Summary
Loss or duplication of chromosome segments can lead to further genomic changes associated with cancer. However, it is not known if only a select subset of genes is responsible for driving further changes. To determine if perturbation of any given gene in a genome suffices to drive subsequent genetic changes, we analyzed the yeast knockout collection for secondary mutations of functional consequence. Unlike wild type, most gene knockout strains were found to have one additional mutant gene affecting nutrient responses and/or heat-stress-induced cell death. Moreover, independent knockouts of the same gene often evolved mutations in the same secondary gene. Genome sequencing identified acquired mutations in several human tumor suppressor homologs. Thus, mutation of any single gene may cause a genomic imbalance with consequences sufficient to drive adaptive genetic changes. This complicates genetic analyses, but is a logical consequence of losing a functional unit originally acquired under pressure during evolution.
Introduction
DNA copy number changes of whole chromosomes, chromosome segments or individual genes can lead to further genomic changes that eventually facilitate beneficial adaptation of the species. However, this process is also thought to underlie cancer (Gordon et al., 2012; Tang and Amon, 2013). Presently, it is not known if a copy number change for all or only the few genes already identified is chiefly responsible for tumorigenesis and other genetic disorders. The phenomenon where one genomic event leads to further genomic changes potentially extends to knockout model organisms bearing single engineered gene deletions designed to gain new insight into gene function. If a functional mutation of any one gene in a genome will drive the selection for new mutations under normal conditions (without deliberately applied selection pressures), the phenotypes of knockout mice, flies and yeast may not directly or fully reflect the deleted gene, potentially confounding interpretations. Yet the full extent of compensatory changes and whether they are genetic or epigenetic are generally unknown. This effort is challenged by difficulties with distinguishing relevant changes from among a larger number of genetic polymorphisms and non-genetic events. These problems are magnified for larger genomes. Human genomics continues to reveal rare mutations unique to individuals and to subsets of cells within an individual (Abecasis et al., 2012). Further from our grasp is a comprehensive understanding of the relative contributions of cumulative genetic changes.
Individual cells from a presumed isogenic population of mammalian or yeast cells can exhibit distinct growth/death rates. Variant behaviors between sister cells can be attributed to stochastic fluctuations in protein expression levels, such as apoptosis mediators in HeLa cells, and yeast master control genes that drive fluctuations in expression of many other genes (Albeck et al., 2008; Levy et al., 2012; Newman et al., 2006; Spencer et al., 2009; Stewart-Ornstein et al., 2012). Consistent with epigenetic causes of cell-cell variation, predicted DNA mutation rates seem insufficient to explain the variant behaviors of two sister cells (Lynch et al., 2008; Qian et al., 2012). This does not however, preclude the possibility that presumed isogenic cell populations have accumulated meaningful genetic differences between cells that are currently underappreciated. A basal error rate of 10−9 per base pair per generation in yeast (corresponding to 107 mutations in 30 generations, or 1 mutation/genome in every 100–200 divisions) quickly leads to every mutation in at least one genome in a large population (Boer et al., 2008; Dunham et al., 2002; Gresham et al., 2008; Lynch et al., 2008). This is consistent with clonal cell mosaicism in human development, in aging and in cancer (Jacobs et al., 2012; Laurie et al., 2012; Liu et al., 2011). However, it can be challenging to pinpoint specific causal genetic differences between cells despite increasingly powerful sequencing technologies (Zong et al., 2012). Furthermore, non-genetic causes of variant phenotypes can persist for several cell divisions, and some perdure even through meiosis (Greer et al., 2011; Nakayama et al., 2000; Pillus and Rine, 1989; Wheeler et al., 2012), although genomes were not sequenced in this case.
Genome-wide knockout collections for Saccharomyces cerevisiae have 80–95% of genes systematically deleted individually and replaced with a bar-coded antibiotic resistance cassette KanMX (Giaever et al., 2002) (Figure 1A). It was originally assumed that deletion strains lacking single non-essential genes would be largely genetically stable under standard laboratory conditions, as there are no obvious growth differences for 85% of single gene knockout strains (Giaever et al., 2002; Goebl and Petes, 1986). However, large genomic perturbations unlinked to the knockout gene were estimated to occur in ~8% of knockout strains of yeast, raising the possibility that genome perturbation is a consequence of engineered gene deletions (Hughes et al., 2000). Sensitive multi-day competition assays detected small fitness defects in 45% of haploid YKO strains, suggesting that individual non-essential genes may be more important than previously assumed (Breslow et al., 2008; Hillenmeyer et al., 2008). If deletion of any non-essential gene has detectable consequences, as suggested by these sensitive fitness tests, then the disturbance caused by any gene knockout may also drive the selection for compensatory genetic alterations even without strong environmental pressures. Thus, just as environmental conditions can drive genome evolution (Barrick et al., 2009; Dettman et al., 2007; Gresham et al., 2008), genome evolution might also be inevitable following mutation/deletion of any single gene, given that genomes have been optimized to coexist under selection for millennia.
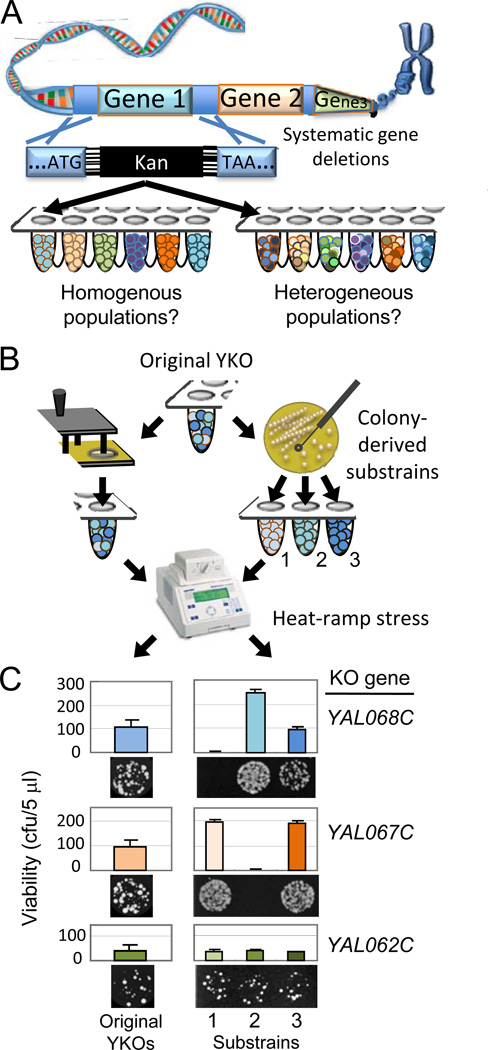
Figure 1. Heat-ramp stress test detects phenotypic variation within knockout strains.
(A) Potential heterogeneity within individual knockout strains in the yeast knockout (YKO) collection consisting of ~5,000 unique single-gene deletion strains in which non-essential gene were replaced with a bar-coded kanamycin resistance cassette.
(B) Original knockout strains (BY MATa) and their single-cell-derived substrains were heat-stressed using a programmable thermocycler.
(C) Yeast viability was assessed as colony-forming units (cfu per 5 µl starting concentration) following heat-ramp treatment. Data are presented as mean ±SD for 3 independent experiments (all data generated are presented). See also Figure S1.
Although suppressor screens applying environmental selection pressures are fundamental tools in genetics (Boer et al., 2008; Dunham et al., 2002; Gresham et al., 2008), compensatory mutations that arise under normal conditions (without deliberate selection pressures) are rarely documented in knockout strains of mice or yeast (Cheng et al., 2008; Game et al., 2003; Lapinskas et al., 1995; Torres et al., 2010; Zheng et al., 2000). Thus, the impact on genome evolution in eukaryotes driven by genome-wide deletions of single genes has not been systematically explored. To determine the consequences of single gene deletions, we used the tractable yeast knockout collection, and found that the majority of haploid knockout strains are phenotypically and genetically heterogeneous. Such heterogeneity is often dismissed as experimental variation, stochastic evolution or erroneous anomalies such as the pressures of kanamycin selection or the mechanics of knockout construction. However, we present evidence indicating that the loss of most individual genes in a genome results in a genomic imbalance capable of driving the selection for new mutations. These new mutations are not general survival mutations that occurred in wild type, but instead appear to be tailored to the original knockout gene, as they often recur in an independent knockout of the same gene. Functionally similar secondary mutations are significantly more likely to recur in another knockout of the same gene than in any other gene knockout strain. If an analogous process exists in tumorigenesis, our findings predict that mutations rarely encountered in other patients drive the selection for prevalent cancer mutations. Unlike human tumors, the first mutation in yeast is known (the engineered knockout gene), facilitating efforts to connect first and evolved secondary mutations.
Results
Heterogeneous stress responses within knockout strains
If deletion of any single gene is sufficient to impact genome evolution in the absence of deliberate selection pressures, then knockout strains may have become genetically heterogeneous (Figure 1A). To investigate this possibility, we developed a sensitive survival assay (heat-ramp delivered by a programmable thermocycler) to detect heterogeneous responses to stress within individual Saccharomyces cerevisiae knockout strains (Figure S1) (Teng et al., 2011; Teng and Hardwick, 2013). The first three verified knockout strains listed in the BY MATa yeast knockout (YKO) collection were streaked onto agar plates, and three single-cell-derived colonies from each were grown in density-matched cultures and heat-ramp treated (Figure 1B). Viability (colony counts) varied greatly between colony-derived substrains for two of the three unique knockout strains (Figure 1C). These variant phenotypes were confirmed by retesting the same substrains, indicating biological rather than technical variation. This variation was masked when analyzing the corresponding (heterogeneous) parental strains (retrieved with a pin tool from the original frozen archive), which yielded a phenotype approximating the average of their substrains, except with larger standard deviations (Figure 1C and Figure S1). In practice, variant phenotypes among clonal substrains are relatively common occurrences with yeast and mammals alike, which often can be avoided by using pooled subjects. However, this strategy is unreliable as it may hide underlying genetic complexity (see below).
To estimate the frequency of heterogeneous knockout strains, a random number generator was used to select 250 unique knockout strains from the YKO collection (BY MATa). Six substrains derived from morphologically indistinguishable colonies of each were tested using the same heat-ramp stress test (Figure 2A, B). Again, we observed striking heterogeneity among substrains derived from the same original knockout strain. Of the 250 unique strains, 105 (42%) had at least one colony-derived substrain with strong phenotypic deviation (Table S1). Unlike stochastic fluctuations in gene expression, these variant phenotypes were remarkably reproducible when frozen stocks of the same six substrains were retested months later (Figure 2B). Substrain variation is not due to unequal starting cell numbers, and was only revealed by applying stress, as the same substrain cultures were indistinguishable prior to treatment. By analyzing only six substrains, some heterogeneous YKO strains will be missed, therefore a statistical model was applied, which estimates that 56% of all YKO strains contain 22–78% variant cells (Figure 2C and Supplemental Experimental Procedures). In contrast to knockout strains, wild type strains from different sources, including parental strains of the YKO collections, had no phenotypic variation among 26 colony-derived substrains tested per strain (Figure 2D and Figure S2). Thus, the heritable variations within individual knockout strains appear to be stable cell-cell differences that arose as a consequence of the engineered knockout and prior to our tests.
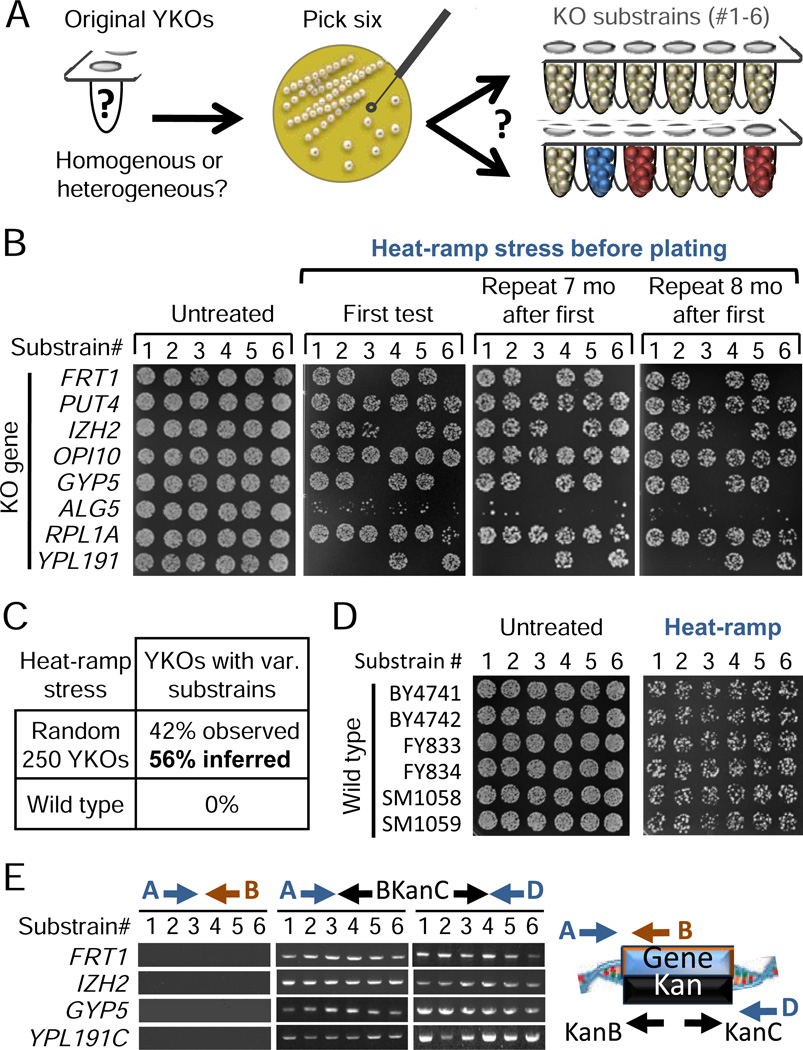
Figure 2. Widespread heterogeneity within knockout strains.
(A) Six morphologically indistinguishable colonies from each of 250 randomly selected knockout strains (BY MATa) were archived as 1,500 substrains.
(B) Example results of heat-ramp stress tests on colony-derived substrains from the 250 randomly selected YKOs before and after treatment in 3 independent experiments; first test performed prior to freezing substrains. Colony counts differed significantly between substrains from BY MATa YKOs Δfrt1, Δizh2, Δgyp5, Δrpl1A, and Δypl191c (ANOVA, p<10−5). Variant Δypl191c substrain #6 is an example false negative as it was below the cut-off threshold (set to avoid false positives in this screen).
(C) Summary of observed results from all experiments performed. Inferred proportion was estimated using a mathematical model. See Supplemental Experimental Procedures.
(D) Wild type substrains were analyzed as for knockout substrains in panel B.
(E) Example PCR genotyping results of variant substrains from panel B.
See also Figure S2, Tables S1 and S2, and Supplemental Experimental Procedures.
PCR genotyping verified that variant phenotypes are not due to experimental mishaps or cross contamination between knockout strains. All 88 substrains (from 20 unique knockout strains), each tested with three primer sets to verify deletion of the correct gene and insertion junctions of the KanMX cassette, yielded the expected results without exception (Figure 2E and Table S2). A wide variety of other assays such as spot sequencing of molecular barcodes has confirmed the identity of >95% of the YKOs in this collection (not shown).
Nutrient-sensing heterogeneity within knockout strains
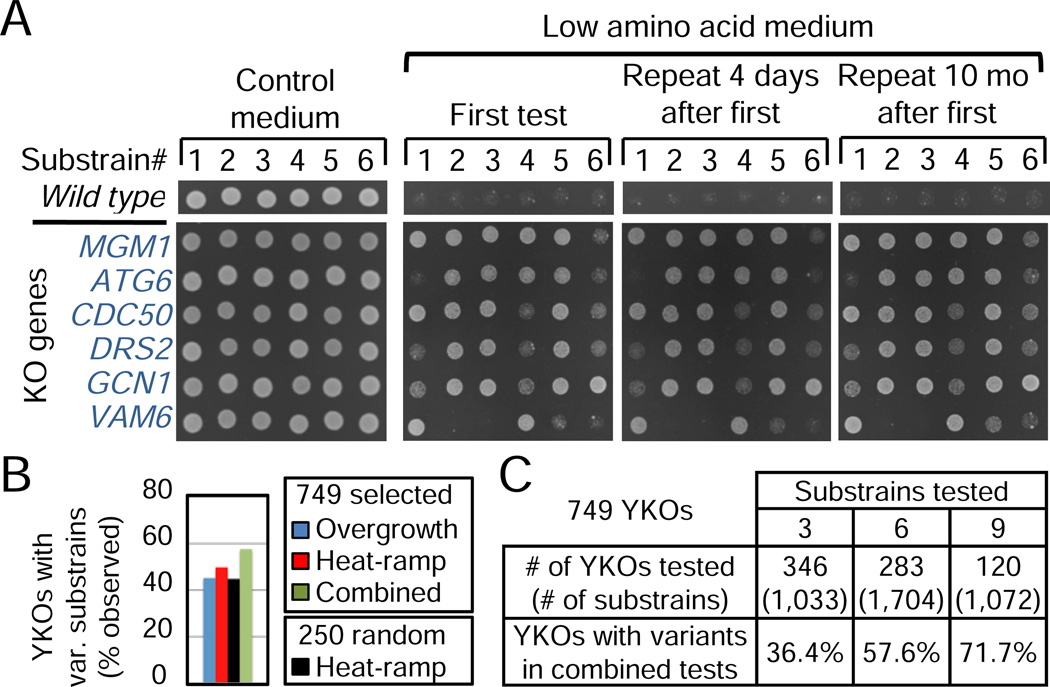
The portion of knockout strains with heterogeneous memberships was likely underestimated above by using a single assay to define variation. To address this caveat, we used a sensitive assay to detect strains having a growth advantage over wild type when nutrients are reduced (Cheng et al., 2008). Overgrowth on low amino acid medium (SCDME, Table S3) is a characteristic of knockouts of the mitochondrial fission gene FIS1, but is actually due to a secondary mutation in the WHI2 gene (Cheng et al., 2008; Fannjiang et al., 2004). Therefore, we screened the entire YKO collection (4,847 BY MATa strains pinned from original frozen stocks) and identified 749 unique strains that overgrow compared to wild type when plated on low amino acid medium without other stimuli. To assess the possibility of secondary mutations in these 749 YKOs, 3–9 colony-derived substrains from each (3,809 substrains) were tested for overgrowth, identifying 44.9% (testing 6 substrains each) with obvious substrain variation (Figure 3A and Table S4). Again, variant phenotypes were highly reproducible in subsequent tests, and this variation was not detected on control medium spotted in parallel (SCDCSH) nor for wild type on either medium (see also Figure 7A).
Figure 3. Variant growth phenotypes of knockout strains in low amino acids.
(A) Variant overgrowth phenotypes among substrains from example YKO strains in the 749 group (BY MATa, see text) plated simultaneously on control (SCDCSH) and low amino acid media (SCDME). The first test was performed before freezing substrain stocks.
(B) Percent (observed) of original YKO strains with at least one substrain among six tested with obvious variation for indicated phenotypes. Observed results from Figure 2C are plotted for direct comparison (black bar).
(C) Summary of results from heat-ramp stress and overgrowth assays for 3,809 substrains of the 749 original YKOs. Rationale for the number of substrains tested is found in Supplemental Experimental Procedures; groupings do not overlap and were grouped without regard to substrain variation. See also Tables S3 and S4.
Figure 7. Model of genome evolution driven by gene mutation.
(A) Most yeast knockout strains are quasispecies harboring prevalent additional mutations.
(B) Though there are potentially several evolutionary paths to compensate for the loss of any one gene, independently constructed knockouts of the same gene tend to evolve similar phenotypes, often by acquiring secondary mutations in the same gene (parallel evolution), indicating a selection process driven by the specific knockout.
(C) Deletion of different genes can drive the selection of mutations in the same genes.
The same 3,809 substrains were simultaneously evaluated in the heat-ramp stress test, revealing variant substrains in 49.8% of the 749 YKOs (testing 6 substrains), only 7.8% higher than for the randomly selected YKO strains (Figure 3B). Both stress and growth phenotypes tended to co-occur, but not concordantly so, and 71.7% of YKOs had variant substrains in one or both types of assays when 9 substrains were tested (Figure 3C). This frequency will still be underestimated if secondary mutations have become fixed in the population, thereby abolishing phenotype variation among substrains (see below).
Genetic changes explain variant phenotypes
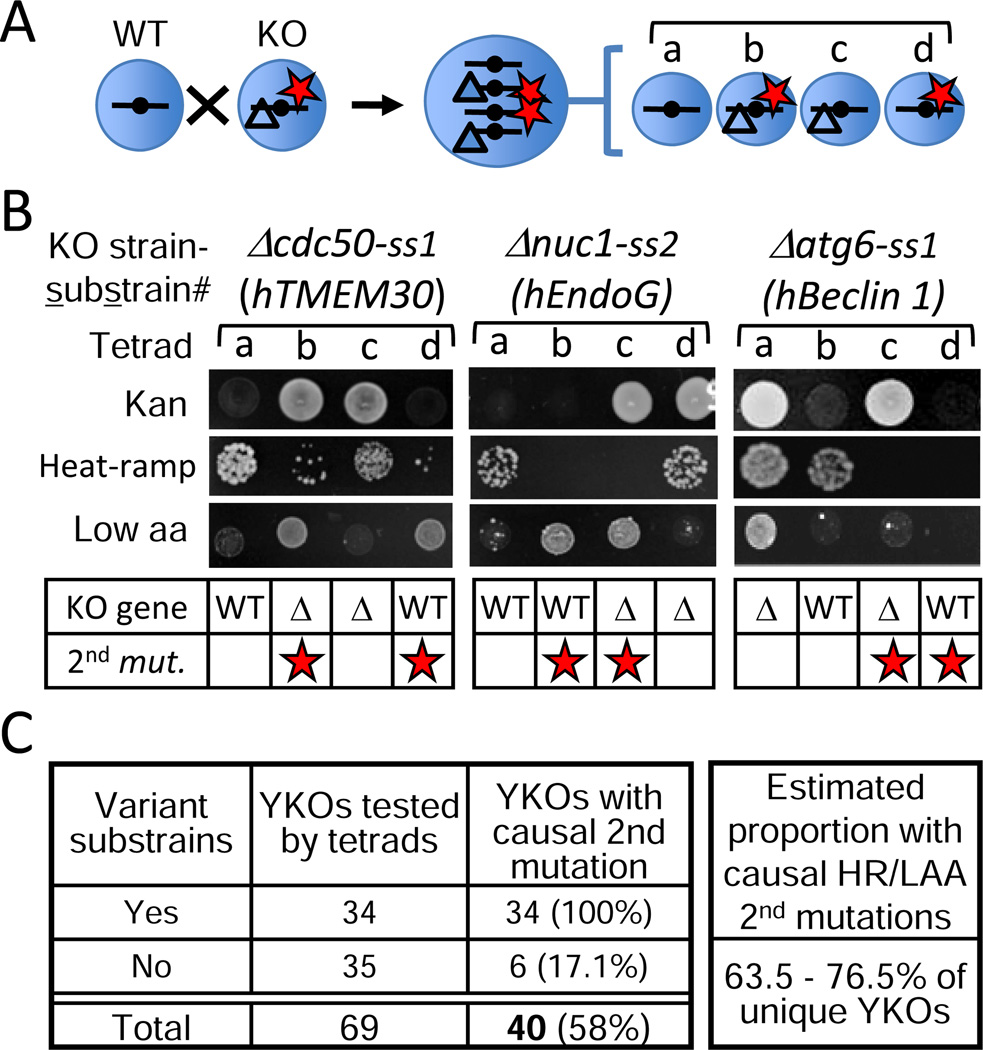
To establish whether substrain variation is caused by genetic changes, backcrossing and tetrad analysis was used to allow secondary mutations to segregate independently from the knockout gene (Figure 4A). Heat-ramp and overgrowth phenotypes were determined for 10–20 validated tetrads for each of 69 YKO strains (BY MATa strains with available tetrads from all other lab projects irrespective of strain variation, including 65 of the 749 YKOs with overgrowth). As predicted, all of the YKOs with substrain variation [34 of 69 unique strains (49%), including 2 of 4 not from the 749 group] had a single unlinked secondary mutation (inferred by 2:2 segregation within tetrads), except four strains with two secondary mutations (independent mutations for heat-stress and overgrowth phenotypes) (Figure 4B and Table S5). Thus, the presence or absence of each phenotype genetically segregated with specific genetic loci distinct from the knockout. Tetrad analysis on parental wild type strains failed to reveal any secondary mutations, although evolution of wild type strains in culture has been documented (Lang et al., 2013; Zeyl, 2005).
Figure 4. Prevalence of partially/fully fixed secondary mutations in knockout populations.
(A) Diagram of a tetratype tetrad in which the knockout (Δ) and second mutant gene (red star) segregate independently.
(B) Example results for tetrads (tetratypes) produced from substrains with second gene mutations affecting overgrowth and heat-stress phenotypes that segregate independently of the knockout locus conferring kanamycin resistance (Kan).
(C) Number of original knockouts (BY MATa) with/without variant substrains that have a second mutation responsible for heat ramp-sensitive (HR) and low amino acid overgrowth (LAA) phenotypes based on tetrad analysis. Final frequency estimates: 17.1% of YKOs with invariant substrains plus the percent of YKOs with variant substrains from Figure 2 [(44%×17.1%)+56%] and Figure 3 [(28.3%×17.1%)+71.7%]. See also Table S5.
While substrain variation reliably identified strains with secondary mutations, this strategy will miss those with secondary mutations that previously became fixed in the whole population. To estimate this proportion of knockout strains, tetrads were analyzed for the remaining 35 of 69 YKO strains that lacked variant substrains, revealing that 17.1% (including one not from the 749 group) have a fixed secondary mutation (Figure 4C and Table S5). This further increases the estimated proportion of knockout strains with meaningful secondary mutations from figures 2C and 3C (4C). This is still likely an underestimate because only two types of assays were used, and because some secondary mutations are masked. For example, overgrowth by autophagy-defective Δatg6 is caused by loss of ATG6 itself (human Beclin 1 homolog), while the secondary mutation restores normal (slower) growth (Figure 4B). This secondary mutation, which lacks obvious benefit, was identified by sequencing the genomes of two substrains exhibiting the overgrowth-suppressor phenotype. Both had the same premature stop near the C-terminus of IRA1, the yeast homolog of human tumor suppressor NF1 (neurofibromin) (Table S6, see Figure 6E).
Figure 6. Knockout-driven evolution by the same and different paths.
(A) Example DNA sequence chromatograms of independently acquired secondary mutations in WHI2 in each of two independently constructed knockout strains of STE20.
(B) Demonstration that the secondary gene/locus and corresponding phenotypes segregate independently from the knockout locus in knockout substrains.
(C) DNA sequence results for strains with WHI2 mutations.
(D) Model depicting influences of gene deletion versus environmental conditions on genome evolution.
(E) Identification of secondary mutations by whole genome sequencing. All mutations unique to the two phenotypically similar spore-derived substrains are shown. Additional intergenic mutations and a mutation in mitochondria-encoded ATP6, which is not expected to segregate 2:2 in tetrads, are unlikely to be of consequence. Presumed passenger mutations identified by genome sequencing are summarized in Table S6.
Knockout-driven parallel evolution in independent strains
Unexpectedly, the tetrad analyses above revealed that heat-sensitivity (with or without overgrowth phenotypes) were typically due to the secondary mutation rather than the knockout (Table S5). Although increased cell death may be a trade-off for some other advantage, inconsequential passenger mutations can also become prominent in a cell population by chance. Therefore, we asked if the biological impact of losing any specific gene can drive specific secondary mutations. If the selection mechanism is specified by the gene deleted, then independently constructed knockouts lacking the same gene would be expected to evolve similarly. To explore this possibility, we analyzed a separate set of 46 independently constructed strain pairs lacking the same gene (BY MATa and BY MATα YKO collections) and observed a significant correlation for the presence or absence of variant substrains using the heat-stress and overgrowth assays (Fisher’s exact test p=0.00124, Table S7). To provide more definitive evidence for parallel evolution, we took advantage of the tetrad analyses above, which distinguished the phenotypes due to knockout versus secondary mutations. Knowing the phenotypes caused by each secondary mutation, we asked if an independently constructed knockout has acquired the same phenotype. For the 40 YKO strains (BY MATa) with secondary mutations (from Figure 4C and Figure 5A), we analyzed independently engineered knockout strains of the same 40 genes (BY MATalpha). Of these, 26 had at least one substrain with phenotypes matching the corresponding secondary mutation (Figure 5B and 5C, and Table S5). Based on the probability of finding these specific phenotypes (e.g. heat-sensitive vs. heat-resistant) in the YKO collection, such co-occurrences are highly improbable by random chance (Figure 5B and Supplemental Experimental Procedures). Furthermore, these 26 knockouts are distributed across the genome, and were constructed using different protocols in 11 different laboratories from 6 different countries, arguing against some types of systematic selection pressures (Table S5).
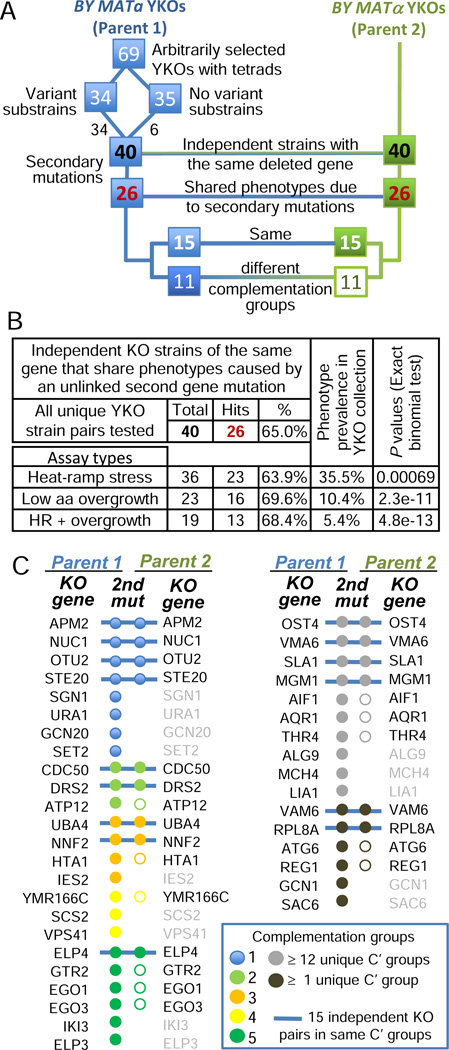
Figure 5. Recurrent secondary mutant genes indicate knockout-driven selection.
(A) Flow chart for the 40 pairs of independent knockout strains tested by complementation.
(B) Frequency of knockout strain pairs with the same deleted gene in which both strains (BY MATa and BY MATα) have substrains bearing the same phenotype constellations known to be caused by a secondary mutation (from Figure 4).
(C) Summary of complementation tests (analysis of diploids after mating substrains with the indicated BY MATa and BY MATα YKOs). Of the 40 YKO pairs with secondary mutations, both partners of 26 pairs share the same secondary phenotypes (black gene names for Parent 2), 15 of which contain secondary mutations in the same gene or complementation group (blue bars), while the remaining 11 define two different complementation groups (open circles). Complementation groups shared by >1 BY MATa strain are color-coded (brightly colored solid circles); secondary mutations in unique complementation groups among all BY MATa strains, or that are shared only by their corresponding MATalpha strain (gray solid circles); overgrowth-suppressor phenotypes not testable by complementation between unique MATa strains (black filled circles). See also Figure S4, Table S5, Table S6, and Supplemental Experimental Procedures.
This analysis was extended to determine if two independent strains lacking the same KO gene have evolved mutations in the same secondary gene. This was assessed by genetic complementation assays (mating the two members of each pair) for the 26 paired YKO strains with matching secondary phenotypes (from Figure 5A). Remarkably, 15 of these 26 knockout pairs failed to complement, suggesting shared secondary mutant genes, or mutations in genes coding for components of a functional complex (blue bars in Figure 5C, Table S5). Importantly, these 15 knockout strains (15 different KO genes, BY MATa) have secondary mutations in at least 10 different unlinked genes (see below). These findings indicate that the specific gene knockout was a critical factor in driving genome evolution (see Figure 7B).
Knockout-driven evolution via distinct paths
While parallel evolution often occurred in strains bearing the same knockout gene (15 of 26 paired YKOs from Figure 5), the remaining 11 YKO pairs have apparently evolved by different paths to arrive at the same phenotypes (see Figure 7B). Similarly, we found that secondary mutations in two different genes explain the overgrowth phenotypes of four different FIS1 knockout strains. Three of these four strains (Δfis1-d1 and Δfis1-d2 generated with KanMX insertion and G418 selection, and Δfis1-d3 with HIS3 selection) belong to the same complementation group and have evolved unique WHI2 mutations (Cheng et al., 2008) (Figure S3). Whole genome sequencing of the fourth FIS1 knockout (Δfis1-d4, with URA3 selection) identified a nonsense mutation in SIN3, which encodes a conserved protein deacetylase that regulates transcription and promotes autophagy (Bartholomew et al., 2012; van Oevelen et al., 2008) (Figure S3, see also Figure 6E). A causal role for this SIN3 mutation was confirmed by wild type SIN3 plasmid rescue, restoring normal (slower) growth rates on low amino acids (Figure S3).
In the converse situation, knockouts of different genes may evolve the same secondary mutant genes, resembling common tumor suppressor genes in cancer (see Figure 7C). To verify this event, we estimated the number of different secondary mutant genes that exist in the 40 YKO strains bearing secondary mutations (BY MATa, from Figure 5) using complementation assays and sequencing. Each strain was crossed with the others, revealing that 24 of 40 unique YKO strains (BY MATa) have secondary mutations belonging to only 5 complementation groups, indicative of convergent evolution (Figure 5C and Figure S4). Furthermore, knockouts of different components of the same protein complexes (Cdc50 and Drs2; Elp3, Elp4 and Iki3/Elp1; Gtr2, Ego1/Meh1 and Ego3/Slm4), all of which are encoded on different chromosomes (except Elp3 and Elp4), have evolved mutations in the same complementation groups, indicating that loss-of-function was an important driver of secondary mutations. Of the 40 unique YKOs tested (BY MATa), 14 have at least one substrain exhibiting whi2-like phenotypes (strong overgrowth on low amino acids and sensitivity to heat-ramp–induced death), 8 of which are in the whi2 complementation group (group 1 of Figure 5C, Table S6). DNA sequencing of the WHI2 gene identified unique mutations in all 8 (deletions of STE20, NUC1, APM2, OTU2, GCN20, SET2, URA1 and SGN1), demonstrating independent evolution (Figure 6A–C, Figure S3B). No WHI2 mutations were found in substrains lacking whi2-like phenotypes, nor in the sequenced genomes of four wild type parental strains (Supplemental Experimental Procedures).
This conglomerate of 8 different gene knockouts harboring secondary WHI2 mutations suggests that environmental pressures have contributed to genome evolution, such as depletion of amino acids during normal culturing. While this is likely an important factor, a role for these specific genes, when deleted, in driving the selection for WHI2 mutations is supported by analysis of independently derived knockouts of the same 8 genes (BY MATalpha). Independent knockouts of 4 of these 8 have acquired additional unique WHI2 mutations (Figure 6C), verifying complementation tests (Figure 5C). Thus, the probability of a WHI2 mutation occurring in a knockout of the same gene (50% in this case) is much greater than the frequency of WHI2 mutations in knockouts of different genes (est. 2.9% per strain). Under comparable environmental conditions, loss of each specific gene function applies unique pressures driving the selection for new mutations, such that the selection coefficient over wild type is greater than currently appreciated under normal laboratory growth conditions (Figure 6D).
Potential correlations in human tumors
Mutations in a limited number of human genes (est. 138 tumor suppressors and oncogenes) are suggested to explain the prominent phenotypes of most all human cancers (Vogelstein et al., 2013). Cancer mutations are generally thought to arise through a progression of clonal cell expansions following advantageous mutations, though few such paths have been delineated, and these are primarily drug-resistance mechanisms. The prospects of identifying potentially prognostic early mutations would be of obvious utility, but the task of identifying these mutations, if such pre-drivers exist, faces many challenges including the difficulty of discerning relevant rare mutations from passengers. Extrapolating from yeast survival and growth pathways to the niches of human tumors is also highly uncertain. Nevertheless, we interrogated the available cancer genome database for possible co-occurrences of mutations in human genes homologous to co-occurring mutations found in our sequenced yeast strains (BY MATa deletion strains of ATG6, FIS1, CDC50 and MGM1) (Figure 6E, Figure S3 and Table S6). Although mutations in human TMEM30A (CDC50) are rare, they tended to co-occur with mutations in human ZFYVE16 (PIB2) in colon cancer and uterine carcinomas (p=0.000124 and p=0.000316, respectively, Fisher’s exact test; Figure S5) but not in other tumor types (Cerami et al., 2012). Although no significant co-occurrences were found between the autophagy factor Beclin 1 (ATG6) and the tumor suppressor NF1 (IRA1, a negative regulator of Ras signaling), cooperating mutations that co-occur with NF1 mutations have been identified in neurofibromatosis patients with aggressive malignancies (Mo et al., 2013). Co-occurrences in tumors of mutations in MX2 (MGM1) with UBE4B (UFD2), and FIS1 (FIS1) with SIN3A/B (SIN3) were also significant (Figure 6E and Figure S5).
Mutations in yeast WHI2 were the most common secondary mutations, arising in several different knockout strains. Although WHI2 is reported to be a fungal-specific gene, our HMM-Pred search readily identified mammalian KCTD (potassium channel tetramerization domain) family proteins as sequence homologs (common ancestor) of the yeast Whi2 protein (probability 98.6, E-value 5.1E-08, P = 8.2E-13) (Figure S6). This analysis was aided by the presence of a BTB (bric-a-brac, tramtrak and broad complex) domain in a solved 3D structure of KCTD5 (Dementieva et al., 2009). Although the 25 human KCTD family proteins are understudied or unstudied, several have been linked to human cancers, most notably KCTD11/Ren, a reported tumor suppressor in medulloblastoma (Ferretti et al., 2005). KCTD11 is encoded near TP53 and is frequently deleted upon TP53 loss-of-heterozygosity. However, loss of a single copy of KCTD11 alone is sufficient to contribute importantly to tumorigenesis based on a mouse model of haploinsufficiency (Scuoppo et al., 2012). Yeast SET2 is one of the 8 gene knockouts having a secondary WHI2 mutation (Figure 5C). Mutations in several human KCTD (WHI2) family members were found to co-occur in sequenced tumors with mutations in human SETD2 (SET2), which is among the 138 human cancer genes, p ≤1.5×10-5 (Figure S5).
Discussion
Any perturbation of a genome may drive genome evolution in a predictable manner. This model is strongly supported by our observation of parallel evolution in yeast, where independently constructed knockouts of the same gene evolve mutations in the same second gene or complementation group (Figure 7). Thus, an important driver of secondary mutations is the original loss-of-function mutation, such that deletion of nearly any gene may apply selection pressure specific to the function of each of the ~4,800 different non-essential genes. A prediction of this model is that the majority of single gene-deletion strains of yeast and other species have acquired meaningful secondary mutations. Thus, much of the contempt directed at the yeast knockout collections for presumed technical mishaps or passage history may be partially attributed to natural consequences of deleting a functional gene from a modern genome, which has arrived at its current composition of interacting genes after millions of years of optimization. We applied two assays designed to detect two critical selection criteria, avoidance of cell death and the ability to grow by ignoring signals that warn of impending nutrient depletion. Although potentially counterintuitive, but consistent with tumor biology, many secondary mutations increased rather than decreased sensitivity to cell death. Mutations identified by these strategies likely represent only a portion of the entire repertoire. The pervasiveness of meaningful secondary mutations in the yeast knockout collections complicates genetic epistasis analyses (which presumes isogenic strains), and encourages further scrutiny when attributing cell-cell differences (any species) to epigenetic or stochastic phenomena. Conversely, secondary mutations may be redeeming if they serve to rescue compromised mutants such as Δfis1 without interfering with the study its mitochondrial fission function. These findings also provide new insight into genome malleability that may enable the delineation of evolutionary paths that possibly occur during tumorigenesis.
The mechanics of deleting a gene, rather than the biological impact of gene deletion, are widely thought to transiently increase mutation rates that tamper with genome integrity though without direct evidence (Supplemental Methods of (Giaever et al., 2002). However, normal basal mutation frequencies provide ample opportunity to accumulate random mutations that dwarf potential experimentally-induced boosts in mutagenesis (Boer et al., 2008; Dunham et al., 2002; Gresham et al., 2008). Another argument against transformation-related mutagenesis in the YKO collections is the single-cell bottleneck. If the secondary mutation came from engineering artifacts, and happened to be in the spore that made the haploid knockout, then these mutations would be fixed in the population. A similar argument applies to YKOs constructed by direct transformation of haploids, which also originated from single colonies. Because most YKO strains are heterogeneous quasispecies, the key secondary mutations presumably arose after formation of the KO genome. However, the 17% of YKOs with fixed secondary mutations could potentially have acquired a rare preexisting mutation from wild type and was selected among the first knockout transformants for its compensatory function. Further passaging without clonal purification (e.g. the YKO collections), can lead to stochastic sweeps of secondary mutations by 200 or more replication cycles even in wild type strains (Lang et al., 2013). Anecdotally, we did not observe variant cell growth/death behaviors among 30 colony-derived substrains in a wild type (BY4741) passaged continuously for 1–2 years (>200 replication cycles), despite an age-dependent decline in viability. However, we assume that the wild type strains used here, which in fact have engineered gene deletions (auxotroph markers), have also undergone compensatory evolution as a consequence of these deletions to reach their current more stable genomic state.
Other factors (beside the gene knockout) may also influence the acquisition of new mutations in knockout strains. Any mutations that impair cell growth may select new mutations for improved growth rates. Indeed strains with compensatory secondary mutations tended to have better reported growth rates (Breslow et al., 2008), though not significantly better (Tables S1 and S5). WHI2 secondary mutations were the most common encountered, yet Δwhi2 itself has particularly slow growth (in rich medium) (Breslow et al., 2008), and is particularly prone to cell death induced by a range of stimuli (Cheng et al., 2008; Ivanovska and Hardwick, 2005). Even though nutrient depletion is an important driver the overgrowth phenotype, secondary mutations in WHI2, SIN3 or PIB2 arose in only a small fraction of knockout strains engineered by the same research teams. The evidence presented supports the model that perhaps any functionally relevant mutation is sufficient to drive genome evolution resulting in the selection for new adaptive mutations even in relatively normal environments.
Experimental Procedures
Clonal substrains
Strain from the yeast knockout (YKO) collections were obtained from frozen glycerol stocks without thawing, streaked onto YPD agar plates and incubated at 30°C for two days. Morphologically indistinguishable colonies were picked for each YKO strain, inoculated into 200 µl liquid YPD in 96-well format, incubated at 30°C for 48 h and frozen with glycerol as archived clonally derived substrain collections. All data generated are presented throughout.
Yeast strains and plasmids
Haploid YKO strains derived from BY MATa (Brachmann et al., 1998) were originally obtained from Research Genetics, replicated once onto YPD, allowed to grow at 30°C for several days and stored as glycerol stocks at −80°C without thawing. BY MATα YKO strains were obtained from R. Rao, Johns Hopkins and D. Lew, Duke University. Substrains used in this study are listed in Tables S1, S2, S4, S5 and S7, other strains and genotypes are listed in Supplemental Experimental Procedures. The SIN3 expression plasmid was provided by David Stillman (University of Utah), and the WHI2 plasmid with native promoter was previously reported (Cheng et al., 2008). The IRA1 expression vector is from the Molecular Barcoded Yeast ORF (MoBY) library (Open Biosystems). PIB2 expression plasmids were generated by inserting PCR amplified coding sequences into the BglII site of pBQ23, a PGK-URA3 plasmid and verified by direct sequencing.
Heat-ramp stress test
Clonal substrains from fresh colonies (first test) were grown to saturation in 200 µl liquid YPD cultures at 30°C for 48 h. Frozen stocks of the same substrains (tests 2–3) and archived original knockout and wild type strains were pinned from frozen stocks onto YPD agar plates. After two days incubation, yeast were pinned into 200 µl liquid YPD and grown at 30°C for 48 h. Saturated 48 h cultures were diluted 10-fold in YPD, and 100 µl of diluted cultures were immediately heat-treated in a thermocycler (Mastercycler, Eppendorf) programmed to ramp the temperature from 30°C to 62°C (or 61°C for diploids) over 20 min and returned to ambient (Teng and Hardwick, 2013). 5 µl of treated cultures were spotted onto YPD agar plates and incubated at 30°C for two days, or enumerated by automation <24 h (Teng et al., 2011). For untreated controls, the same saturated 48 h cultures were diluted 5000-fold (1:500 relative to treated) in sterile ddH2O and 5 µl of the diluted cultures were spotted onto YPD agar plates for two days at 30°C.
Low amino acid overgrowth assay
Genome-wide screen for growth on low amino acid medium (SCDME) was performed by pinning the original YKO strains and wild type strains from frozen stocks onto YPD agar plates and then grown to saturation in liquid YPD at 30°C for 48 h. 5 µl of saturated 48 h cultures were diluted 50-fold in sterile ddH2O, spotted onto agar plates and allowed to grow into buttons by incubating at 30°C for 2 days on control SCDCSH (Burke et al., 2000) and 3 days on low amino acid SCDME (Sherman, 2002), which contains 25% reduced amino acid levels (molar), primarily due to lower leucine levels (Table S3) (Cheng et al., 2008). No other conditions were applied.
PCR genotyping
Primer sequences for each unique knockout strain were as reported for the Saccharomyces genome deletion project
(http://www-sequence.stanford.edu:16080/group/yeast_deletion_project/).
Tetrad analysis
Colony-derived substrains of YKO strains (MATa BY4741) were mated to wild type (MATα BY4742) and resulting heterozygous diploids were sporulated. Spores from 10–20 tetrads per substrain were validated using auxotrophic markers LYS2 and MET15, mating types and KanMX cassette, and subsequently analyzed for heat-stress and low amino acid overgrowth phenotypes. For the 34 YKO strains with variant substrains (Figure 4C), if the phenotype of the first substrain tested by tetrad analysis was found to segregate with the knockout gene, then a second substrain with the alternate phenotype was tested. Either the first or second substrain always identified a secondary mutation.
Complementation tests
Colony-derived substrains from the BY MATa collection were mated with either BY MATα (BY4742) strains deleted of the same gene, or MATα ascospore segregants generated from BY MATa substrains (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0; see Supplemental Experimental Procedures) bearing the secondary mutations from wild type backcrosses (Figure 4). Two or three diploids from each cross were subsequently analyzed for heat stress and low amino acid overgrowth phenotypes.
Whole genome sequencing
For each knockout strain, two spore-derived strains generated from characterized substrains with secondary mutations were analyzed by whole genome sequencing and all mutation(s) shared only by both sequenced strains are listed in Figure 6E. Sequencing was performed at the Next Generation Sequencing Center, Johns Hopkins Oncology Center, or by high density tiling array for Δfis1-d4, as described (Cheng et al., 2008; Gresham et al., 2008) and confirmed by direct sequencing of PCR products. Statistical correlations with human cancer genomes were obtained at http://www.cbioportal.org/public-portal/index.do.
Supplementary Material
Highlights.
Knockouts of the same gene evolve similarly without applied environmental pressure
Yeast knockouts harbor meaningful second mutations that challenge genetic analyses
Common assumptions about problems with yeast knockout collections are unlikely
First/engineered and evolved yeast mutations can also co-occur in human tumors
Acknowledgments
This project was supported by the National Institutes of Health NS083373, GM077875 and NS037402 (J.M.H.), P50-GM071508 (M.J.D.) and U54 GM103520 (J.D.B.). We thank the Next Generation Sequencing Center at the Sidney Kimmel Comprehensive Cancer Center of Johns Hopkins, Cheryl Tucker, Ari Hakimi, James Hsieh and Steven Santos for genome sequencing and tetrad analyses.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Information
Supplemental information contains six figures, seven tables, and experimental procedures.
Accession Numbers
Read data for whole genome sequencing are publically available at NCBI, SRA Accession number SRP030480.
The authors declare no conflicts of interest.
ProjectID number from NCBI: BioProject ID: PRJNA221721
REFERENCES
- Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albeck JG, Burke JM, Aldridge BB, Zhang M, Lauffenburger DA, Sorger PK. Quantitative analysis of pathways controlling extrinsic apoptosis in single cells. Mol Cell. 2008;30:11–25. doi: 10.1016/j.molcel.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick JE, Yu DS, Yoon SH, Jeong H, Oh TK, Schneider D, Lenski RE, Kim JF. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature. 2009;461:1243–1247. doi: 10.1038/nature08480. [DOI] [PubMed] [Google Scholar]
- Bartholomew CR, Suzuki T, Du Z, Backues SK, Jin M, Lynch-Day MA, Umekawa M, Kamath A, Zhao M, Xie Z, et al. Ume6 transcription factor is part of a signaling cascade that regulates autophagy. Proc Natl Acad Sci U S A. 2012;109:11206–11210. doi: 10.1073/pnas.1200313109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boer VM, Amini S, Botstein D. Influence of genotype and nutrition on survival and metabolism of starving yeast. Proc Natl Acad Sci U S A. 2008;105:6930–6935. doi: 10.1073/pnas.0802601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Breslow DK, Cameron DM, Collins SR, Schuldiner M, Stewart-Ornstein J, Newman HW, Braun S, Madhani HD, Krogan NJ, Weissman JS. A comprehensive strategy enabling high-resolution functional analysis of the yeast genome. Nat Methods. 2008;5:711–718. doi: 10.1038/nmeth.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke D, Dawson D, Stearns T. Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual. 2000 edn. xvii. Plainview, N.Y: Cold Spring Harbor Press; 2000. p. 205. [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng WC, Teng X, Park HK, Tucker CM, Dunham MJ, Hardwick JM. Fis1 deficiency selects for compensatory mutations responsible for cell death and growth control defects. Cell Death Differ. 2008;15:1838–1846. doi: 10.1038/cdd.2008.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dementieva IS, Tereshko V, McCrossan ZA, Solomaha E, Araki D, Xu C, Grigorieff N, Goldstein SA. Pentameric assembly of potassium channel tetramerization domain-containing protein 5. J Mol Biol. 2009;387:175–191. doi: 10.1016/j.jmb.2009.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettman JR, Sirjusingh C, Kohn LM, Anderson JB. Incipient speciation by divergent adaptation and antagonistic epistasis in yeast. Nature. 2007;447:585–588. doi: 10.1038/nature05856. [DOI] [PubMed] [Google Scholar]
- Dunham MJ, Badrane H, Ferea T, Adams J, Brown PO, Rosenzweig F, Botstein D. Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2002;99:16144–16149. doi: 10.1073/pnas.242624799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fannjiang Y, Cheng WC, Lee SJ, Qi B, Pevsner J, McCaffery JM, Hill RB, Basañez G, Hardwick JM. Mitochondrial fission proteins regulate programmed cell death in yeast. Genes Dev. 2004;18:2785–2797. doi: 10.1101/gad.1247904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti E, De Smaele E, Di Marcotullio L, Screpanti I, Gulino A. Hedgehog checkpoints in medulloblastoma: the chromosome 17p deletion paradigm. Trends Mol Med. 2005;11:537–545. doi: 10.1016/j.molmed.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Game JC, Birrell GW, Brown JA, Shibata T, Baccari C, Chu AM, Williamson MS, Brown JM. Use of a genome-wide approach to identify new genes that control resistance of Saccharomyces cerevisiae to ionizing radiation. Radiat Res. 2003;160:14–24. doi: 10.1667/rr3019. [DOI] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- Goebl MG, Petes TD. Most of the yeast genomic sequences are not essential for cell growth and division. Cell. 1986;46:983–992. doi: 10.1016/0092-8674(86)90697-5. [DOI] [PubMed] [Google Scholar]
- Gordon DJ, Resio B, Pellman D. Causes and consequences of aneuploidy in cancer. Nat Rev Genet. 2012;13:189–203. doi: 10.1038/nrg3123. [DOI] [PubMed] [Google Scholar]
- Greer EL, Maures TJ, Ucar D, Hauswirth AG, Mancini E, Lim JP, Benayoun BA, Shi Y, Brunet A. Transgenerational epigenetic inheritance of longevity in Caenorhabditis elegans. Nature. 2011;479:365–371. doi: 10.1038/nature10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gresham D, Desai MM, Tucker CM, Jenq HT, Pai DA, Ward A, DeSevo CG, Botstein D, Dunham MJ. The repertoire and dynamics of evolutionary adaptations to controlled nutrient-limited environments in yeast. PLoS Genet. 2008;4:e1000303. doi: 10.1371/journal.pgen.1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillenmeyer ME, Fung E, Wildenhain J, Pierce SE, Hoon S, Lee W, Proctor M, St Onge RP, Tyers M, Koller D, et al. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science. 2008;320:362–365. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes TR, Roberts CJ, Dai H, Jones AR, Meyer MR, Slade D, Burchard J, Dow S, Ward TR, Kidd MJ, et al. Widespread aneuploidy revealed by DNA microarray expression profiling. Nat Genet. 2000;25:333–337. doi: 10.1038/77116. [DOI] [PubMed] [Google Scholar]
- Ivanovska I, Hardwick JM. Viruses activate a genetically conserved cell death pathway in a unicellular organism. J Cell Biol. 2005;170:391–399. doi: 10.1083/jcb.200503069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs KB, Yeager M, Zhou W, Wacholder S, Wang Z, Rodriguez-Santiago B, Hutchinson A, Deng X, Liu C, Horner MJ, et al. Detectable clonal mosaicism and its relationship to aging and cancer. Nat Genet. 2012;44:651–658. doi: 10.1038/ng.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang GI, Rice DP, Hickman MJ, Sodergren E, Weinstock GM, Botstein D, Desai MM. Pervasive genetic hitchhiking and clonal interference in forty evolving yeast populations. Nature. 2013 doi: 10.1038/nature12344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapinskas PJ, Cunningham KW, Liu XF, Fink GR, Culotta VC. Mutations in PMR1 suppress oxidative damage in yeast cells lacking superoxide dismutase. Mol Cell Biol. 1995;15:1382–1388. doi: 10.1128/mcb.15.3.1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie CC, Laurie CA, Rice K, Doheny KF, Zelnick LR, McHugh CP, Ling H, Hetrick KN, Pugh EW, Amos C, et al. Detectable clonal mosaicism from birth to old age and its relationship to cancer. Nat Genet. 2012;44:642–650. doi: 10.1038/ng.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy SF, Ziv N, Siegal ML. Bet hedging in yeast by heterogeneous, age-correlated expression of a stress protectant. PLoS Biol. 2012;10:e1001325. doi: 10.1371/journal.pbio.1001325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Sage JC, Miller MR, Verhaak RG, Hippenmeyer S, Vogel H, Foreman O, Bronson RT, Nishiyama A, Luo L, et al. Mosaic analysis with double markers reveals tumor cell of origin in glioma. Cell. 2011;146:209–221. doi: 10.1016/j.cell.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Sung W, Morris K, Coffey N, Landry CR, Dopman EB, Dickinson WJ, Okamoto K, Kulkarni S, Hartl DL, et al. A genome-wide view of the spectrum of spontaneous mutations in yeast. Proc Natl Acad Sci U S A. 2008;105:9272–9277. doi: 10.1073/pnas.0803466105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo W, Chen J, Patel A, Zhang L, Chau V, Li Y, Cho W, Lim K, Xu J, Lazar AJ, et al. CXCR4/CXCL12 mediate autocrine cell- cycle progression in NF1-associated malignant peripheral nerve sheath tumors. Cell. 2013;152:1077–1090. doi: 10.1016/j.cell.2013.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J, Klar AJ, Grewal SI. A chromodomain protein, Swi6, performs imprinting functions in fission yeast during mitosis and meiosis. Cell. 2000;101:307–317. doi: 10.1016/s0092-8674(00)80840-5. [DOI] [PubMed] [Google Scholar]
- Newman JR, Ghaemmaghami S, Ihmels J, Breslow DK, Noble M, DeRisi JL, Weissman JS. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature. 2006;441:840–846. doi: 10.1038/nature04785. [DOI] [PubMed] [Google Scholar]
- Pillus L, Rine J. Epigenetic inheritance of transcriptional states in S. cerevisiae. Cell. 1989;59:637–647. doi: 10.1016/0092-8674(89)90009-3. [DOI] [PubMed] [Google Scholar]
- Qian W, Ma D, Xiao C, Wang Z, Zhang J. The genomic landscape and evolutionary resolution of antagonistic pleiotropy in yeast. Cell Rep. 2012;2:1399–1410. doi: 10.1016/j.celrep.2012.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scuoppo C, Miething C, Lindqvist L, Reyes J, Ruse C, Appelmann I, Yoon S, Krasnitz A, Teruya-Feldstein J, Pappin D, et al. A tumour suppressor network relying on the polyamine-hypusine axis. Nature. 2012;487:244–248. doi: 10.1038/nature11126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. In: Guide to yeast genetics and molecular biology, Part A. In Methods in Enzymology. Guthrie C, Fink GR, editors. Vol. 194. 2002. pp. 3–41. [Google Scholar]
- Spencer SL, Gaudet S, Albeck JG, Burke JM, Sorger PK. Non-genetic origins of cell-to-cell variability in TRAIL-induced apoptosis. Nature. 2009;459:428–432. doi: 10.1038/nature08012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart-Ornstein J, Weissman JS, El-Samad H. Cellular noise regulons underlie fluctuations in Saccharomyces cerevisiae. Mol Cell. 2012;45:483–493. doi: 10.1016/j.molcel.2011.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang YC, Amon A. Gene copy-number alterations: a cost-benefit analysis. Cell. 2013;152:394–405. doi: 10.1016/j.cell.2012.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng X, Cheng WC, Qi B, Yu TX, Ramachandran K, Boersma MD, Hattier T, Lehmann PV, Pineda FJ, Hardwick JM. Gene-dependent cell death in yeast. Cell Death Dis. 2011;2(1–9):e188. doi: 10.1038/cddis.2011.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng X, Hardwick JM. Quantification of genetically controlled cell death in budding yeast. Methods Mol Biol. 2013;1004:161–170. doi: 10.1007/978-1-62703-383-1_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres EM, Dephoure N, Panneerselvam A, Tucker CM, Whittaker CA, Gygi SP, Dunham MJ, Amon A. Identification of aneuploidy-tolerating mutations. Cell. 2010;143:71–83. doi: 10.1016/j.cell.2010.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oevelen C, Wang J, Asp P, Yan Q, Kaelin WG, Jr., Kluger Y, Dynlacht BD. A role for mammalian Sin3 in permanent gene silencing. Mol Cell. 2008;32:359–370. doi: 10.1016/j.molcel.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr., Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler BS, Ruderman BT, Willard HF, Scott KC. Uncoupling of genomic and epigenetic signals in the maintenance and inheritance of heterochromatin domains in fission yeast. Genetics. 2012;190:549–557. doi: 10.1534/genetics.111.137083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeyl C. The number of mutations selected during adaptation in a laboratory population of Saccharomyces cerevisiae. Genetics. 2005;169:1825–1831. doi: 10.1534/genetics.104.027102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng TS, Hunot S, Kuida K, Momoi T, Srinivasan A, Nicholson DW, Lazebnik Y, Flavell RA. Deficiency in caspase-9 or caspase-3 induces compensatory caspase activation. Nat Med. 2000;6:1241–1247. doi: 10.1038/81343. [DOI] [PubMed] [Google Scholar]
- Zong C, Lu S, Chapman AR, Xie XS. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science. 2012;338:1622–1626. doi: 10.1126/science.1229164. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.