Abstract
Yeast are the foremost genetic model system. With relative ease, entire chemical libraries can be screened for effects on essentially every gene in the yeast genome. Until recently, researchers focused only on whether yeast were killed by the conditions applied, irrespective of the mechanisms by which they died. In contrast, considerable effort has been devoted to understanding the mechanisms of mammalian cell death. However, most of the methodologies for detecting programmed apoptotic and necrotic death of mammalian cells have not been applicable to yeast. Therefore, we developed a cell death assay for baker’s yeast Saccharomyces cerevisiae to identify genes involved in the mechanisms of yeast cell death. Small volumes of yeast suspensions are subjected to a precisely controlled heat ramp, allowing sufficient time for yeast cell factors to suppress or facilitate death, which can be quantified by high-throughput automated analyses. This assay produces remarkably reliable results that typically reflect results with other death stimuli. Here we describe the protocol and its caveats, which can be easily overcome.
Keywords: Yeast, Cell death, Heat ramp, Necrosis, Apoptosis
1 Introduction
The fact that yeast lack the canonical apoptosis machinery of meta-zoans led to widespread skepticism concerning the existence of programmed cell death in yeast. However, animal cells also undergo non-apoptotic cell death mechanisms ending in cell morphologies that are generally lumped into the broad category of necrosis [1–3]. This raised the possibility that alternative non-apoptotic cell death programs could have arisen in unicellular species [4–11]. When considering how and why programmed cell suicide arose during evolution, there are a number of compelling arguments to support the model that programmed death arose in unicellular species, and that programmed cell death in multicellular species may stem from these earlier versions [12]. The ability of unicellular species to control infectious pathogens [13–15], to nurture younger cells [16], to adjust their population size in response to nutrient deprivation before supplies are completely consumed [17, 18], to respond to other environmental stresses [19–23], and potentially to stratify functional cell layers [24] are all consistent with the evolution of an inducible program of cell suicide that ensures the survival of the species (rather than the individual). The identification of yeast cell death genes that are also involved in other cellular processes such as actin dynamics [25–28], the mating process [7], histone modification [29, 30], aging [31, 32], mRNA stability [33, 34], DNA damage response [23, 35], and mitochondrial dynamics and bioenergetics [21, 36, 37] suggests that cell death could be linked to many cellular processes gone awry.
Delineation of phylogenetically conserved death pathways in yeast may be revealing about non-apoptotic mechanisms in mammals, or could lead to novel strategies for controlling pathogenic yeast infections. However, these pathways in yeast have not been extensively explored as programmed death mechanisms, partially due to the lack of suitable cell death assays. We developed an effective yeast cell death assay by applying principles from mammalian models to distinguish cell death that occurs by an active process rather than by direct assault [38]. For consistent results, we apply a precisely controlled death stimulus that uniformly treats all cells in the sample simultaneously. Assay parameters can be easily adjusted to accommodate different metabolic states and strain backgrounds [38]. High-throughput BioSpot counting technology with image recognition software adds additional power to this assay. The BioSpot Analyzer counts up to 500 microscopic colonies per spot in a 96-place format. This strategy reduces plate incubation times and avoids errors inherent to serial dilutions required for visual inspection of minimally merged colonies [38]. Here we provide a detailed protocol for the heat ramp yeast cell death assay.
2 Materials
2.1 Yeast Cultures and Heat Ramp Treatment
Yeast strains: Wild-type BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) and knockout strains from the BY MATa YKO collection (see Note 1).
YPD liquid medium: 2 % peptone (Peptone-Y, MP Biomedicals), 1 % yeast extract (Fisher Scientific), and 2 % glucose (J.T. Baker).
Roller drum (TC-7, New Brunswick Scientific).
Glass culture tubes (18 × 150 mm) with loose-fitting caps.
0.2 ml PCR tubes.
Thermocycler/PCR machine with a narrow temperature variance and that can be programmed to slowly ramp/step up the temperature (e.g., Mastercycler gradient, Eppendorf).
2.2 Colony Forming Assay
96-well microplates (V-bottom, 200 ml).
Omni trays (128 × 86 cm).
YPD agar plates: Liquid YPD with 2 % agar (BD).
12-channel micropipette.
2.3 FUN1 Staining Viability Assay
Live/Dead Yeast Viability Kit (Molecular Probes) containing FUN1 cell stain [10 mM solution in anhydrous dimethylsulfoxide (DMSO)] and Calcofluor White M2R (5 mM solution in water).
Fluorescence microscope (Nikon Eclipse E800, 100× objective lens, multipass filter sets appropriate for viewing DAPI/ fluorescein/rhodamine).
Micro slides (25 × 75 × 1 mm) and micro cover glass (18 × 18 mm) (VWR Scientific).
2.4 High-Throughput Heat Ramp Assay
96-pin tool (96 Pin Replicator Model 140500, Boekel Scientific).
96-well microplates (flat-bottom, 200 ml).
Plate reader (Kinetic microplate reader, Molecular Devices).
96-well PCR tubes (TempPlate PCR plate, 0.2 ml thin-wall standard wells, USA Scientific).
BioSpot S5 Micro Analyzer (CTL/Cellular Technology Limited).
BioSpot Academic Software Version 5.0 (CTL).
3 Methods
3.1 Low-Throughput Heat Ramp Cell Death Assay
WT and mutant yeast strains are streaked out from frozen −80 °C glycerol stocks (without thawing) onto YPD agar plates and incubated at 30 °C for ~2 days.
Single colonies are transferred from the plate with a sterile implement into 2 ml YPD in yeast culture tubes (see Note 2). Cultures are incubated at 30 °C on a roller drum (~35 rotations/min) for the times specified below to test yeast in log phase and post-diauxic growth phases.
For mid-log-phase yeast, each overnight culture is diluted to OD600 = 0.20 in fresh YPD (with at least 1 ml more than the amount required for experiments), and further incubated at 30 °C until mid-log phase, OD600 = ≤0.5. Before treatment, all cultures are readjusted to the same OD600 (for example the OD600 of the least dense culture) at 30 min before reaching OD600 = 0.5 by diluting the slightly overgrown samples with YPD, and then continue incubation until all samples reach the same density simultaneously, OD600 = ~0.5. Further volume adjustments may be necessary to make all samples equal in cell density before treatment (see Note 3).
100 μl of each mid-log-phase culture (OD600 = ~0.5) are transferred in 0.2 ml PCR tubes and incubated in a thermocycler/PCR machine programmed to ramp the temperature from RT to 30 °C immediately, held at 30 °C for 1 min, ramped from 30 to 40 °C in 2 min, then ramped from 40 to 51 °C in 10 min, and held at 51 °C for 10 min (see Note 4). The remaining untreated OD-adjusted cultures are plated as controls to verify equal cell numbers prior to treatment.
For post-diauxic cultures, the 2 ml YPD yeast culture tubes are incubated at 30 °C on a roller drum continuously for 16 or 48 h.
The 16- or 48-h cultures are directly diluted to OD600 = 0.5 in fresh YPD, and 100 μl of each culture are transferred in 0.2 ml PCR tubes and treated in the thermocycler with appropriate parameters. For 16-h cultures, the temperature is ramped from RT to 30 °C immediately, held at 30 °C for 1 min, then ramped from 30 to 55 °C in 10 min, and held at 55 °C for 5 min. For 48-h cultures, the temperature is ramped from RT to 30 °C immediately, held at 30 °C for 1 min, and then ramped from 30 to 62 °C in 20 min without holding (see Note 5). The remainder of each diluted culture is plated as untreated control. Dilutions may be required (e.g., 1:5,000) to accurately compare strains.
3.2 Viability Measured by Colony-Forming Units for Low-Throughput Assays
For both the untreated controls and the heat ramp-treated aliquots, 100 μl of each culture (previously adjusted to OD600 = 0.5) are transferred to a 96-well microplate and five 1:5 serial dilutions are performed using a 12-channel pipette (20 μl culture is added to 80 μl ddH2O and mixed well by pipetting 5–6 times) (see Note 6). 5 μl from each dilution are plated (highest to lowest) onto YPD agar plates.
Agar plates are incubated at 30 °C for 2 days to visualize colonies. Colonies within the plated spots containing countable numbers (less than 50) can be counted by hand, or plates can be counted at earlier times with the BioSpot Analyzer (see below). Final colony-forming units (CFUs) are calculated by correcting for the dilution factor.
3.3 Viability Measured by FUN1 Staining
Before heat ramp treatment (see Note 7), 0.5 μl FUN1 cell stain and 5 μl Calcofluor White M2R are added to 1 ml yeast culture (OD600 adjusted) with final concentrations of 5 and 25 μM, respectively.
100 μl of each culture is treated with milder heat ramp conditions (e.g., ramp the temperature from RT to 30 °C immediately, held at 30 °C for 1 min, ramped from 30 to 51 °C in 30 min, and held at 51 °C for 10 min) (see Note 8). After heat ramp treatment, yeast cells are pelleted and the supernatant is removed except for ~50 μl. Yeast cell pellets are resuspended and 1.5 μl of stained yeast suspension is trapped between a slide and cover glass for observation by fluorescence microscopy.
Dead cells stain diffusely green, while healthy cells stain blue and contain a red bar inside. At least 500 cells are counted from representative fields (see Note 8).
3.4 High-Throughput Heat Ramp Cell Death Assay
Frozen stocks in 96-well format of yeast strains [e.g., the yeast knockout (YKO) collections] are pinned (without thawing) onto square YPD agar plates using a 96-pin tool, and grown at 30 °C for 2 days.
Yeast from these solid cultures are transferred into 200 μl liquid YPD using the 96-pin tool and incubated at 30 °C for 48 h to saturation. OD600 of each well on the plate are determined with the plate reader (see Note 9).
Saturated cultures are diluted directly to OD600 = ~0.5 in fresh YPD, and 100 μl from each diluted/mixed sample are immediately transferred to 96-well PCR tubes using a 12-channel pipette for treatment in the thermocycler (see Note 6). Ramp the temperature from 30 to 62 °C over 20 min without holding at maximum temperature.
Following treatment, 5 μl of each sample (undiluted and a 1:5 dilution) are spotted on YPD agar plates in four replicates with a 12-channel pipette (see Note 10).
Plates are incubated at 30 °C for about 18 h and analyzed using the BioSpot Reader (see Note 11).
3.5 Automated High-Throughput Colony Counting
Images of each YPD agar plate with yeast colonies are captured by the BioSpot Analyzer. Plate position is manually adjusted to ensure that all spots are within the camera field. Highest resolution settings (1,024 by 768 pixels) are chosen for all the plate images taken.
The BioSpot Analyzer collects colony counts as well as other information such as the density of yeast growth, which can be useful for other studies. However, for accurate assessment of cell death, only cell counts and not cell density are accurate measures of clonogenic cell viability (Fig. 1).
Colony counting is performed using the BioSpot counting software. Plate images are counted using the parameters in Table 1 (see Note 12), but images can be recounted with alternative parameters. Data can be extracted as both Text and Excel files.
Fig. 1.
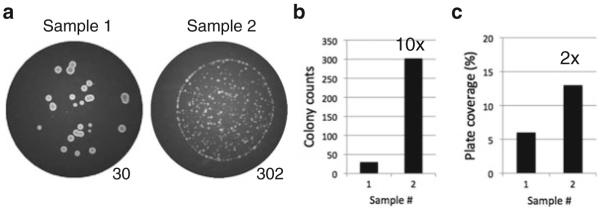
Colony number rather than growth density accurately reflects clonogenic survival. (a) BioSpot images of two wells from a 96-well plate revealing strain-dependent and/or colony density-dependent differences in colony size. Machine colony counts are indicated. (b) Survival difference between samples based on colony counts is tenfold. (c) Survival difference between samples based on growth density is inaccurately estimated at twofold, analogous to colony-forming clonogenic survival assays for mammalian cells
Table 1.
BioSpot analyzer colony counting parameters
| Parameter | Setting |
|---|---|
| Sensitivity | 216 |
| Background balance | 40 |
| Spot separation | 8 |
| Diffuseness | Large |
| Min–max spot size | 0.0001–9.6466 mm 2 |
| Fill holes | Off |
| Hair removal | On |
| Counting mask size (%) | 98, normalization off |
Fig. 2.
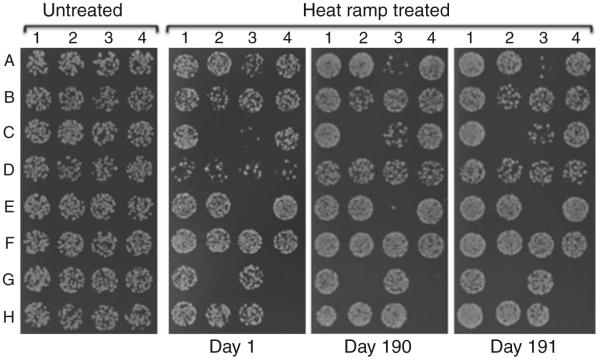
Heat ramp cell death assay results are highly reproducible. Results from three independent experiments, each initiated from the same frozen strains. Daily variation is likely due to differences in the precise timing, differences in specific lots of media components, or other trivial explanation, as the relative differences between strains are uniform, with the potential exception of strain C-3
Acknowledgment
This work was supported by NIH grants GM077875 and NS037402 (J.M.H.).
4 Notes
This cell death assay can be readily adapted for other yeast strains by adjusting the intensity of the death stimulus. Strains grown on minimal media and those bearing plasmids are generally more sensitive to cell death, requiring parallel strains bearing empty plasmid vectors as controls. Diploid strains and older haploid strains (passaged multiple times) can be much more sensitive to cell death than newly sporulated strains of the same. However, using the same conditions yields highly reproducible results in independent experiments (Fig. 2).
Different results obtained with different colonies from the same strain could indicate genetic variation and alternative approaches are required to evaluate the true phenotype. To determine if genetic variability is a possibility, prepare frozen stocks of overnight cultures for each of the original colonies picked for analysis, and repeat the cell death assay on these archived substrains to determine if their cell death phenotypes are stable.
Differences in maximum OD of individual cultures prior to treatment can influence outcome, but can be avoided by not allowing cultures to reach OD600 = ~0.6.
The key to inducing programmed cell death in yeast is to set up sufficiently mild conditions to allow time for gene-dependent death to occur [38]. Ramping from ambient to maximum temperature over a specified period accomplishes this goal. The heat ramp conditions listed here have been optimized, but the ramp rate(s), maximum temperature, and holding time can be adjusted with relative latitude to accommodate different metabolic states, background strains, media types, and culture densities. Cell death sensitivity can differ for individual strains depending on their metabolic state. Heat ramp-induced cell death usually results in similar phenotypes when compared to unrelated death stimuli [38].
The yeast cell death phenotype is highly dependent on metabolic state. Post-diauxic cultures grown over 1 night (~16 h) or over 2 nights (~48 h) are far more resistant to heat ramp-induced death compared to mid-log-phase cultures and require higher maximum temperatures. However, single heat ramp conditions can simultaneously distinguish death-sensitive and death-resistant strains from WT grown under the same conditions [38].
Delays between sample dilution and heat ramp treatment, or between treatment and plating, can affect outcomes.
Unlike other yeast cell death stimuli such as acetic acid treatment for which the FUN1 dye is usually added after the death stimulus [39], the dye must be added before the death stimulus in the heat ramp assay, because heat treatment impairs uptake of the dye.
Absolute values for cell viabilities obtained with vital dyes such as FUN1 are proportionately higher than the corresponding CFUs for the same samples, apparently because not all cells have died at the earlier time after treatment when staining for vital dyes is evaluated. Lower heat ramp doses may be needed to obtain viabilities in the linear (rather than log) scale.
One critical parameter is to standardize the starting number of cells prior to treatment. Although untreated cells could be plated and counted, the increased workload and extensive dilutions that themselves introduce inaccuracies were found to exceed beneficial output. To maximally equalize cell number for high-throughput assays, 48-h cultures are used to allow yeast to grow to saturation. To minimize day-to-day variation, exact 48-h incubation time should be performed.
The BioSpot Analyzer (BioSpot™ S5 Micro Analyzer, CTL) automatically captures plate images in a 96-place format with fixed magnification. To ensure that each spot of yeast growth is within the field of the camera lens, all the spots need to be aligned uniformly. A template with uniformly aligned 96 spots can be put underneath the agar plate for guidance. In our experience, pinning tools cannot deliver sufficiently accurate volumes of liquid, or of cells in suspension to agar plates. Agar surface tensions and plastic plate types can affect plating accuracies.
For visual enumeration of colony number, yeast are typically grown on agar plates for 2 days. The BioSpot Analyzer has much higher resolution than human acuity, distinguishing objects of 25 μm in diameter, thereby reducing the required yeast growth time to only 18 h [38]. This capability allows accurate counts of 300–500 colonies per spot while reducing colony overlap and limiting the number of error-prone dilution steps required before plating.
The most important counting parameter settings on the BioSpot Analyzer are “sensitivity” and “background balance,” which adjust the sensitivity required to distinguish real object signals from background noise. Higher “sensitivity” settings result in higher colony counts, but also increase false positives. Lower “background balance” settings accordingly reduce noise, but can also fail to detect real colonies. Therefore, a balance needs to be achieved between signal sensitivity and background noise. Machine counting parameters are customized by visual inspection of the counted plate images. Here, a high sensitivity setting of 216 enables small colonies to be recognized as real subject, while the low background balance setting of 40 minimizes the noise effect. The contrast of plate images is highly dependent on the thickness of the agar plates. To minimize plate-to-plate and spot-to-spot variation, plates should be poured with equal volumes of media on a surface adjusted with a leveler for best results.
References
- 1.Degterev A, Hitomi J, Germscheid M, Ch’en IL, Korkina O, Teng X, Abbott D, Cuny GD, Yuan C, Wagner G, Hedrick SM, Gerber SA, Lugovskoy A, Yuan J. Identification of RIP1 kinase as a specific cellular target of necrostatins. Nat Chem Biol. 2008;4:313–321. doi: 10.1038/nchembio.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vanlangenakker N, Berghe TV, Krysko DV, Festjens N, Vandenabeele P. Molecular mechanisms and pathophysiology of necrotic cell death. Curr Mol Med. 2008;8:207–220. doi: 10.2174/156652408784221306. [DOI] [PubMed] [Google Scholar]
- 3.Kirkegaard T, Jaattela M. Lysosomal involvement in cell death and cancer. Biochim Biophys Acta. 2009;1793:746–754. doi: 10.1016/j.bbamcr.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 4.Jin C, Reed JC. Yeast and apoptosis. Nat Rev Mol Cell Biol. 2002;3:453–459. doi: 10.1038/nrm832. [DOI] [PubMed] [Google Scholar]
- 5.Gourlay CW, Du W, Ayscough KR. Apoptosis in yeast–mechanisms and benefits to a unicellular organism. Mol Microbiol. 2006;62:1515–1521. doi: 10.1111/j.1365-2958.2006.05486.x. [DOI] [PubMed] [Google Scholar]
- 6.Frohlich KU, Fussi H, Ruckenstuhl C. Yeast apoptosis–from genes to pathways. Semin Cancer Biol. 2007;17:112–121. doi: 10.1016/j.semcancer.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 7.Zhang NN, Dudgeon DD, Paliwal S, Levchenko A, Grote E, Cunningham KW. Multiple signaling pathways regulate yeast cell death during the response to mating pheromones. Mol Biol Cell. 2006;17:3409–3422. doi: 10.1091/mbc.E06-03-0177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng WC, Teng X, Park HK, Tucker CM, Dunham MJ, Hardwick JM. Fis1 deficiency selects for compensatory mutations responsible for cell death and growth control defects. Cell Death Differ. 2008;15:1838–1846. doi: 10.1038/cdd.2008.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abdelwahid E, Rolland S, Teng X, Conradt B, Hardwick JM, White K. Mitochondrial involvement in cell death of non-mammalian eukaryotes. Biochim Biophys Acta. 2011;1813:597–607. doi: 10.1016/j.bbamcr.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng WC, Leach KM, Hardwick JM. Mitochondrial death pathways in yeast and mammalian cells. Biochim Biophys Acta. 2008;1783:1272–1279. doi: 10.1016/j.bbamcr.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vachova L, Palkova Z. Caspases in yeast apoptosis-like death: facts and artefacts. FEMS Yeast Res. 2007;7:12–21. doi: 10.1111/j.1567-1364.2006.00137.x. [DOI] [PubMed] [Google Scholar]
- 12.Ameisen JC. On the origin, evolution, and nature of programmed cell death: a time-line of four billion years. Cell Death Differ. 2002;9:367–393. doi: 10.1038/sj.cdd.4400950. [DOI] [PubMed] [Google Scholar]
- 13.Ivanovska I, Hardwick JM. Viruses activate a genetically conserved cell death pathway in a unicellular organism. J Cell Biol. 2005;170:391–399. doi: 10.1083/jcb.200503069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reiter J, Herker E, Madeo F, Schmitt MJ. Viral killer toxins induce caspase-mediated apoptosis in yeast. J Cell Biol. 2005;168:353–358. doi: 10.1083/jcb.200408071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmitt MJ, Reiter J. Viral induced yeast apoptosis. Biochim Biophys Acta. 2008;1783:1413–1417. doi: 10.1016/j.bbamcr.2008.01.017. [DOI] [PubMed] [Google Scholar]
- 16.Vachova L, Palkova Z. Physiological regulation of yeast cell death in multicellular colonies is triggered by ammonia. J Cell Biol. 2005;169:711–717. doi: 10.1083/jcb.200410064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fabrizio P, Battistella L, Vardavas R, Gattazzo C, Liou LL, Diaspro A, Dossen JW, Gralla EB, Longo VD. Superoxide is a mediator of an altruistic aging program in Saccharomyces cerevisiae. J Cell Biol. 2004;166:1055–1067. doi: 10.1083/jcb.200404002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laun P, Pichova A, Madeo F, Fuchs J, Ellinger A, Kohlwein S, Dawes I, Frohlich KU, Breitenbach M. Aged mother cells of Saccharomyces cerevisiae show markers of oxidative stress and apoptosis. Mol Microbiol. 2001;39:1166–1173. [PubMed] [Google Scholar]
- 19.Almeida B, Buttner S, Ohlmeier S, Silva A, Mesquita A, Sampaio-Marques B, Osorio NS, Kollau A, Mayer B, Leao C, Laranjinha J, Rodrigues F, Madeo F, Ludovico P. NO-mediated apoptosis in yeast. J Cell Sci. 2007;120:3279–3288. doi: 10.1242/jcs.010926. [DOI] [PubMed] [Google Scholar]
- 20.Eisler H, Frohlich KU, Heidenreich E. Starvation for an essential amino acid induces apoptosis and oxidative stress in yeast. Exp Cell Res. 2004;300:345–353. doi: 10.1016/j.yexcr.2004.07.025. [DOI] [PubMed] [Google Scholar]
- 21.Pereira C, Chaves S, Alves S, Salin B, Camougrand N, Manon S, Sousa MJ, Corte-Real M. Mitochondrial degradation in acetic acid-induced yeast apoptosis: the role of Pep4 and the ADP/ATP carrier. Mol Microbiol. 2010;76(6):1398–410. doi: 10.1111/j.1365-2958.2010.07122.x. [DOI] [PubMed] [Google Scholar]
- 22.Du L, Su Y, Sun D, Zhu W, Wang J, Zhuang X, Zhou S, Lu Y. Formic acid induces Yca1p-independent apoptosis-like cell death in the yeast Saccharomyces cerevisiae. FEMS Yeast Res. 2008;8:531–539. doi: 10.1111/j.1567-1364.2008.00375.x. [DOI] [PubMed] [Google Scholar]
- 23.Burhans WC, Weinberger M, Marchetti MA, Ramachandran L, D’Urso G, Huberman JA. Apoptosis-like yeast cell death in response to DNA damage and replication defects. Mutat Res. 2003;532:227–243. doi: 10.1016/j.mrfmmm.2003.08.019. [DOI] [PubMed] [Google Scholar]
- 24.Cap M, Stepanek L, Harant K, Vachova L, Palkova Z. Cell differentiation within a yeast colony: metabolic and regulatory parallels with a tumor-affected organism. Mol Cell. 2012;46:436–448. doi: 10.1016/j.molcel.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 25.Gourlay CW, Ayscough KR. Actin-induced hyperactivation of the Ras signaling pathway leads to apoptosis in Saccharomyces cerevisiae. Mol Cell Biol. 2006;26:6487–6501. doi: 10.1128/MCB.00117-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leadsham JE, Miller K, Ayscough KR, Colombo S, Martegani E, Sudbery P, Gourlay CW. Whi2p links nutritional sensing to actin-dependent Ras-cAMP-PKA regulation and apoptosis in yeast. J Cell Sci. 2009;122:706–715. doi: 10.1242/jcs.042424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gourlay CW, Carpp LN, Timpson P, Winder SJ, Ayscough KR. A role for the actin cytoskeleton in cell death and aging in yeast. J Cell Biol. 2004;164:803–809. doi: 10.1083/jcb.200310148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gourlay CW, Ayscough KR. Identification of an upstream regulatory pathway controlling actin-mediated apoptosis in yeast. J Cell Sci. 2005;118:2119–2132. doi: 10.1242/jcs.02337. [DOI] [PubMed] [Google Scholar]
- 29.Ahn SH, Cheung WL, Hsu JY, Diaz RL, Smith MM, Allis CD. Sterile 20 kinase phosphorylates histone H2B at serine 10 during hydrogen peroxide-induced apoptosis in S. cerevisiae. Cell. 2005;120:25–36. doi: 10.1016/j.cell.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 30.Ahn SH, Diaz RL, Grunstein M, Allis CD. Histone H2B deacetylation at lysine 11 is required for yeast apoptosis induced by phosphorylation of H2B at serine 10. Mol Cell. 2006;24:211–220. doi: 10.1016/j.molcel.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 31.Fabrizio P, Longo VD. Chronological aging-induced apoptosis in yeast. Biochim Biophys Acta. 2008;1783:1280–1285. doi: 10.1016/j.bbamcr.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laun P, Heeren G, Rinnerthaler M, Rid R, Kossler S, Koller L, Breitenbach M. Senescence and apoptosis in yeast mother cell-specific aging and in higher cells: a short review. Biochim Biophys Acta. 2008;1783:1328–1334. doi: 10.1016/j.bbamcr.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 33.Mazzoni C, Mancini P, Verdone L, Madeo F, Serafini A, Herker E, Falcone C. A truncated form of KlLsm4p and the absence of factors involved in mRNA decapping trigger apoptosis in yeast. Mol Biol Cell. 2003;14:721–729. doi: 10.1091/mbc.E02-05-0258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mazzoni C, Herker E, Palermo V, Jungwirth H, Eisenberg T, Madeo F, Falcone C. Yeast caspase 1 links messenger RNA stability to apoptosis in yeast. EMBO Rep. 2005;6:1076–1081. doi: 10.1038/sj.embor.7400514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weinberger M, Ramachandran L, Feng L, Sharma K, Sun X, Marchetti M, Huberman JA, Burhans WC. Apoptosis in budding yeast caused by defects in initiation of DNA replication. J Cell Sci. 2005;118:3543–3553. doi: 10.1242/jcs.02477. [DOI] [PubMed] [Google Scholar]
- 36.Fannjiang Y, Cheng WC, Lee SJ, Qi B, Pevsner J, McCaffery JM, Hill RB, Basanez G, Hardwick JM. Mitochondrial fission proteins regulate programmed cell death in yeast. Genes Dev. 2004;18:2785–2797. doi: 10.1101/gad.1247904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aerts AM, Zabrocki P, Govaert G, Mathys J, Carmona-Gutierrez D, Madeo F, Winderickx J, Cammue BP, Thevissen K. Mitochondrial dysfunction leads to reduced chronological lifespan and increased apoptosis in yeast. FEBS Lett. 2009;583:113–117. doi: 10.1016/j.febslet.2008.11.028. [DOI] [PubMed] [Google Scholar]
- 38.Teng X, Cheng WC, Qi B, Yu TX, Ramachandran K, Boersma MD, Hattier T, Lehmann PV, Pineda FJ, Hardwick JM. Gene-dependent cell death in yeast. Cell Death Dis. 2011;2:e188. doi: 10.1038/cddis.2011.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Teng X, Hardwick JM. Reliable method for detection of programmed cell death in yeast. Methods Mol Biol. 2009;559:335–342. doi: 10.1007/978-1-60327-017-5_23. [DOI] [PMC free article] [PubMed] [Google Scholar]


