoted as “other” which was significant for CD8+ T and B cells (Table S1). Cell viability after isolation was not significantly different between the methods for each cell type as determined by a non-parametric paired t-test (data not shown).
Figure 1. Cell purity.
Purities of each cell subset following isolation by positive selection, negative selection or FACS. Average percentages of each cell subtype (CD4+ T cells, CD8+ T cells, B cells, Monocytes, and Other) across donors are shown for each isolation method except for B cells and monocytes isolated by FACS. Only one B-cell sample isolated by FACS was assessed for purity and the other B-cell as well as the monocyte samples did not yield sufficient numbers of cells to perform both purity assessments and microarray analysis, so samples were prioritized for the latter.
Clustering approaches reveal that isolation methods influence gene expression
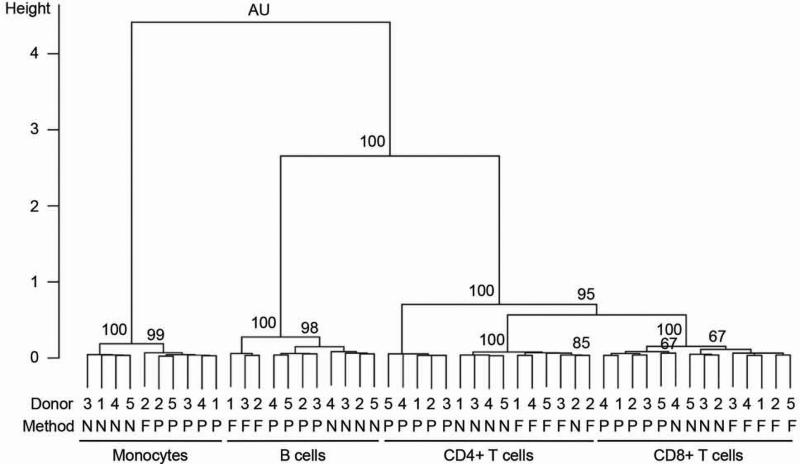
Unsupervised clustering was employed in order to elucidate whether gene expression was affected by contaminating cell subsets or by isolation method. If isolation method did not affect gene expression, then cells of a given cell type (e.g., CD4+ T cells) isolated from a particular donor (e.g., Donor 1) should cluster together regardless of isolation method. Following examination of RNA yields and quality control of microarray data the original 60 sample design was reduced to 50 samples as indicated in Table 1. For initial analyses, 5,843 genes were used that resulted from filtering the genes based on their IQR being >0.7. Unsupervised clustering of samples clearly demonstrated that samples clustered primarily by cell type but then by isolation method and not by donor. The only exception was the clustering together of CD4+ T cell samples isolated by negative selection and FACS for Donor 2 (Fig. 2). The four major clusters associated with cell type were all significant as assessed by AU p-values (95% corresponding to p<0.05). Subclusters associated with isolation method were significant for all cell types except for CD8+ T cells where samples isolated by negative selection and FACS generally could not be discriminated from each other (formed one major cluster), and one sample isolated by negative selection from Donor 4 clustered with cells isolated by positive selection (Fig. 2).
Table 1.
Samples included in the analyses of the microarray data for each cell subset and cell isolation method. Samples were excluded based on poor RNA yields and outlier microarrays as determined using microarray data quality control measures.
| Positive Selection | Negative Selection | FACS | |
|---|---|---|---|
| CD4+ T cells | Donors 1,2,3,4,5 | Donors 1,2,3,4,5 | Donors 1,2,3,4,5 |
| CD8+ T cells | Donors 1,2,3,4,5 | Donors _,2,3,4,5 | Donors 1,2,3,4,5 |
| B cells | Donors _,2,3,4,5 | Donors _,2,3,4,5 | Donors 1,2,3,_,_ |
| Monocytes | Donors 1,2,3,4,5 | Donors 1,_,3,4,5 | Donor _,2,_,_,_ |
Figure 2. Unsupervised clustering of samples based on gene expression.
Unsupervised clustering performed using pvclust package in statistical computing environment R version 2.14.1 using the entire filtered gene set (N=5,843). Pearson correlation was used to measure distances between the samples. Ward's minimum variance method was used for clustering. AU refers to the approximately unbiased p-value. P-values are indicated as percentages (e.g. 95 corresponds to p<0.05). Donor ID (from 1 to 5), isolation methods and cell types are indicated. P, N and F denote positive immunomagnetic selection, negative immunomagnetic selection, and FACS, respectively.
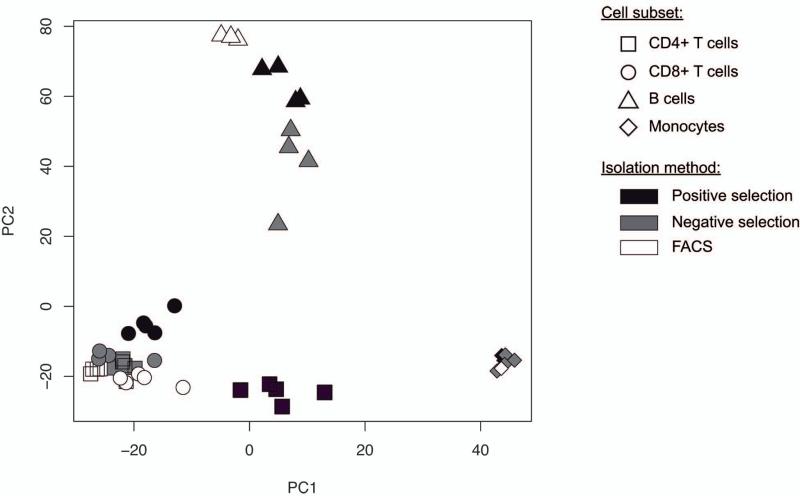
Results from PCA corroborated those from unsupervised clustering. Approximately 70% of the variation in gene expression was derived from the first two principal components (data not shown), associated primarily with cell type (principal component 1) and then secondarily with isolation method (principal component 2). As expected, all the T lymphocyte samples clustered close to each other and further away from cells of other lineages (i.e., B cells and monocytes) (Fig 3). Of particular note, CD4+ T cells isolated by positive selection gravitated towards the monocyte cluster along principal component 1 (Fig. 3) consistent with results from purity assessments, which demonstrated that CD4+ T cells isolated by this method were contaminated with monocytes. B cell samples isolated by different methods appeared to have the largest spread along principal component 2 compared to the spread exhibited by samples of other cell types. CD8+ T cell samples and monocytes in particular formed much tighter clusters within each principal component suggesting that both contamination and isolation method may affect gene expression to a lesser extent in these two cell types.
Figure 3. Principal component analysis.
Variation in gene expression summarized along the first two primary components is depicted. Information for cell type and isolation method was superimposed onto this plot. Although cell type is the major factor clustering samples along principal component 1, it is clear that cell isolation method and not donor is the signal for clustering in the 2nd component. PC1, principal component 1; PC2, principal component 2.
Identification of genes whose expression is affected by isolation method
Genes differentially expressed between the 3 different isolation methods were then identified for each cell type in turn. Samples from all cell types were then clustered based on the expression of each set of cell type specific differentially expressed genes. If samples continued to cluster based on cell type and then isolation method, this would indicate that the signal for differential gene expression was introduced primarily due to isolation method. In contrast, if cells of a specific type isolated by one of three methods clustered with cells of another type, then this would reflect contamination. An RM ANOVA or a paired t-test was used depending on a number of isolation methods compared: RM ANOVAs to compare positive, negative selection and FACS for CD4+ T cells and CD8+ T cells (Table S2A and B) and paired t-tests to compare positive and negative selection for B cells and monocytes (Table S2C and D).
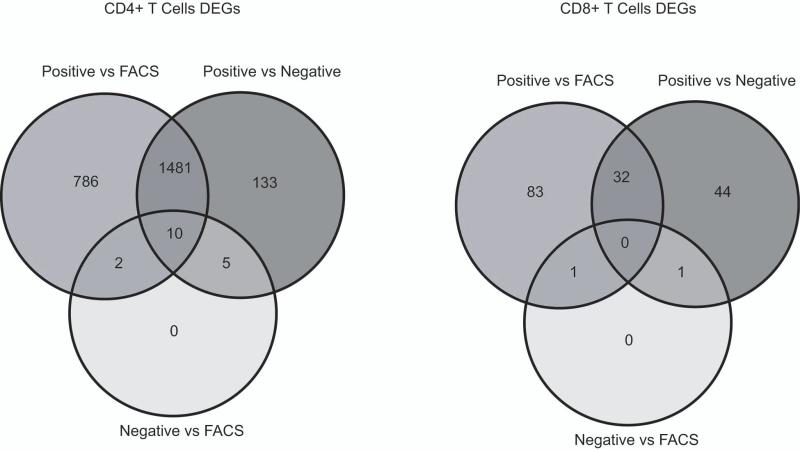
An RM ANOVA identified 2,417 DEGs (41% of the 5,843 genes tested that survived unsupervised filtering, Table S2A) between isolation methods in the CD4+ T cell subset. Post hoc Tukey test demonstrated that 2,279 (39%) genes were differentially expressed between positive selection and FACS, 1,629 genes (28%) between positive and negative selection and only 17 genes (0.3%) between negative selection and FACS (Fig 4). The clustergram (Fig. S3A) for genes differentially expressed in CD4+ T cells showed a similar pattern of cluster formation as the original clustergram constructed based on the entire initial gene set (N=5,843). CD4+ T cell samples isolated by positive selection clustered separately from other T cells in a significant cluster (AU=100% corresponding to p<0.01, Fig S3A). This result can be explained by monocyte contamination and the fact that enough of the DEGs identified for CD4+ T cells were monocyte specific genes that forced these samples to cluster outside of the main T cell cluster. Thus, in the case of CD4+ T cells the observed differential gene expression is mainly due to cell contamination.
Figure 4. Differentially expressed genes in CD4+ and CD8+ T cells.
Venn diagram of differentially expressed genes (DEGs, false discovery rate <0.05) identified for CD4+ and CD8+ T cells using an RM ANOVA with a post hoc Tukey test. Overlaps for the subsets of genes differentially expressed for each of the 3 comparisons (positive selection vs FACS, positive vs negative selection and negative selection vs FACS). The data indicates similarities in gene expression signatures in cells isolated by negative selection and FACS since the number of DEGs between these methods is low for both CD4+ and CD8+ T cells. On the other hand, gene expression signature in cells isolated by positive selection is different from both negative selection and FACS.
An RM ANOVA identified 164 DEGs (2.8% of all genes tested) between isolation methods in the CD8+ T cell subset (Table S2B). Post hoc Tukey test demonstrated that 116 genes (2%) were differentially expressed between positive selection and FACS, 77 genes (1.3%) between positive and negative selection and 2 genes (0.03%) between negative selection and FACS (Fig. 4). CD8+ T cell samples isolated by positive selection clustered with monocytes (Fig. S3B), however, this cluster was not significant (AU=70%), neither was the cluster separating CD8+ T cells isolated by FACS from other T cells (AU=75%). However, CD8+ and CD4+ T cells isolated by negative selection formed a significant cluster separating them from other CD4+ T cells (AU=99%). This would suggest that CD8+ T cells isolated by negative selection may be contaminated with CD4+ T cells, but the purity assessment indicated that it was not the case (Fig 1). Therefore, the DEGs identified may be the result of isolation method, in particular, the effect of negative selection. Alternatively, this clustering pattern may reflect the comparable contamination of CD8+ and CD4+ T cells isolated by negative selection by cells designated as “other” that introduce similar changes in their gene expression pattern compared to CD4+ and CD8+ T cells isolated by other methods.
Paired t-tests identified 310 DEGs (5.3% of all genes tested) between negatively and positively isolated B cells (Table S2C) and 269 DEGs (4.6%) for monocytes (Table S2D). Clustergrams built based on the expression of DEGs for either B cells or monocytes revealed that all samples clustered by cell type and then by isolation method (Fig. S3C and D) indicating that these DEGs are likely the result of the isolation method and not contamination. In summary, DEGs for each cell type were identified, but it appeared that only in cases of B cells and monocytes was the signal due to the effect of isolation method alone, while in case of T cells contamination also caused an effect, which was particularly robust for CD4+ T cells.
Genes associated with stress have the lowest expression in cells isolated by FACS
GO term analysis was performed to identify functional categories affected by isolation method for each cell type. Four hundred ninety six significant terms (p<0.05 after FDR) were identified for CD4+ T cells, two terms for CD8+ T cells, 170 terms for B cells and 58 terms for monocytes (data not shown). The initial hypothesis was that FACS induces the expression of genes related to stress due to the physical stresses associated with sorting cells. To test this hypothesis, the GO term “Response to stress” (Accession GO:0006950) was examined for the significant over-representation of DEGs identified when isolation methods were compared within each of the 4 cell subsets. This GO term was significantly enriched for DEGs in case of CD4+ T cells (354 genes, p=3.64E-14), B cells (53 genes, p=1.43E-4) and monocytes (41 genes, p=7.00E-4), but not in the case of CD8+ T cells. Even though the subset of DEGs that mapped to this GO term was different for each cell type, the majority of these stress-related genes had the highest expression in B cells (49 of 53 genes) and monocytes (34 of 41 genes) isolated by negative selection (Table S3). In contrast, in the case of CD4+ T cells, the majority of these genes had the highest expression in cells isolated by positive selection (270 of 354 genes, Table S3). Average gene expression values for B cells for available donors and a single donor value for monocytes isolated by FACS were compared with average gene expression values for positive and negative selection (Table S3). In all cases, the majority of all the stress related genes had the lowest expression value for cells isolated by FACS (CD4+ T cells, 286 of 354; B cells, 53 of 53; monocytes, 40 of 41). There was an overlap of 7 DEGs that were associated with stress across all three cell types (i.e., CD4+ T cells, B cells and monocytes), and the expression of these genes was generally the highest in cells isolated by negative selection for each cell type (Table S4).
Platelet contamination of monocytes isolated by negative selection
Surprisingly, the GO term “Platelet activation” was also identified as significantly over-represented (p=2.60E-4) for DEGs identified when comparing positive and negative selection in the monocyte subset. Therefore, we hypothesized that monocytes isolated by either positive or negative selection might be contaminated with platelets. The mean expression of the genes that mapped to the “Platelet activation” GO term was the highest in monocytes isolated by negative selection compared to monocytes isolated by positive selection or compared to the values from a single donor tested by FACS. Moreover, a number of genes predominantly or exclusively expressed by platelets, MPL, SPDR, RSG18 and SEPT5 (31-34), were expressed at higher levels (up to 16-fold) in monocytes isolated by negative selection. McFarland et al. (35) previously demonstrated that in the low forward and side scatter area platelets comprise up to 80% of all events that also include dead cells, debris and red blood cells. Therefore, the percentage of events detected by forward and side light scatter in monocyte samples isolated by positive and negative selection was evaluated. There was 70.38 ±12.72% of the events detected in this low scatter area for monocytes isolated by negative selection compared to 7.00±2.9% for monocytes isolated by positive selection, indicating that negatively isolated monocytes were contaminated with 10 times as many platelets.
Stress-related genes identified as the result of differences in protocols and platelet contamination
To better understand why many of the stress-related genes were differentially expressed, we performed a careful comparison of the isolation protocols. Generally, cells were kept on ice during all the procedures, but time of incubation with antibodies performed at room temperature varied. In the case of FACS, cells were exposed to room temperature for a total of 30 minutes, negative selection - 25 minutes and positive selection - 55 minutes for B cells and monocytes, and 110 minutes for CD4+ and CD8+ T cells. Thus, different times at room temperature vs ice exposure for each of the isolation procedures could have contributed to stress response.
To test the hypothesis that different times at room temperature vs ice exposure contributed to stress response, we analyzed the distribution of DEGs related to stress among Response to stress child GO terms. Regardless of the cell type, most of the genes were found in the GO terms Defense Response and Response to wounding (Table S5). These GO terms encompass genes whose expression is modified by any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating the organism is under an exogenous stress (e.g. temperature, humidity, ionizing radiation) or associated with a damage to the organism. Different incubation time on ice and/or the room temperature during the procedure, as well as different cell-susceptibility to different stimuli, might lead to enrichment of the Defense Response and Response to wounding terms by DEGs. On the other hand, the GO term Response to wounding is overrepresented by genes expressed in platelets. Remarkably, among the 7 genes that overlapped for all three cell types, three genes (SEPT5, PRKCQ and F13A1) were mapped to GO terms relevant to platelet function in wounding, which is activated in response to stress (Blood coagulation, intrinsic pathway; Platelet activation; and Blood coagulation, respectively). Thus, both differences in the cell isolation protocols between different methods and contamination with platelets may contribute to the enrichment of the GO term Response to stress with DEGs.
Validation of gene expression by ddPCR
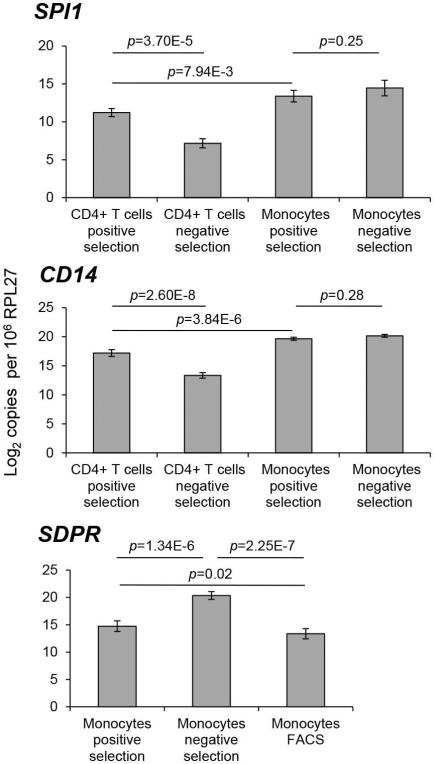
To validate gene expression by a method independent of microarrays, the following three genes were selected for ddPCR confirmation: SPI1, CD14and SDPR. SPI1 and CD14 are monocyte-specific genes and were selected to confirm that CD4+ T cells isolated by positive selection were contaminated with monocytes. Both SPI1 and CD14 were significantly differentially expressed by ddPCR between positively and negatively selected CD4+ T cells (SPI1, p=3.70E-5; CD14, p=2.60E-8), both having higher expression in positively selected compared to negatively selected cells (Fig 5 top and middle). In contrast, as expected, neither gene was differentially expressed between positively and negatively selected monocytes (SPI1, p=0.25; CD14, p=0.28). Therefore, the pattern of expression of these monocyte markers initially identified using microarray data was confirmed by ddPCR and reaffirms that positively selected CD4+ T cells were contaminated with monocytes. Reassuringly, the expression of these monocyte markers was significantly higher in monocytes themselves compared to CD4+ T cells contaminated with monocytes (SPI1, p=7.94E-3; CD14, p=3.84E-6, Fig 5).
Figure 5. Gene expression validation by droplet digital PCR.
Number of molecules of each target mRNA was normalized to the number of molecules of the normalizer mRNA (i.e., RPL27) in the 5 ng of total RNA. The resulting values were then used to calculate the number of each mRNA per one million molecules of RPL27. The data was log2 transformed and analyzed using a RM ANOVA with post hoc Tukey test. Expression of SPI1 and CD14 was compared between CD4+ T cells and monocytes isolated by positive and negative selection. All the between-group comparisons were significant expect comparing monocytes isolated by positive selection to monocytes isolated by negative selection. For display purposes, only three p-values are presented. Expression of SDPR in monocytes was compared between all three isolation methods. Error bars represent standard deviation of the mean calculated for normalized mRNA expression values for the 5 donors.
SDPR is a platelet specific gene (33) and was selected to confirm that negatively selected monocytes were contaminated with platelets. Since ddPCR requires only a very small amount of RNA (i.e., 5 ng) compared to microarrays, monocyte samples isolated by FACS for all five donors could be analyzed. SDPR was expressed at significantly higher levels in monocytes isolated by negative selection compared to the other two methods (i.e., positive selection and FACS) (Fig. 5, bottom). In addition, SPDR was expressed at a significantly higher level in monocytes isolated by positive selection compared to FACS. Therefore, this ddPCR data confirms that negatively isolated monocytes were contaminated with platelets but extends the microarray findings by identifying platelet contamination in positively selected monocytes as well. Although monocyte yields are low with FACS, this method appears superior when pure populations of monocytes are required.
DISCUSSION
This study aimed to identify the best method to isolate blood cell subtypes when the gene expression signature reflective of the disease state or therapeutic intervention at time of sampling is of interest. The performance of three isolation methods (i.e., FACS, and positive and negative immunomagnetic selection) and their effect on gene expression in four blood cell types (i.e., CD4+, CD8+ T cells, B cells and monocytes) was compared. While positive and negative selection of CD4+ T cells, CD8+ T cells and monocytes have been compared previously (19), the effects of FACS on gene expression was not previously investigated. FACS was of particular interest since this method allows the separation of multiple cell types from the same sample. While FACS is known to result in high cell purity, we hypothesized that this method would be similar in gene expression signature to positive selection because of the use of antibodies to bind cell receptors. Receptor cross-linking resulted in T cell activation (14-16) and inhibition of interferon-α production by plasmacytoid dendritic cells in response to viral stimulation (18). Furthermore, it was hypothesized that genes related to the stress response would be modulated due to the high pressure exerted on cells during sorting.
Cell purity measurements indicated that FACS resulted in the purest isolation for each cell type. With respect to the other isolation methods analyzed here, Lyons et al. (19) previously reported that cells isolated by negative selection were contaminated to a greater extent than cells isolated by positive selection. Specifically, CD4+ T cells were contaminated with CD3- as well as CD3+CD4- cells, monocytes were contaminated with CD14- cells some of which were CD4+and CD8+ T cells were contaminated with CD3- as well as CD3+CD8- cells. Inversely, in the current study, positively selected cells of a particular subtype had the highest contamination when combining percentages of all contaminating cell types (CD4+, CD8+ T, B cells and monocytes) compared to the same cells isolated by negative selection (Fig. 1). In case of positive selection each cell subset was contaminated by multiple different cell types except for monocytes which were contaminated with cells not stained in our experiment and denoted as “other”. All cell types isolated by negative selection were contaminated with “other” cells and not as much with cells that were stained in our experiment (CD4+, CD8+ T, B cells and monocytes). These discrepancies with results reported by Lyons et al. (19) may be explained by usage of different amounts of cells and isolation kits that were produced by different manufacturers (Miltenyi Biotec in Lyons et al. (19) vs StemCell Technologies in this study) and differed in magnetic bead size and antibody cocktail composition, as well as subtle experiment-to-experiment and donor-to-donor differences.
Besides contamination with other types of PBMCs, monocyte samples isolated by negative selection had considerably higher percentages of platelets than monocyte samples isolated by positive selection. This was best reflected by ddPCR result of the expression of a platelet-specific gene SDPR which had higher expression in negatively selected compared to positively selected monocytes (Fig. 5). Similar observations related to greater contamination of negatively isolated monocytes with platelets compared to positively isolated monocytes was noted in a recent study by Zhou and colleagues that compared isolation methods of monocytes and granulocytes for purity and functional assays (36). Additional washes following isolation of PBMC or cell subsets following incubation with antibodies or the addition of anti-CD41 antibodies (37) may help remove more platelets with the caveat of additional cell loss.
To investigate the effects of isolation methods and contaminating cell subsets on gene expression in isolated cells, a microarray analysis was performed. In comparison to the study performed by Lyons et al. (19), an attempt was made to segregate the effects of isolation methods on gene expression from the effects of contamination with other cell subsets. This was best exemplified by CD8+ T cells and B cells. In case of B cells, based on unsupervised clustering (Fig. S3A) and PCA (Fig. 3) analyses, the identified DEGs were more likely reflective of the effect of isolation methods as evidenced by wider spread among B cells samples along principal component 2 (isolation method, Y axis). CD8+ T cells were affected by contamination or isolation method the least (samples not widely spread along either principal component), with the fewest DEGs (N=164) identified (Table S2B). CD4+ T cells appeared to be more prone to the effect of contamination based on the spread of CD4+ T cells samples along principal component 1 (cell type, X axis). In addition, the number of DEGs identified (N=2,417) was an order of magnitude higher compared to the other three cell types investigated (Fig. S2). In particular, monocyte-specific DEGs (e.g., SPI1 and CD14) had higher expression values in CD4+ T cells isolated by positive selection compared to the other two methods, as detected by ddPCR (Fig. 5). The data obtained for CD4+ T cells suggest that the effect of contamination dominates for this cell type and obscures the identification of DEGs that are affected by isolation method. In the case of monocytes, the effects of contamination or isolation method were not obvious based on the tight cluster of monocyte samples on the PCA plot (Fig. 3). However, DEGs were identified and a platelets-related GO term was enriched for DEGs. Since platelets have very small amount of RNA, the remaining DEGs were likely the effect of isolation methods.
One of the primary goals of the current study was to assess the effect of FACS on gene expression, particularly with respect to genes related to stress response. When DEGs identified between isolation methods for each cell type were mapped to gene ontologies, the term Response to stress was significantly enriched for DEGs in case of CD4+ T cells, B cells and monocytes, but not CD8+ T cells. Among the children terms of Response to stress, Defense Response and Response to wounding contained the majority of genes that mapped to this term. Identification of these terms may point to response to any of the exogenous stress such as temperature, which would be the case in our experiment due to varying times of cell incubation at room temperature vs on ice during the isolation procedure. However, Response to wounding has children terms that relate to blood coagulation and platelet activation, platelet contamination may be the reason of its enrichment by DEGs in our experiment at least in case of monocytes isolated by negative selection. Overall gene expression signature in CD4+ and CD8+ T cells, for which all three methods were compared, was similar between FACS and negative selection compared to positive selection (Fig. 4). In case of positively isolated CD4+ T cells it could be explained by monocyte contamination while in case of CD8+ T cells it appeared to be cell contamination independent and was likely due to the effect of isolation method.
Our data suggests based on purity and gene expression results that FACS is the optimal method for PBMC subset isolation for gene expression profiling studies when blood samples from patients are used and the gene expression signature inherent to a disease or therapeutic treatment is desired. FACS allows isolation of multiple cell subsets from the same sample and exclusion of platelets and dead cells. Our findings suggest the use of FACS has little impact on cellular transcription and allows capturing gene expression signature directly relevant to a disease or the effect of a medication. Specifically, none of the stress-related genes common across cell types (N=7) identified were upregulated in samples isolated by FACS. Monocytes may represent one exception from this general recommendation since low yields were obtained by FACS when starting with ten million PBMCs. Starting with higher cell numbers or using positive selection would provide superior monocyte yields. Our experiments did not focus on long-term effects of isolation methods on gene expression, therefore, future studies will be required to determine the optimal isolation method if the isolated cells are intended for long-term in vitro experiments. In summary, this study demonstrated that both cell contamination and isolation method affect gene expression when isolating a number of different cell subsets (CD4+ T cells, CD8+ T cells, B cells and monocytes) using positive and negative immunomagnetic selection, and FACS.
Supplementary Material
ACKNOWLEDGEMENTS
This work was primarily supported by an R01 grant to CHW from the NIH (AI087164). These studies were also performed using equipment provided by the Pendleton Trust, and with the support of the Genomics Core and Flow Cytometry Core at the UCSD Center for AIDS Research (AI36214). MM was supported by Agència de Gestió d'Ajuts Universitaris i de Recerca from Generalitat de Catalunya and European Social Fund. This material is based upon work supported in part by the Department of Veterans Affairs (VA), Veterans Health Administration, Office of Research and Development. The views expressed in this article are those of the authors and do not necessarily reflect the position or policy of the Department of Veterans Affairs or the United States government. We thank Dr. Rachel Schrier for her suggestions on the strategy of the cell sorting.
Footnotes
Authors declare no conflict of interest.
LITERATURE CITED
- 1.Reis-Filho JS, Pusztai L. Gene expression profiling in breast cancer: classification, prognostication, and prediction. The Lancet. 2011;378:1812–1823. doi: 10.1016/S0140-6736(11)61539-0. [DOI] [PubMed] [Google Scholar]
- 2.Campbell JD, Spira A, Lenburg ME. Applying gene expression microarrays to pulmonary disease. Respirology. 2011;16:407–418. doi: 10.1111/j.1440-1843.2011.01942.x. [DOI] [PubMed] [Google Scholar]
- 3.Bates S. The role of gene expression profiling in drug discovery. Current Opinion in Pharmacology. 2011;11:549–556. doi: 10.1016/j.coph.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 4.Woelk C, Singhania A, Pérez-Santiago J, Glatt S, Tsuang M. The utility of gene expression in blood cells for diagnosing neuropsychiatric disorders. Int Rev Neurobiol. 2011;101:41–63. doi: 10.1016/B978-0-12-387718-5.00003-1. [DOI] [PubMed] [Google Scholar]
- 5.Woelk CH, Burczynski ME. The clinical relevance of gene expression profiles in peripheral blood mononuclear cells. In: Columbus F, editor. Oligonucleotide Array Sequence Analysis. NOVA publishing; Hauppage, NY: 2008. pp. 38–50. [Google Scholar]
- 6.Felger JC, Cole SW, Pace TWW, Hu F, Woolwine BJ, Doho GH, Raison CL, Miller AH. Molecular signatures of peripheral blood mononuclear cells during chronic interferon-α treatment: relationship with depression and fatigue. Psychological Medicine. 2011:1–13. doi: 10.1017/S0033291711002868. FirstView. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Montano M, Rarick M, Sebastiani P, Brinkmann P, Russell M, Navis A, Wester C, Thior I, Essex M. Gene-expression profiling of HIV-1 infection and perinatal transmission in Botswana. Genes Immun. 2006;7:298–309. doi: 10.1038/sj.gene.6364297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lambotte O, Taoufik Y, Goër MGd, Wallon C, Goujard C, Delfraissy JF. Detection of Infectious HIV in Circulating Monocytes From Patients on Prolonged Highly Active Antiretroviral Therapy. JAIDS Journal of Acquired Immune Deficiency Syndromes. 2000;23:114–119. doi: 10.1097/00126334-200002010-00002. [DOI] [PubMed] [Google Scholar]
- 9.Sonza S, Mutimer HP, Oelrichs R, Jardine D, Harvey K, Dunne A, Purcell DF, Birch C, Crowe SM. Monocytes harbour replication-competent, non-latent HIV-1 in patients on highly active antiretroviral therapy. AIDS. 2001;15:17–22. doi: 10.1097/00002030-200101050-00005. [DOI] [PubMed] [Google Scholar]
- 10.Kedzierska K, Crowe SM. The role of monocytes and macrophages in the pathogenesis of HIV-1 infection. Curr. Med. Chem. 2002;9:1893–903. doi: 10.2174/0929867023368935. [DOI] [PubMed] [Google Scholar]
- 11.Zhu T. HIV-1 in peripheral blood monocytes: an underrated viral source. Journal of Antimicrobial Chemotherapy. 2002;50:309–311. doi: 10.1093/jac/dkf143. [DOI] [PubMed] [Google Scholar]
- 12.Bennett L, Palucka AK, Arce E, Cantrell V, Borvak J, Banchereau J, Pascual V. Interferon and granulopoiesis signatures in systemic lupus erythematosus blood. J Exp Med. 2003;197:711–23. doi: 10.1084/jem.20021553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stanciu LA, Shute J, Holgate ST, Djukanović R. Production of IL-8 and IL-4 by positively and negatively selected CD4+ and CD8+ human T cells following a four-step cell separation method including magnetic cell sorting ( MACS). Journal of Immunological Methods. 1996;189:107–115. doi: 10.1016/0022-1759(95)00240-5. [DOI] [PubMed] [Google Scholar]
- 14.Ledbetter JA, June CH, Grosmaire LS, Rabinovitch PS. Crosslinking of surface antigens causes mobilization of intracellular ionized calcium in T lynphocytes. Immunology. 1987;84:1384–1388. doi: 10.1073/pnas.84.5.1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dixon JF, Law JL, Favero JJ. Activation of human T lymphocytes by crosslinking of anti-CD3 monoclonal antibodies. Journal of Leukocyte Biology. 1989;46:214–20. doi: 10.1002/jlb.46.3.214. [DOI] [PubMed] [Google Scholar]
- 16.Tsoukas CD, Landgraf B, Bentin J, Valentine M, Lotz M, Vaughan JH, Carson DA. Activation of resting T lymphocytes by anti-CD3 (T3) antibodies in the absence of monocytes. The Journal of Immunology. 1985;135:1719–23. [PubMed] [Google Scholar]
- 17.Elkord E, Williams PE, Kynaston H, Rowbottom AW. Human monocyte isolation methods influence cytokine production from in vitro generated dendritic cells. Immunology. 2005;114:204–212. doi: 10.1111/j.1365-2567.2004.02076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fanning SL, George TC, Feng D, Feldman SB, Megjugorac NJ, Izaguirre AG, Fitzgerald-Bocarsly P. Receptor Cross-Linking on Human Plasmacytoid Dendritic Cells Leads to the Regulation of IFN-α Production. The Journal of Immunology. 2006;177:5829–5839. doi: 10.4049/jimmunol.177.9.5829. [DOI] [PubMed] [Google Scholar]
- 19.Lyons PA, Koukoulaki M, Hatton A, Doggett K, Woffendin HB, Chaudhry AN, Smith KGC. Microarray analysis of human leukocyte subsets: the advantages of positive selection and rapid purification. BMC Genomics. 2007:8. doi: 10.1186/1471-2164-8-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miltenyi S, Muller W, Weichel W, Radbruch A. High gradient magnetic cell separation with MACS. Cytometry. 1990;11:231–238. doi: 10.1002/cyto.990110203. [DOI] [PubMed] [Google Scholar]
- 21.Bonner WA, Hulett HR, Sweet RG, Herzenberg LA. Fluorescnece activated cell sorting. Rev. Sci. Instrum. 1972;43:404–409. doi: 10.1063/1.1685647. [DOI] [PubMed] [Google Scholar]
- 22.Basu S, Campbell HM, Dittel BN, Ray A. Purification of Specific Cell Population by Fluorescence Activated Cell Sorting (FACS). J Vis Exp. 2010:e1546. doi: 10.3791/1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bolstad BM, Collin F, Brettschneider J, Simpson K, Cope L, Irizarry R, Speed TP. Quality Assessment of Affymetrix GeneChip Data. In: Gentleman R, Carey V, Huber W, Irizarry R, Dutoit S, editors. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Springer; Heidelberg: 2005. pp. 33–47. [Google Scholar]
- 24.Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–8. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 25.Gentleman RC, VJ, Huber W, Hahne F. Genefilter: Methods for filtering genes from microarray experiments. R package: Version 1.24.2. 2009 [Google Scholar]
- 26.Suzuki R, Shimodaira H. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics. 2006;22:1540–1542. doi: 10.1093/bioinformatics/btl117. [DOI] [PubMed] [Google Scholar]
- 27.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy Stat Soc, B MET. 1995;57:289–300. [Google Scholar]
- 28.Simon R, Lam A, Li MC, Ngan M, Menenzes S, Zhao Y. Analysis of gene expression data using BRB-Array Tools. Cancer Informatics. 2007;3:11–17. [PMC free article] [PubMed] [Google Scholar]
- 29.Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of Gene Ontology categories in Biological Networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 30.De Jonge HJ, Fehrmann RS, de Bont ES, Hofstra RM, Gerbens F, Kamps WA, de Vries EG, van der Zee AG, te Meerman GJ, ter Elst A. Evidence based selection of housekeeping genes. PLoS ONE. 2007;2:e898. doi: 10.1371/journal.pone.0000898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Methia N, Louache F, Vainchenker W, Wendling F. Oligodeoxynucleotides antisense to the proto-oncogene c-mpl specifically inhibit in vitro megakaryocytopoiesis. Blood. 1993;82:1395–1401. [PubMed] [Google Scholar]
- 32.Gagnon AW, Murray DL, Leadley RJ., Jr Cloning and characterization of a novel regulator of G protein signalling in human platelets. Cellular Signalling. 2002;14:595–606. doi: 10.1016/s0898-6568(02)00012-8. [DOI] [PubMed] [Google Scholar]
- 33.Burgener R, Wolf M, Ganz T, Baggiolini M. Purification and characterization of a major phosphatydilserine-binding phosphoprotein from human platelets. Biochem J. 1990;269:729–734. doi: 10.1042/bj2690729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yagi M, Zieger B, Roth GJ, Ware J. Structure and expression of the human septin gene HCDCREL-1. Gene. 1998;212:229–236. doi: 10.1016/s0378-1119(98)00146-2. [DOI] [PubMed] [Google Scholar]
- 35.McFarland DC, Zhang C, Thomas HC, Tl R. Confounding effects of platelets on flow cytometric analysis and cell-sorting experiments using blood-derived cells. Cytometry Part A. 2006;69A:86–94. doi: 10.1002/cyto.a.20207. [DOI] [PubMed] [Google Scholar]
- 36.Zhou L, Somasundaram R, Nederhof RF, Dijkstra G, Faber KN, Peppelenbosch MP, Fuhler GM. Impact of human granulocyte and monocyte isolation procedures on functional studies. Clinical Vaccine and Immunology. 2012;19:1065. doi: 10.1128/CVI.05715-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coma G, Peña R, Blanco J, Rosell A, Borras FE, Esté JA, Clotet B, Ruiz L, Parkhouse RME, Bofill M. Treatment of monocytes with interleukin (IL)-12 plus IL-18 stimulates survival, differentiation and the production of CXC chemokine ligands (CXCL)8, CXCL9 and CXCL10. Clin Exp Immunol. 2006;145:535–544. doi: 10.1111/j.1365-2249.2006.03145.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.