Abstract
Despite considerable differences in morphology and complexity of body plans among animals, a great part of the gene set is shared among Bilateria and their basally branching sister group, the Cnidaria. This suggests that the common ancestor of eumetazoans already had a highly complex gene repertoire. At present it is therefore unclear how morphological diversification is encoded in the genome. Here we address the possibility that differences in gene regulation could contribute to the large morphological divergence between cnidarians and bilaterians. To this end, we generated the first genome-wide map of gene regulatory elements in a nonbilaterian animal, the sea anemone Nematostella vectensis. Using chromatin immunoprecipitation followed by deep sequencing of five chromatin modifications and a transcriptional cofactor, we identified over 5000 enhancers in the Nematostella genome and could validate 75% of the tested enhancers in vivo. We found that in Nematostella, but not in yeast, enhancers are characterized by the same combination of histone modifications as in bilaterians, and these enhancers preferentially target developmental regulatory genes. Surprisingly, the distribution and abundance of gene regulatory elements relative to these genes are shared between Nematostella and bilaterian model organisms. Our results suggest that complex gene regulation originated at least 600 million yr ago, predating the common ancestor of eumetazoans.
It is currently not known to what extent the evolution of morphological complexity was driven by changes in gene content or differences in gene regulation. The sea anemone Nematostella vectensis belongs to cnidarians, the sister group to all bilaterians, which diverged from bilaterians more than 600 million yr ago. Cnidarians consist of only two germ layers (endoderm and ectoderm) and have roughly 12–20 morphologically distinct somatic cell types. Yet, the Nematostella gene repertoire is surprisingly similar to that of vertebrates (Putnam et al. 2007). Despite the simplicity of its morphology and of gene expression patterns (Steele et al. 2011; Technau and Steele 2011), the complexity of developmental gene families (e.g., Wnt, homeobox transcription factors) in Nematostella is similar to that found in vertebrates (Kusserow et al. 2005; Chourrout et al. 2006; Ryan et al. 2007), and its genome harbors many genes important for the development of key bilaterian traits such as mesoderm and bilaterality (Finnerty 2004; Fritzenwanker et al. 2004; Martindale et al. 2004; Technau et al. 2005; Steele et al. 2011; Technau and Steele 2011). The simple body plan of an animal with such a complex gene repertoire led to the assumption that the complexity of gene regulation may be different between bilaterians and nonbilaterians and that these differences might underlie apparent morphological differences. Indeed, the evolution of new body plans is often driven by changes in the regulation of gene expression (Carroll 2008; Wittkopp and Kalay 2011) and many differences between closely related species are caused by changes in cis-regulatory elements that regulate the expression patterns of their target genes in a modular way (Sucena et al. 2003; Jeong et al. 2008; Rebeiz et al. 2009; Chan et al. 2010; Frankel et al. 2012).
In bilaterians, genes are regulated not only by proximal promoters, but also by a combination of distal cis-regulatory elements. Each gene can have multiple enhancer elements, which are often found at large distances from the gene they regulate (Spitz and Furlong 2012; Yáñez-Cuna et al. 2012). Therefore, an important initial step in comparing gene regulation between evolutionary distant species is to identify enhancer elements throughout their genomes. Despite their importance, enhancers have been very difficult to locate until recently. However, with the advent of microarray and next-generation sequencing technologies, genome-wide maps of enhancer elements have been predicted in human cell lines (Heintzman et al. 2007, 2009; The ENCODE Project Consortium 2012) and several major bilaterian model organisms (Visel et al. 2009a; Gerstein et al. 2010; The modENCODE Consortium et al. 2010; May et al. 2011; Nègre et al. 2011; Shen et al. 2012). This was achieved not only by mapping the binding sites of a number of tissue-specific transcription factors (Gerstein et al. 2010), but also based on characteristic histone modifications and the binding of transcriptional cofactors (Visel et al. 2009a; The modENCODE Consortium et al. 2010; May et al. 2011; Nègre et al. 2011; Shen et al. 2012). Initially, it was shown in mammalian cell lines that while H3K4me3 levels are higher at promoters, H3K4me1 is more enriched at enhancer elements (Heintzman et al. 2007, 2009; Supplemental Table S1). This observation has recently been extended to Drosophila as well as vertebrate model organisms (Nègre et al. 2011; Bogdanovic et al. 2012; Bonn et al. 2012). It has been suggested that inactive enhancers can be distinguished from active ones due to the fact that active enhancers are also associated with acetylated histones (Creyghton et al. 2010; Rada-Iglesias et al. 2010). One of the enzymes that catalyzes the acetylation of histones, the transcriptional cofactor EP300 (also known as p300), has been shown to bind to enhancers, and has recently been used in several studies to predict enhancer elements genome wide (Visel et al. 2009b; May et al. 2011; Nègre et al. 2011; Shen et al. 2012). Thus, while these studies underlined the complexity of cis-regulatory landscapes in vertebrates and insects, their evolutionary origin remains unclear, as no studies of nonbilaterian animals have been performed.
Here, we profiled five histone modifications (Supplemental Table S1) and the binding sites of the Nematostella histone acetylase p300 throughout the Nematostella genome. Our analysis revealed that the distribution of histone modifications across the Nematostella genome, relative to genes, as well as their colocalization with DNA methylation (Zemach et al. 2010), were remarkably similar to what has been shown in bilaterian model organisms. This allowed us to use this data to predict more than 5000 enhancer elements across two developmental stages of Nematostella. A total of 75% of the predicted enhancers tested in vivo drove tissue-specific expression, suggesting that they represent important gene regulatory elements. Upon comparison with predicted enhancer maps in bilaterians, we could show that the genomic distribution and location relative to genes is shared between cnidarians and bilaterians.
Results
Distribution of histone modifications across Nematostella genes
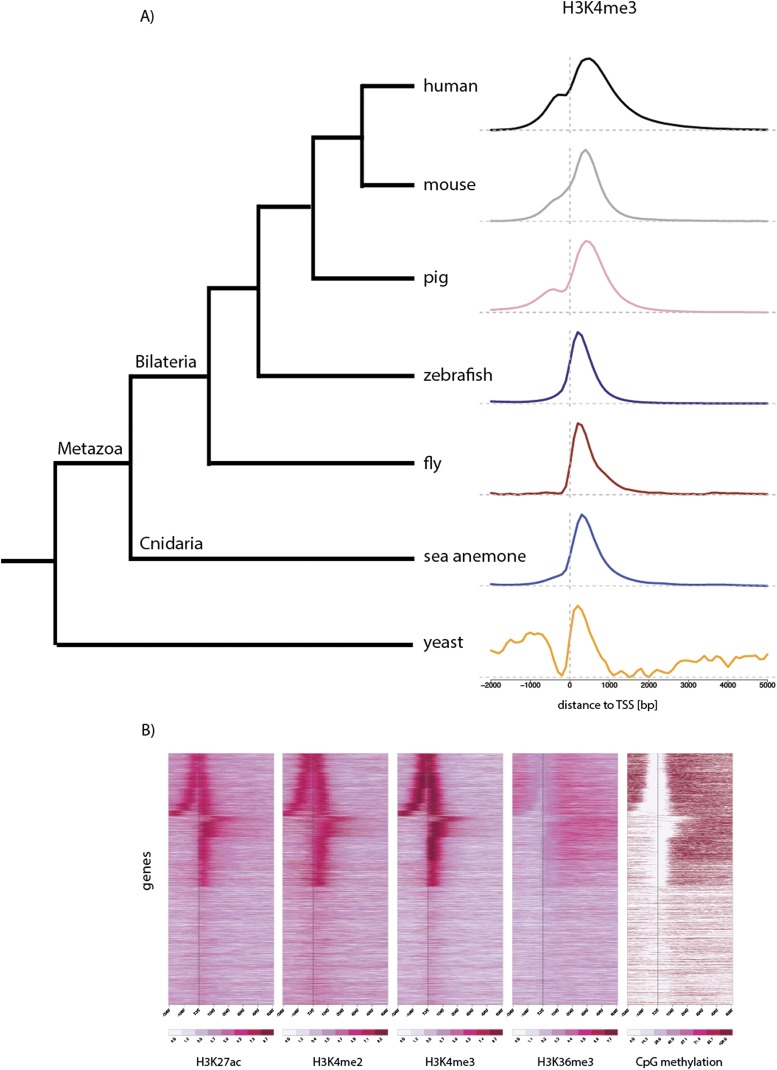
Using antibodies recognizing modified histone H3 tails, we established a chromatin immunoprecipitation (ChIP) protocol to determine the distribution of several “active” chromatin modifications in Nematostella vectensis adult female polyps (whole animals). ChIP-enriched and input DNA was subjected to deep sequencing and the reads were aligned to the Nematostella genome (Putnam et al. 2007), resulting in highly reproducible data sets of histone H3 di- and trimethylation of Lysine 4, trimethylation of lysine 36, and acetylation of Lysine 27 (Supplemental Tables S1, S2). We calculated and compared the average enrichment of H3K4me3 relative to the transcription start sites (TSSs) of genes in Nematostella and several other model organisms. As expected, H3K4me3 showed the strongest enrichments slightly downstream from TSSs in all species, including Nematostella (Fig. 1A). This suggests that its distribution is shared among most eukaryotic genomes, which was also the case for the other tested histone modifications (Liu et al. 2005; Barski et al. 2007; Rando and Chang 2009; The modENCODE Consortium et al. 2010; Supplemental Fig. S1). However, we noted that while in Nematostella and Drosophila H3K4me3 peaks downstream from the TSS, mammalian genes show additional H3K4me3 just upstream of the TSS. This H3K4me3 enrichment just upstream of TSSs could stem from additional transcription in these regions (Supplemental Fig. 2).
Figure 1.
Conserved distribution of chromatin marks across genes. (A) The distribution of H3K4me3 ChIP-seq reads (y-axis) is shown around genes of different species; human (The ENCODE Project Consortium 2012), mouse (Xiao et al. 2012), pig (Xiao et al. 2012), fish (Bogdanovic et al. 2012), fly (Bonn et al. 2012), sea anemone (adult female polyps), and yeast (Maltby et al. 2012) aligned at their TSSs. TSSs were obtained from Ensembl Biomart, except for Nematostella, where they are based on RNA-seq data (D Fredman, M Schwaiger, F Rentzsch, and U Technau, in prep.). The x-axis spans −2 kb to +5 kb around TSSs. Genes with TSSs closer than 2 kb (yeast: 1 kb) to each other and shorter than 5 kb (yeast: 2 kb) were excluded from the analysis. (B) Heatmaps of histone modifications and CpG methylation (Zemach et al. 2010) in adult female Nematostella polyps around genes aligned at their TSSs, as in A. Each line of the heatmap represents a single gene (y-axis); only nonoverlapping genes longer than 5 kb were plotted. The colors indicate the number of reads on a log-scale (histone modifications) or the average percentage of CpG methylation. Note that many genes are transcribed in the opposite direction from nearby TSSs.
H3K4me3 and CpG methylation target mutually exclusive regions
In vertebrates, DNA methylation at CpG dinucleotides is found throughout large parts of the genome, but is excluded from sites of H3K4 methylation (Meissner et al. 2008; Suzuki and Bird 2008; Jones 2012; Long et al. 2012). While the invertebrate model organisms Drosophila melanogaster and Caenorhabditis elegans lack DNA methylation, it has recently been shown that other invertebrates, including Nematostella vectensis, methylate CpGs throughout many active gene bodies (Feng et al. 2010; Zemach et al. 2010). To test whether the mutually exclusive location of H3K4 methylation and DNA methylation is shared between vertebrates and cnidarians, we sorted all genes based on their distribution of H3K4me3 around the TSS (Fig. 1B). While the distributions of H3K4me3, H3K4me2, and H3K27ac are very similar (Fig. 1B; Supplemental Fig. S1), H3K36me3 is enriched within gene bodies of the same genes, starting immediately after the TSS. CpG methylation localizes to gene bodies (Zemach et al. 2010) and its levels correlate with H3K36me3 (Fig. 1B; Supplemental Fig. S3) (P < 2.2 × 10−16). In contrast to H3K36me3, CpG methylation begins only about 1 kb after the TSS, a position where H3K4me3 is no longer enriched, thus suggesting a mutually exclusive location of H3K4me3 and CpG methylation in Nematostella.
Enhancer chromatin modifications at promoter-distal p300 peaks
The transcriptional cofactor p300 has been shown to be associated with enhancer regions in vertebrates (Visel et al. 2009a; May et al. 2011). To find gene regulatory elements throughout the genome of Nematostella, we generated an antibody specifically recognizing the Nematostella ortholog of the p300/CBP protein, hereafter referred to as p300 (Supplemental Fig. S4). We determined the genome-wide binding sites of Nematostella p300 in gastrulae (data not shown) and planulae (Fig. 2). Gastrulation is an important point during development. Yet many developmental regulators only start to be expressed after gastrulation, during the development of the planula larva. We found 3354 (55%) and 6460 (60%) p300 peaks distal (>300 bp) from the nearest TSS in gastrulae and planulae, respectively (data not shown). This suggests that p300 identifies thousands of putative enhancer regions in the Nematostella genome.
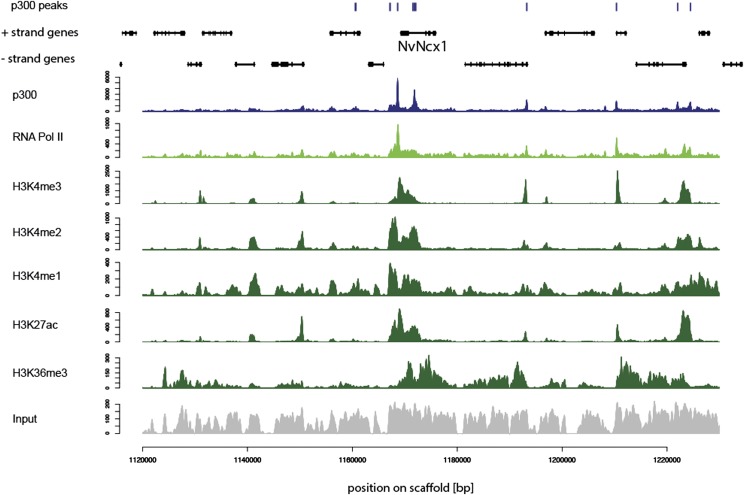
Figure 2.
Many p300 peaks overlap with sites of open chromatin. Region surrounding the NvNcx1 gene showing the distribution of p300 peaks (top, blue), gene models (black), p300 (blue), RNA Pol II (light green), several histone modifications (dark green), and input. (x-axis) Position on the scaffold; (y-axis) number of reads. The data are derived from planula larvae.
In bilaterian model organisms, distal enhancers and promoters have different compositions of chromatin marks surrounding them: H3K4me3 is located at promoters and H3K4me1 preferentially surrounds enhancers (Heintzman et al. 2007; The modENCODE Consortium et al. 2010; Bonn et al. 2012). We therefore asked whether the distribution of chromatin marks around such distal p300 peaks is different from the distribution of chromatin marks around TSSs in Nematostella embryos. To this end, we performed ChIP-seq with H3K4me3, H3K4me2, H3K4me1, H3K27ac, and H3K36me3 antibodies in Nematostella gastrulae and planula larvae. In addition, we performed RNA-seq to determine expression levels of all genes and ChIP-seq using an antibody recognizing the unphosphorylated C-terminal repeat of RNA polymerase II (RNA Pol II), which recognized RNA Pol II bound at the TSSs of active genes in gastrulae (Supplemental Fig. S5) and planulae (Figs. 2, 3).
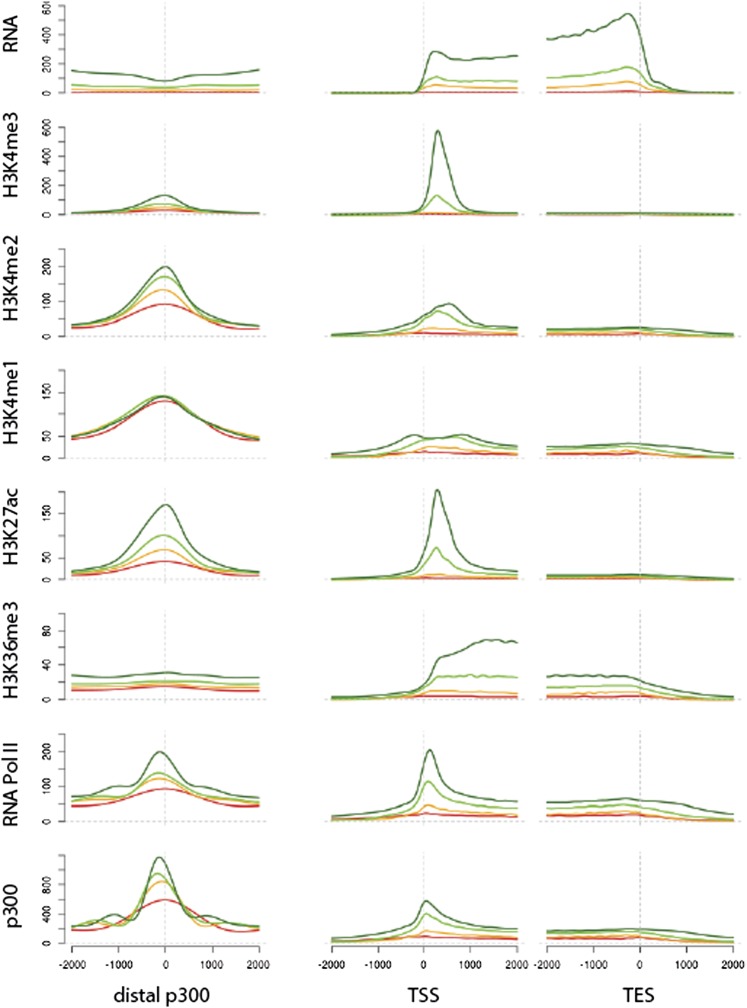
Figure 3.
Enhancer-related chromatin modifications are associated with distal p300 peaks. Distribution of chromatin marks, RNA polymerase II, and p300 across distal p300 peaks and genes. Planula p300 peaks that do not overlap with TSSs were aligned relative to their peak summit (left plots), and genes were aligned relative to their annotated transcription start (middle plots) and end (right plots). The x-axis in each plot represents the position within the gene relative to peak summits, transcription start sites, and 3′ ends. The y-axis in each plot represents the relative enrichment for epigenomic variables such as several histone modifications in the planula stage. (Red line) Nonexpressed genes (FPKM <1.5). (Orange line) Lowly expressed genes. (Green line) Medium expressed genes. (Dark green line) Highly expressed genes. (Expressed genes) FPKM >2. The expressed genes were divided into three bins of an equal number of genes according to their FPKM values.
To test whether this pattern is conserved in cnidarians, we plotted the distribution of histone modifications, RNA Pol II and p300, across distal p300 peaks aligned at their summits and genes aligned at their TSSs and transcription end sites (TESs) (Fig. 3; Supplemental Fig. S5A). Indeed we found the distribution of chromatin marks to be highly similar to mammalian cells or Drosophila (Heintzman et al. 2007, 2009; The modENCODE Consortium et al. 2010; Nègre et al. 2011): H3K4me3 was located over TSSs of active genes, but hardly detected at distal p300 peaks, while H3K4me1 was found at higher levels at distal p300 peaks (Figs. 3, 4A,B; Supplemental Fig. S5A,B). H3K4me2 and H3K27ac were found both at promoters and distal p300 peaks (Fig. 3; Supplemental Figs. S5A,B, S7A,B). RNA Pol II was mostly enriched at TSSs, but also found at distal p300 peaks (Fig. 3; Supplemental Fig. S5A). As expected, H3K36me3 was only located over transcribed gene bodies, which are also covered by RNA-seq reads (Fig. 3; Supplemental Fig. S5A). Overall, active genes contained higher levels of p300 and active histone modifications at their TSSs and distal p300 peaks (Fig. 3; Supplemental Fig. S5A). This is in line with previously published analyses in bilaterians, where H3K4me1 marks all enhancers, while active enhancers are distinguished from inactive ones by the presence of H3K27ac (Bonn and Furlong 2008; Creyghton et al. 2010; Rada-Iglesias et al. 2010). Indeed we find H3K27ac peaks preferentially around active genes in the Nematostella as well as in the Drosophila genome (Supplemental Fig. S7D–G). Together, the distribution of chromatin marks across TSSs and distal p300 peaks suggests that not only promoter, but also enhancer modifications are highly conserved between bilaterians and cnidarians. To test whether this is specific to metazoans, we looked at the distribution of H3K4me3 and H3K4me1 (Kirmizis et al. 2007) in the genome of Saccharomyces cerevisiae. While we find an enrichment of H3K4me3 compared with H3K4me1 around TSSs in yeast, there is no enrichment of H3K4me1 compared with H3K4me3 at TSS distal transcription factor peaks (Fig. 4C). Instead, H3K4me1 is enriched downstream from TSSs within gene bodies (Liu et al. 2005). This suggests that distal enhancers as described in bilaterians exist also in Nematostella, but not in yeast.
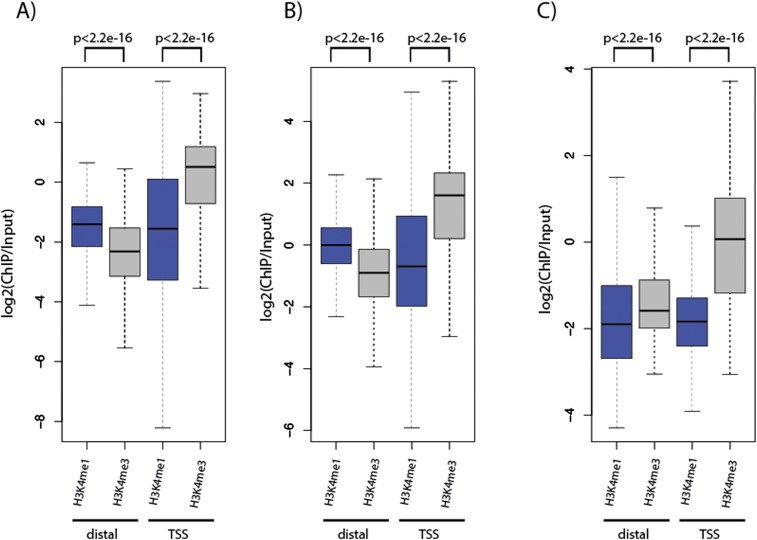
Figure 4.
H3K4me1 is enriched at distal p300 peaks in Nematostella. (A,B) Gastrula (A) or planula (B) p300 peaks were split into peaks >300 bp distal to transcription start sites (TSSs) (distal, left) and peaks within 300 bp around TSSs (TSS, right). On the y-axis, the enrichment of H3K4me1 (blue boxes) or H3K4me3 (gray boxes) normalized to input is plotted. Distal p300 peaks have higher H3K4me1 than H3K4me3 levels, a characteristic of enhancers. As expected, p300 peaks around TSSs are more enriched in H3K4me3. (C) Peaks of three different transcription factors (Reb1, Gal4, Phd1) derived from ChIP-exo experiments (Rhee and Pugh 2011) in Saccharomyces cerevisiae were split into distal and TSS overlapping peaks as in A and B. A total of 70% of peaks overlapped TSSs, and the remaining peaks were still within 2 kb around the TSS. On the y-axis, the enrichment of H3K4me1 (blue boxes) or H3K4me3 (gray boxes) normalized to input (Kirmizis et al. 2007) is plotted. P-values were calculated using the Wilcoxon rank sum test.
Prediction of gene regulatory elements in Nematostella
In order to identify enhancer regions on a genome-wide level, we combined the distal p300 peaks and the information provided by the chromatin mark distributions to predict enhancers throughout the Nematostella genome. To predict chromatin states throughout the genome in an unbiased manner, we used ChromHMM (Ernst and Kellis 2012), a hidden Markov model-based classifier, to separate the genome into six different chromatin states based on the five different histone modifications we assayed (Supplemental Fig. S6A,B). State 1 is defined by enrichment of H3K4me2, H3K4me3, and H3K27ac (Supplemental Fig. S6A), suggesting that it marks promoters. Indeed, we find it enriched over TSSs (Supplemental Fig. S6C,D). State 2 shares the same promoter-like modifications, but also contains H3K36me3 (Supplemental Fig. S6A), which suggests that it lies in gene bodies just downstream from the TSS (Supplemental Fig. S6C,D). State 3 contains only H3K36me3 (Supplemental Fig. S6A) and is located over transcribed gene bodies (Supplemental Fig. S6C,D). State 4 contains all histone modifications except H3K36me3, and low levels of H3K4me3 (Supplemental Fig. S6A), suggesting that it marks mostly active enhancers. State 5 contains high levels of H3K4me1, low levels of H3K4me2, and lacks H3K4me3, a characteristic of enhancers (Supplemental Fig. S6A). State 6 covers the rest of the genome lacking any of the active modifications we profiled (Supplemental Fig. S6A).
To predict enhancer elements, we selected only those distal p300 peaks, which overlapped chromatin states 4 or 5. This resulted in 2558 predicted enhancers in gastrulae and 4732 in planulae. Of those, 1543 were shared between both developmental stages. A similar overlap was found for p300 peaks between the two different developmental stages (Supplemental Fig. S7C). The genomic location of all predicted enhancers and their distance to the closest TSS (on average 3898 bp) is provided in Supplemental Table S3.
Interestingly, some predicted enhancers are occupied by RNA Pol II (Supplemental Fig. S9A). This has also been observed in bilaterians, where many enhancers are TSSs for diverse classes of lowly expressed noncoding RNAs (De Santa et al. 2010; Kim et al. 2010; Wang et al. 2011; Chen et al. 2013). It may also indicate that enhancers are physically interacting with the transcription initiation complex at the TSS of their target gene. By looking for the TSSs located closest to each predicted enhancer, we identified 2967 potential target genes. As expected, genes associated with gastrula-specific enhancers tend to be expressed at slightly but significantly higher levels in gastrulae compared with planulae, while planula-specific enhancers are located close to genes more highly transcribed in planulae than in gastrulae (Supplemental Fig. S9B).
To investigate the dynamics of enhancer activity in Nematostella, we compared the levels of the two enhancer modifications that we have profiled in three stages (gastrula, planula, and adult female polyp). Enrichments for H3K27ac and H3K4me2 at predicted enhancers were highly correlated between the gastrula and planula stages, but revealed a larger amount of enhancers with dynamic activity when compared with adult animals (Supplemental Fig. S8). A total of 440 enhancers were acetylated in planulae, and therefore likely active during early development, but inactive (significantly less acetylated) in adults (Supplemental Fig. S8C). Some of these enhancers were associated with genes involved in developmental transcriptional regulation (Supplemental Fig. S8G). On the other hand, 397 enhancers with significantly higher H3K27ac in adult polyps compared with planulae had a preference for association with neuronal genes (Supplemental Fig. S8C,G).
Complex regulation of genes encoding transcription factors in Nematostella
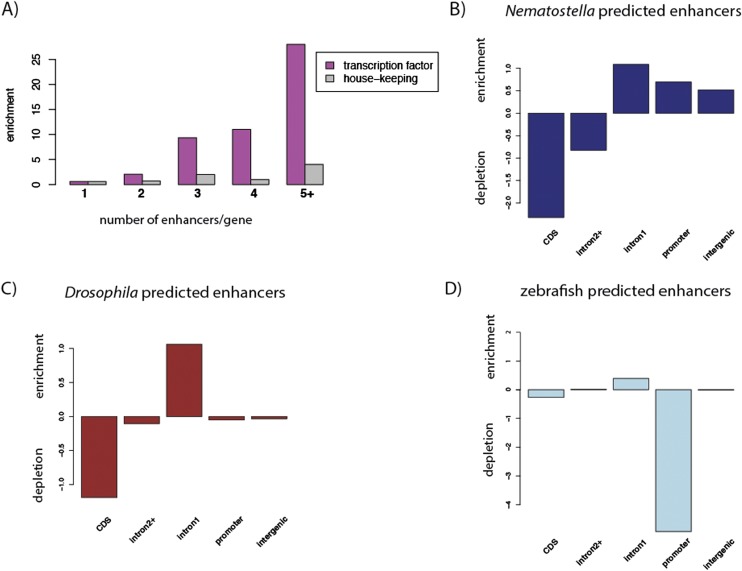
Having predicted thousands of gene regulatory elements throughout the Nematostella genome, we determined their distribution relative to annotated genes. We found that gene ontology terms for transcriptional regulation, signaling, and developmental processes were significantly enriched among genes in the vicinity of predicted enhancers (Supplemental Fig. 10A). A total of 934 genes are potentially regulated by multiple enhancer elements (Supplemental Fig. S10B). Interestingly, genes encoding transcription factors are associated with multiple enhancers more often than housekeeping genes (Fig. 5A). Despite apparently simple spatial and temporal expression patterns of most transcription factor genes in Nematostella (Steele et al. 2011; Technau and Steele 2011), this is similar to the situation in bilaterians, where many developmental regulatory genes are regulated by a complex landscape of gene regulatory elements (Bejerano et al. 2004; Woolfe et al. 2005; Hong et al. 2008; Visel et al. 2008, 2009a; Heintzman et al. 2009; Arnold et al. 2013). Indeed, we found that in Drosophila, a protostome model organism in which enhancers have been predicted in embryos genome wide (Nègre et al. 2011), and to a slightly lesser extent in zebrafish, a vertebrate in which embryonic enhancers have recently been predicted (Bogdanovic et al. 2012), transcriptional regulatory genes tend to be associated with multiple enhancers (Supplemental Fig. 11).
Figure 5.
Similar genomic distribution of predicted enhancers in different eumetazoans. (A) The number of genes associated with 1, 2, 3, 4, and 5 or more predicted enhancers in Nematostella is plotted for genes encoding transcription factors (pink) and housekeeping genes (gray). The counts of genes with a given number of predicted enhancers have been normalized to the counts of genes associated with a given number of shuffled predicted enhancers. (B–D) Distribution of predicted enhancer regions normalized to shuffled predicted enhancers across genomic annotations in Nematostella (B), Drosophila (C), and zebrafish (D). Positive numbers indicate enrichment, and negative numbers indicate depletion of predicted enhancers in a certain genomic region compared with the random expectation. Promoter regions are defined from the TSS to 1 kb upstream of the TSS.
Genomic landscape of gene regulation
In comparison with the Drosophila genome, the Nematostella genome is about twice as large and contains an increased proportion of introns rather than intergenic regions (Putnam et al. 2007; Supplemental Fig. S12A,B). On the other hand, the zebrafish genome is about five times larger than the Nematostella genome (Putnam et al. 2007; Howe et al. 2013) and contains 70% intergenic sequence, where most predicted enhancers are located (Supplemental Fig. S12C). In contrast, in Nematostella and Drosophila, more enhancers are located in introns (Supplemental Fig. S12A,B). As expected, enhancers are depleted from the coding sequence in Nematostella (Fig. 5B, P-value <0.001), just like in Drosophila (Fig. 5C, P-value <0.001) (Arnold et al. 2013) and zebrafish (Fig. 5D, P-value <0.001). However, only the large zebrafish genome often lacks predicted enhancers within 1 kb upstream of transcription start sites (Fig. 5D; Supplemental Fig. S12C). All three species show an enrichment of regulatory sequences in the initial introns of different genes (Fig. 5B–D, all P-values <0.001). Predicted enhancers are distributed at equal distances from TSSs in Nematostella and Drosophila, while they are distributed more broadly in zebrafish (Supplemental Fig. S12D). Together, our data suggest that the overall genomic distribution of enhancers is similar between Nematostella and Drosophila. Since the evolutionary distance between Nematostella and bilaterian species precludes genome-wide alignment of their nucleotide sequences, we tested whether there was any trace of conservation between enhancers in orthologous regions by comparing the 6-mer DNA sequence content (Göke et al. 2012) between Nematostella and Drosophila or zebrafish-predicted enhancers associated with gene orthologs, compared with predicted enhancers associated with random cross-species gene pairs. We could not detect any increased conservation, implying that these enhancers either do not share any common ancestry or have diverged beyond recognition by sequence similarity and even motif content (data not shown).
In vivo validation of predicted enhancers
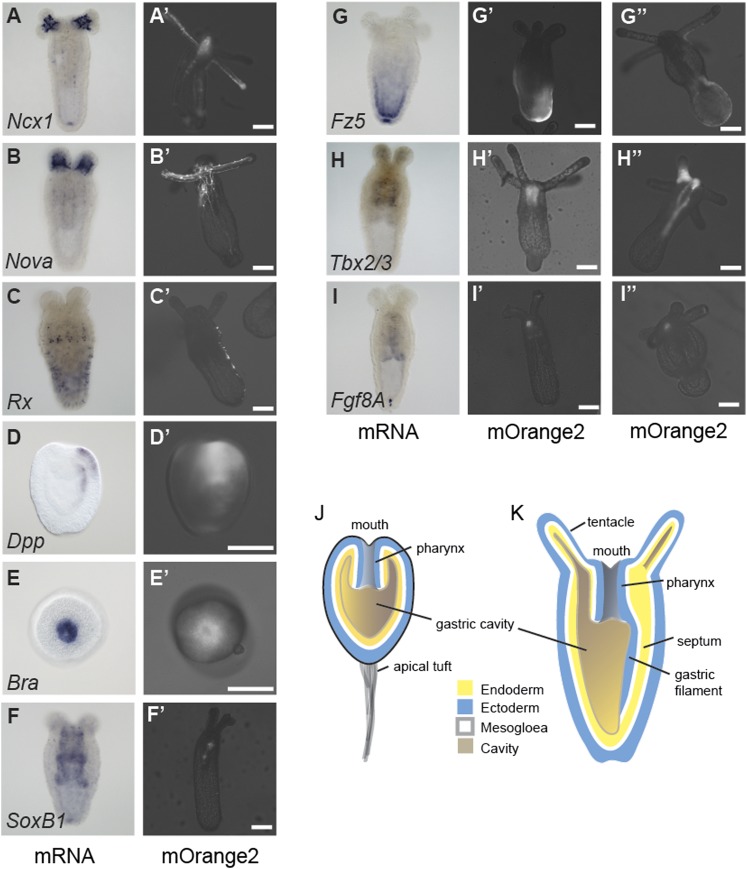
To test the activity of distal enhancer elements we isolated a minimal promoter of a Nematostella hsp70a gene, and confirmed that it does not induce the expression of a reporter gene on its own (Supplemental Table S4). We then tested 16 regions containing predicted enhancers for their ability to induce tissue-specific expression of the reporter gene via the hsp70a or their endogenous promoter (Supplemental Table S4). We found that 12 of the tested regions drove expression in a mosaic and tissue-specific pattern at least partially reflecting the in situ hybridization expression pattern of the gene closest to the predicted enhancer (Fig. 6; Supplemental Table S4). Four predicted enhancers did not drive any reporter gene expression (Supplemental Table S4). The expression patterns of three genes were recovered with two independent enhancer regions for each gene (Fig. 6G–I). Their expression domains appear to be at least partially overlapping, reminiscent of the redundant function of enhancers observed around some bilaterian genes (Hong et al. 2008). Reporter gene expression was mostly independent of the orientation or placement of the predicted enhancer upstream of or downstream from the reporter gene (Supplemental Table S4). The only exception is the NvNcx1 enhancer (Fig. 2), which drove expression only in reverse orientation (Supplemental Table S4). Interestingly, another predicted enhancer region >5 kb upstream of the nearest gene (Supplemental Fig. S13) drove expression specifically in neurons, with fluorescent neurites clearly visible (Supplemental Fig. S13). Taken together, at least 75% of Nematostella predicted enhancers likely represent true regulatory elements.
Figure 6.
Predicted enhancer elements activate transcription in vivo. (A–I) Whole-mount in situ hybridizations of Nematostella embryos and primary polyps. (A′–I′) Fluorescent mOrange2 signal of live embryos or primary polyps injected with a construct where a predicted enhancer region of the indicated Nematostella gene was driving mOrange2 expression. (G″–I″) Fluorescent mOrange2 signal of live primary polyps injected with a construct where a second predicted enhancer region (different from the region in G′–I′) of the indicated Nematostella gene was driving mOrange2 expression. The Nematostella gene names are indicated inside the in situ hybridization pictures (Nv in the beginning of the gene name was omitted due to space constraints). The white scale bars represent 100 µm. All pictures were taken at the primary polyp stage (>8 d post-fertilization; lateral view) except D and E, which depict planula larvae (lateral view: D,D′; oral view: E,E′). (J,K) Schematic representation of a Nematostella planula larva (J) and primary polyp (K).
In summary, our genome-wide analyses as well as the in vivo validated examples suggest that despite their rather simple expression patterns and anatomy, Nematostella genes are subject to complex regulation by multiple enhancer elements.
Discussion
Gene regulatory elements are crucial for the regulation of gene expression in multicellular organisms (Levine 2010). Despite their importance for development, gene regulatory elements have so far only been studied in major bilaterian model organisms (Heintzman et al. 2007, 2009; Visel et al. 2009, 2013; The modENCODE Consortium et al. 2010; May et al. 2011; Nègre et al. 2011; Bogdanovic et al. 2012; Shen et al. 2012; Chen et al. 2013; The ENCODE Project Consortium 2012; Neph et al. 2013). To obtain insights into the evolution of gene regulation in multicellular animals, we compared gene regulatory mechanisms in bilaterians and cnidarians, which split at least 600 million yr ago. Here we report the first comprehensive map of gene regulatory elements as well as chromatin modifications in a nonbilaterian animal, the sea anemone Nematostella vectensis. We find a high degree of conservation in the genomic distribution of chromatin modifications and gene regulatory elements between cnidarians and bilaterians. Promoter histone modifications are shared in most eukaryotes, and enhancers are marked by the same histone modifications in Nematostella and bilaterian model organisms. While the function of these histone modifications at regulatory elements is still unclear (Calo and Wysocka 2013), it suggests conserved targeting mechanisms of the histone modifying enzymes to chromatin.
Studies in mammalian cells have shown that H3K4me3 at CpG islands inhibits the methylation of CpGs (Ooi et al. 2007; Thomson et al. 2010). Since Drosophila melanogaster and Caenorhabditis elegans, the most common invertebrate model organisms, lack significant amounts of DNA methylation, it was not clear whether this is specific to vertebrates or whether this feature has been lost in Drosophila and Caenorhabditis (Zemach and Zilberman 2010). Comparisons between species showed that invertebrate DNA methylation is found at the same genes as active histone modifications in Drosophila (Nanty et al. 2011; Sarda et al. 2012; Hunt et al. 2013). Nematostella is the first invertebrate where both DNA methylation (Zemach et al. 2010) and histone modifications (this study) have been analyzed genome wide. We could show that in Nematostella, H3K4me3 and DNA methylation are found in mutually exclusive regions of the same genes. This suggests that the inhibition of DNA methylation by H3K4me3 might be occurring not only in mammals but also in cnidarians. Detailed mechanistic studies of the targeting of DNA methylation in Nematostella, as well as in protostomes containing DNA methylation, will be required to determine whether an inhibitory regulation of DNA methylation by H3K4me3 is ancestral to eumetazoans. Since DNA methylation and active histone modifications are found at the same genes in Nematostella, we suspect that they share a common targeting mechanism, most likely transcriptional activity.
The analysis of the Nematostella genome revealed that it is more similar to vertebrate genomes in terms of gene content and genomic organization (exon–intron structures, broad gene synteny) than to the genomes of common invertebrate model organisms (Putnam et al. 2007). This suggested that bilaterians, rather than acquiring many novel genes, mostly evolved more elaborate mechanisms to regulate gene expression. This might be the case for post-transcriptional gene regulation mediated by microRNAs, which we found to differ substantially in cnidarians and bilaterians (Moran et al. 2014). Thus, we expected also the gene regulatory landscape of Nematostella to be less complex than that of the investigated bilaterian model species.
Surprisingly, our findings do not support this view. Instead, careful analysis indicates that despite lacking CTCF (Heger et al. 2012), a factor implicated in gene regulation by distal enhancers (Merkenschlager and Odom 2013), the gene regulatory landscape of Nematostella shows a similar complexity as that of bilaterians. In all animals studied so far, genes encoding tissue-specific transcription factors are regulated by a high number of enhancers (Bejerano et al. 2004; Woolfe et al. 2005; Hong et al. 2008; Visel et al. 2008; Heintzman et al. 2009; Visel et al. 2009a). Furthermore, predicted enhancers are enriched in the same genomic regions and at a similar distance from transcription start sites in Nematostella and Drosophila. While noncoding transcription at enhancers will need to be investigated in detail, the enrichment of RNA Pol II at predicted enhancers already suggests that some enhancers in Nematostella could transcribe noncoding RNAs, as is the case in bilaterians (De Santa et al. 2010; Kim et al. 2010; Wang et al. 2011; Chen et al. 2013). One example could be the NvNcx1 enhancer, which activates transcription in an orientation-dependent manner and which is bound by low levels of RNA Pol II. This could reflect a common mechanism of transcriptional activation by these enhancers. It is possible that this mechanism does not involve chromatin looping through CTCF, but read-through low-level transcription from the enhancer to the downstream promoter. However, most enhancers we tested in vivo also activated transcription when placed downstream from the hsp70 basal promoter and the reporter gene, suggesting that this is not the only mechanism of activation in Nematostella. Instead, just like in bilaterians, most Nematostella distal enhancers can activate transcription from a promoter independently of the position and orientation relative to that promoter. This suggests that chromatin looping of enhancers to their target promoters in Nematostella occurs through CTCF-independent cohesin binding, which plays a role in enhancer looping in bilaterians (Pauli et al. 2008; Schuldiner et al. 2008; Phillips and Corces 2009; Kagey et al. 2010; Schmidt et al. 2010; Seitan et al. 2011; Faure et al. 2012; Merkenschlager and Odom 2013). The enrichment of RNA Pol II at enhancers could also be an indication for physical interaction of enhancers and their target promoters in Nematostella. In the future, detailed mechanistic studies of chromatin looping and noncoding RNA function in Nematostella will identify the factors required for enhancer promoter interaction in this organism. In addition, examining histone modifications associated with inactive or repressed chromatin will give a more complete picture of the cnidarian gene regulatory landscape.
Taken together, we provide the first genome-wide analysis of gene regulation in a nonbilaterian animal and thereby shed light on the evolution of morphological complexity in metazoans. Our results suggest that a complex gene regulatory architecture as well as epigenomic chromatin signatures predate the divergence of bilaterians and cnidarians from their last common ancestor.
Methods
Antibodies
We used a 146-aa peptide of the C terminus of Nematostella p300 protein, including a N-terminal 10× His tag for immunization of two rabbits each. p300 antisera and purified p300 antibodies were generated by Primm (www.primmbiotech.com). We used a mouse monoclonal antibody against the unphosphorylated C-terminal repeat of RNA polymerase II (8WG16, kind gift from C. Wirbelauer), as well as polyclonal antibodies against H3K4me3 (Diagenode, pAb-003-050), H3K4me2 (Diagenode, pAb-035-050), H3K4me1 (Abcam, ab8895), H3K36me3 (Diagenode, pAb-058-050), and H3K27ac (Abcam, ab4729). The entire N-terminal tail of histone H3 is perfectly conserved between Nematostella and model organisms where these antibodies have been used successfully.
Chromatin immunoprecipitation
Fertilized eggs from ∼500 female Nematostella polyps were grown to the mid-gastrula or planula stage as described (Fritzenwanker and Technau 2002). After removal from jelly, eggs were fixed in 2% formaldehyde for 12 min at room temperature. Cross-linking was stopped using glycine, and embryos were homogenized in nuclei buffer 1 (50 mM HEPES at pH 7.5, 140 mM NaCl, 1 mM EDTA, 10% Glycerol, 0.5% NP40, 0.25% Triton X-100, 1 mM DTT) and centrifuged at 500g for 3 min. After another centrifugation of the supernatant at 2000g for 10 min, the pelleted nuclei were washed with nuclei buffer 2 (50 mM HEPES at pH 7.5, 140 mM NaCl, 1 mM EDTA, 10% Glycerol, 0.5% NP40, 0.25% Triton X-100) and finally resuspended in Lysis buffer (50 mM HEPES at pH 7.5, 500 mM NaCl, 1 mM EDTA, 0.1% DOC, 0.1% Triton X-100, 0.1% SDS). After shearing the chromatin to 100–200 bp using a Covaris instrument and removing the nonsoluble material, we blocked the chromatin with preblocked (1 mg/mL tRNA, 1 mg/mL BSA) protein A beads CL-4B (GE Healthcare 71-7090-00 AE). At this point, we removed an aliquot of chromatin for input DNA and incubated the remaining chromatin with the antibodies, rotating at 4°C overnight. We then added preblocked protein A beads and washed the beads twice with Lysis buffer, twice with DOC buffer (10 mM Tris at pH 8, 0.25 M LiCl, 0.5% NP-40, 0.5% DOC, 1 mM EDTA), and once with TE buffer. Chromatin was eluted from the beads twice with elution buffer (1% SDS, 0.1 M NaHCO3) and de-crosslinked. Libraries of immunoprecipitated DNA or input DNA were prepared according to Illumina’s instructions (catalog number IP-102-1001) and deep sequencing was performed at the CSF NGS Unit (http://csf.ac.at/) with 36 bp GA II or 50 bp HiSeq reads.
RNA-seq
RNA of Nematostella embryos was isolated using TRIzol followed by DNase I treatment, at the same developmental time points as described for ChIP. For two planula and one gastrula sample, poly(A) RNA was isolated and libraries were prepared according to Illumina’s instructions. The second gastrula sample was subjected to ribosomal RNA depletion (RiboMinus) and strand-specific library preparation using the dUTP method (Levin et al. 2010). Deep sequencing by Illumina HiSeq and GA IIx was performed at the CSF NGS Unit with 76-bp paired end reads.
Data analysis
ChIP-seq reads were mapped to the genome using BWA (Li and Durbin 2009) with default settings and those with a mapping quality score <30 were discarded. We used the following genome versions: Nematostella: Nemvec1; Drosophila: dm3; zebrafish: zv8; pig: SusScr2; mouse: mm9; human: hg19; yeast: SacCer3. Further analysis was performed using the BamTools and BEDTools suites (Quinlan and Hall 2010; Barnett et al. 2011) as well as custom perl and R (R Development Core Team 2012) scripts. K-means clustering was performed using Cluster 3.0 (Eisen et al. 1998). Chromatin states were predicted across the genome using ChromHMM (Ernst and Kellis 2012). p300 peaks were detected using the peakzilla software (Bardet et al. 2013). See Supplemental Methods for details.
In vivo analysis of regulatory elements
Predicted gene regulatory elements were tested for their activity to induce expression of mOrange2 in developing Nematostella embryos and polyps as described (Renfer et al. 2010). In situ hybridizations on Nematostella embryos and primary polyps were performed as described (Genikhovich and Technau 2009).
Data access
All data sets have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo) (Edgar et al. 2002) under accession number GSE46488.
Acknowledgments
This work was supported by a postdoctoral fellowship from EMBO (ALTF 81-2009) and a Hertha Firnberg Austrian Science Fund (FWF) grant (T496-B17) to M.S. and a FWF grant (P248-58) to U.T. We thank Hanna Kraus for illustrations, Christiane Wirbelauer and Dirk Schübeler for assistance with Western blotting and providing antibodies, as well as members of the Technau lab for sharing reagents. We thank Rabih Murr and Andreas Sommer for advice on next-generation sequencing library preparation, Anaïs Bardet for the peakzilla software, and Alex Stark for critically reading the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.162529.113.
Freely available online through the Genome Research Open Access option.
References
- Arnold CD, Gerlach D, Stelzer C, Boryn LM, Rath M, Stark A 2013. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science 339: 1074–1077 [DOI] [PubMed] [Google Scholar]
- Bardet AF, Steinmann J, Bafna S, Knoblich JA, Zeitlinger J, Stark A 2013. Identification of transcription factor binding sites from ChIP-seq data at high resolution. Bioinformatics 29: 2705–2713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett DW, Garrison EK, Quinlan AR, Strömberg MP, Marth GT 2011. BamTools: A C++ API and toolkit for analyzing and managing BAM files. Bioinformatics 27: 1691–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh T-Y, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, Haussler D 2004. Ultraconserved elements in the human genome. Science 304: 1321–1325 [DOI] [PubMed] [Google Scholar]
- Bogdanovic O, Fernandez-Minan A, Tena JJ, de la Calle-Mustienes E, Hidalgo C, van Kruysbergen I, van Heeringen SJ, Veenstra GJC, Gomez-Skarmeta JL 2012. Dynamics of enhancer chromatin signatures mark the transition from pluripotency to cell specification during embryogenesis. Genome Res 22: 2043–2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonn S, Furlong EEM 2008. cis-Regulatory networks during development: A view of Drosophila. Curr Opin Genet Dev 18: 513–520 [DOI] [PubMed] [Google Scholar]
- Bonn S, Zinzen RP, Girardot C, Gustafson EH, Perez-Gonzalez A, Delhomme N, Ghavi-Helm Y, Wilczyński B, Riddell A, Furlong EEM 2012. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat Genet 44: 148–156 [DOI] [PubMed] [Google Scholar]
- Calo E, Wysocka J 2013. Modification of enhancer chromatin: What, how, and why? Mol Cell 49: 825–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB 2008. Evo-devo and an expanding evolutionary synthesis: A genetic theory of morphological evolution. Cell 134: 25–36 [DOI] [PubMed] [Google Scholar]
- Chan YF, Marks ME, Jones FC, Villarreal G, Shapiro MD, Brady SD, Southwick AM, Absher DM, Grimwood J, Schmutz J, et al. 2010. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science 327: 302–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RAJ, Down TA, Stempor P, Chen QB, Egelhofer TA, Hillier LW, Jeffers TE, Ahringer J 2013. The landscape of RNA polymerase II transcription initiation in C. elegans reveals promoter and enhancer architectures. Genome Res 23: 1339–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chourrout D, Delsuc F, Chourrout P, Edvardsen RB, Rentzsch F, Renfer E, Jensen MF, Zhu B, de Jong P, Steele RE, et al. 2006. Minimal ProtoHox cluster inferred from bilaterian and cnidarian Hox complements. Nature 442: 684–687 [DOI] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. 2010. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci 107: 21931–21936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, Muller H, Ragoussis J, Wei C-L, Natoli G 2010. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol 8: e1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D 1998. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kellis M 2012. ChromHMM: Automating chromatin-state discovery and characterization. Nat Methods 9: 215–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure AJ, Schmidt D, Watt S, Schwalie PC, Wilson MD, Xu H, Ramsay RG, Odom DT, Flicek P 2012. Cohesin regulates tissue-specific expression by stabilizing highly occupied cis-regulatory modules. Genome Res 22: 2163–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Cokus SJ, Zhang X, Chen PY, Bostick M, Goll MG, Hetzel J, Jain J, Strauss SH, Halpern ME, et al. 2010. Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci 107: 8689–8694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnerty JR 2004. Origins of bilateral symmetry: Hox and dpp expression in a sea anemone. Science 304: 1335–1337 [DOI] [PubMed] [Google Scholar]
- Frankel N, Erezyilmaz DF, McGregor AP, Wang S, Payre F, Stern DL 2012. Morphological evolution caused by many subtle-effect substitutions in regulatory DNA. Nature 474: 598–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzenwanker JH, Technau U 2002. Induction of gametogenesis in the basal cnidarian Nematostella vectensis (Anthozoa). Dev Genes Evol 212: 99–103 [DOI] [PubMed] [Google Scholar]
- Fritzenwanker JH, Saina M, Technau U 2004. Analysis of forkhead and snail expression reveals epithelial-mesenchymal transitions during embryonic and larval development of Nematostella vectensis. Dev Biol 275: 389–402 [DOI] [PubMed] [Google Scholar]
- Genikhovich G, Technau U 2009. In situ hybridization of starlet sea anemone (Nematostella vectensis) embryos, larvae, and polyps. Cold Spring Harb Protoc 10.1101/pdb.prot5281 [DOI] [PubMed]
- Gerstein MB, Lu ZJ, Van Nostrand EL, Cheng C, Arshinoff BI, Liu T, Yip KY, Robilotto R, Rechtsteiner A, Ikegami K, et al. 2010. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE Project. Science 330: 1775–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göke J, Schulz MH, Lasserre J, Vingron M 2012. Estimation of pairwise sequence similarity of mammalian enhancers with word neighbourhood counts. Bioinformatics 28: 656–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heger P, Marin B, Bartkuhn M, Schierenberg E, Wiehe T 2012. The chromatin insulator CTCF and the emergence of metazoan diversity. 109: 17507–17512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. 2007. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39: 311–318 [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. 2009. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459: 108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong J-W, Hendrix DA, Levine MS 2008. Shadow enhancers as a source of evolutionary novelty. Science 321: 1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe K, Clark MD, Torroja CF, Torrance J, Berthelot C, Muffato M, Collins JE, Humphray S, McLaren K, Matthews L, et al. 2013. The zebrafish reference genome sequence and its relationship to the human genome. Nature 496: 498–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt BG, Glastad KM, Yi SV, Goodisman MAD 2013. Patterning and regulatory associations of DNA methylation are mirrored by histone modifications in insects. Genome Biol Evol 5: 591–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong S, Rebeiz M, Andolfatto P, Werner T, True J, Carroll SB 2008. The evolution of gene regulation underlies a morphological difference between two Drosophila sister species. Cell 132: 783–793 [DOI] [PubMed] [Google Scholar]
- Jones PA 2012. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat Rev Genet 13: 484–492 [DOI] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. 2010. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467: 430–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM 2010. Widespread transcription at neuronal activity-regulated enhancers. Nature 465: 182–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirmizis A, Santos-Rosa H, Penkett CJ, Singer MA, Vermeulen M, Mann M, Bähler J, Green RD, Kouzarides T 2007. Arginine methylation at histone H3R2 controls deposition of H3K4 trimethylation. Nature 449: 928–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusserow A, Pang K, Sturm C, Hrouda M, Lentfer J, Schmidt HA, Technau U, von Haeseler A, Hobmayer B, Martindale MQ, et al. 2005. Unexpected complexity of the Wnt gene family in a sea anemone. Nature 433: 156–160 [DOI] [PubMed] [Google Scholar]
- Levin JZ, Yassour M, Adiconis X, Nusbaum C, Thompson DA, Friedman N, Gnirke A, Regev A 2010. Comprehensive comparative analysis of strand-specific RNA sequencing methods. Nat Methods 7: 709–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M 2010. Transcriptional enhancers in animal development and evolution. Curr Biol 20: R754–R763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CL, Kaplan T, Kim M, Buratowski S, Schreiber SL, Friedman N, Rando OJ 2005. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol 3: e328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long HK, Sims D, Heger A, Blackledge NP, Kutter C, Wright ML, Grützner F, Odom DT, Patient R, Ponting CP, et al. 2012. Epigenetic conservation at gene regulatory elements revealed by non-methylated DNA profiling in seven vertebrates. eLife 2: e00348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maltby VE, Martin BJE, Brind’amour J, Chruscicki AT, McBurney KL, Schulze JM, Johnson IJ, Hills M, Hentrich T, Kobor MS, et al. 2012. Histone H3K4 demethylation is negatively regulated by histone H3 acetylation in Saccharomyces cerevisiae. Proc Natl Acad Sci 109: 18505–18510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martindale MQ, Pang K, Finnerty JR 2004. Investigating the origins of triploblasty: ‘mesodermal’ gene expression in a diploblastic animal, the sea anemone Nematostella vectensis (phylum, Cnidaria; class, Anthozoa). Development 131: 2463–2474 [DOI] [PubMed] [Google Scholar]
- May D, Blow MJ, Kaplan T, McCulley DJ, Jensen BC, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, et al. 2011. Large-scale discovery of enhancers from human heart tissue. Nat Genet 44: 89–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. 2008. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454: 766–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkenschlager M, Odom DT 2013. CTCF and cohesin: Linking gene regulatory elements with their targets. Cell 152: 1285–1297 [DOI] [PubMed] [Google Scholar]
- The modENCODE Consortium, Roy S, Ernst J, Kharchenko PV, Kheradpour P, Negre N, Eaton ML, Landolin JM, Bristow CA, Ma L, et al. 2010. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330: 1787–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran Y, Fredman D, Praher D, Li XZ, Wee LM, Rentzsch F, Zamore PD, Technau U, Seitz H 2014. Cnidarian microRNAs frequently regulate targets by cleavage. Genome Res (this issue). doi: 10.1101/gr.162503.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanty L, Carbajosa G, Heap GA, Ratnieks F, van Heel DA, Down TA, Rakyan VK 2011. Comparative methylomics reveals gene-body H3K36me3 in Drosophila predicts DNA methylation and CpG landscapes in other invertebrates. Genome Res 21: 1841–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nègre N, Brown CD, Ma L, Bristow CA, Miller SW, Wagner U, Kheradpour P, Eaton ML, Loriaux P, Sealfon R, et al. 2011. A cis-regulatory map of the Drosophila genome. Nature 471: 527–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neph S, Vierstra J, Stergachis AB, Reynolds AP, Haugen E, Vernot B, Thurman RE, John S, Sandstrom R, Johnson AK, et al. 2013. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature 488: 83–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi SKT, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin S-P, Allis CD, et al. 2007. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448: 714–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli A, Althoff F, Oliveira RA, Heidmann S, Schuldiner O, Lehner CF, Dickson BJ, Nasmyth K 2008. Cell-type-specific TEV protease cleavage reveals cohesin functions in Drosophila neurons. Dev Cell 14: 239–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JE, Corces VG 2009. CTCF: Master weaver of the genome. Cell 137: 1194–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam NH, Srivastava M, Hellsten U, Dirks B, Chapman J, Salamov A, Terry A, Shapiro H, Lindquist E, Kapitonov VV, et al. 2007. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science 317: 86–94 [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM 2010. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team 2012. R: A language and environment for statistical computing R Foundation for Statistical Computing, Vienna, Austria.
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J 2010. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470: 279–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando OJ, Chang HY 2009. Genome-wide views of chromatin structure. Annu Rev Biochem 78: 245–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebeiz M, Pool JE, Kassner VA, Aquadro CF, Carroll SB 2009. Stepwise modification of a modular enhancer underlies adaptation in a Drosophila population. Science 326: 1663–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renfer E, Amon-Hassenzahl A, Steinmetz PRH, Technau U 2010. A muscle-specific transgenic reporter line of the sea anemone, Nematostella vectensis. Proc Natl Acad Sci 107: 104–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee HS, Pugh BF 2011. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell 147: 1408–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan JF, Mazza ME, Pang K, Matus DQ, Baxevanis AD, Martindale MQ, Finnerty JR 2007. Pre-bilaterian origins of the Hox cluster and the Hox code: Evidence from the sea anemone, Nematostella vectensis. PLoS ONE 2: e153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarda S, Zeng J, Hunt BG, Yi SV 2012. The evolution of invertebrate gene body methylation. Mol Biol Evol 29: 1907–1916 [DOI] [PubMed] [Google Scholar]
- Schmidt D, Schwalie PC, Ross-Innes CS, Hurtado A, Brown GD, Carroll JS, Flicek P, Odom DT 2010. A CTCF-independent role for cohesin in tissue-specific transcription. Genome Res 20: 578–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner O, Berdnik D, Levy JM, Wu JS, Luginbuhl D, Gontang AC, Luo L 2008. piggyBac-based mosaic screen identifies a postmitotic function for cohesin in regulating developmental axon pruning. Dev Cell 14: 227–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitan VC, Hao B, Tachibana-Konwalski K, Lavagnolli T, Mira-Bontenbal H, Brown KE, Teng G, Carroll T, Terry A, Horan K, et al. 2011. A role for cohesin in T-cell-receptor rearrangement and thymocyte differentiation. Nature 476: 467–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, et al. 2012. A map of the cis-regulatory sequences in the mouse genome. Nature 488: 116–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F, Furlong EEM 2012. Transcription factors: From enhancer binding to developmental control. Nat Rev Genet 13: 613–626 [DOI] [PubMed] [Google Scholar]
- Steele RE, David CN, Technau U 2011. A genomic view of 500 million years of cnidarian evolution. Trends Genet 27: 7–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sucena E, Delon I, Jones I, Payre F, Stern DL 2003. Regulatory evolution of shavenbaby/ovo underlies multiple cases of morphological parallelism. Nature 424: 935–938 [DOI] [PubMed] [Google Scholar]
- Suzuki MM, Bird A 2008. DNA methylation landscapes: Provocative insights from epigenomics. Nat Rev Genet 9: 465–476 [DOI] [PubMed] [Google Scholar]
- Technau U, Steele RE 2011. Evolutionary crossroads in developmental biology: Cnidaria. Development 138: 1447–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Technau U, Rudd S, Maxwell P, Gordon PMK, Saina M, Grasso LC, Hayward DC, Sensen CW, Saint R, Holstein TW, et al. 2005. Maintenance of ancestral complexity and non-metazoan genes in two basal cnidarians. Trends Genet 21: 633–639 [DOI] [PubMed] [Google Scholar]
- Thomson JP, Skene PJ, Selfridge J, Clouaire T, Guy J, Webb S, Kerr ARW, Deaton A, Andrews R, James KD, et al. 2010. CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature 464: 1082–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Prabhakar S, Akiyama JA, Shoukry M, Lewis KD, Holt A, Plajzer-Frick I, Afzal V, Rubin EM, Pennacchio LA 2008. Ultraconservation identifies a small subset of extremely constrained developmental enhancers. Nat Genet 40: 158–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. 2009a. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457: 854–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Rubin EM, Pennacchio LA 2009b. Genomic views of distant-acting enhancers. Nature 461: 199–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Taher L, Girgis H, May D, Golonzhka O, Hoch RV, McKinsey GL, Pattabiraman K, Silberberg SN, Blow MJ, et al. 2013. A high-resolution enhancer atlas of the developing telencephalon. Cell 152: 895–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, et al. 2011. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472: 120–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp PJ, Kalay G 2011. Cis-regulatory elements: Molecular mechanisms and evolutionary processes underlying divergence. Nat Rev Genet 13: 59–69 [DOI] [PubMed] [Google Scholar]
- Woolfe A, Goodson M, Goode DK, Snell P, McEwen GK, Vavouri T, Smith SF, North P, Callaway H, Kelly K, et al. 2005. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol 3: e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S, Xie D, Cao X, Yu P, Xing X, Chen C-C, Musselman M, Xie M, West FD, Lewin HA, et al. 2012. Comparative epigenomic annotation of regulatory DNA. Cell 149: 1381–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yáñez-Cuna JO, Kvon EZ, Stark A 2012. Deciphering the transcriptional cis-regulatory code. Trends Genet 28: 11–22 [DOI] [PubMed] [Google Scholar]
- Zemach A, Zilberman D 2010. Evolution of eukaryotic DNA methylation and the pursuit of safer sex. Curr Biol 20: R780–R785 [DOI] [PubMed] [Google Scholar]
- Zemach A, McDaniel IE, Silva P, Zilberman D 2010. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 328: 916–919 [DOI] [PubMed] [Google Scholar]