Abstract
Background. Escherichia coli–bearing Dr-adhesins (Dr+ E. coli) cause chronic pyelonephritis in pregnant women and animal models. This chronic renal infection correlates with the capacity of bacteria to invade epithelial cells expressing CD55. The mechanism of infection remains unknown.
Methods. CD55 amino acids in the vicinity of binding pocket-Ser155 for Dr-adhesin were mutated to alanine and subjected to temporal gentamicin-invasion/gentamicin-survival assay in Chinese hamster ovary cells. CD55/microtubule (MT) responses were studied using confocal/electron microscopy, and 3-dimensional structure analysis.
Results. Mutant analysis revealed that complement-protective CD55-Ser165 and CD55-Phe154 epitopes control E. coli invasion by coregulating CD55–MT complex expression. Single-point CD55 mutations changed E. coli to either a minimally invasive (Ser165Ala) or a hypervirulent pathogen (Phe154Ala). Thus, single amino acid modifications with no impact on CD55 structure and bacterial attachment can have a profound impact on E. coli virulence. While CD55-Ser165Ala decreased E. coli invasion and led to dormant intracellular persistence, intracellular E. coli in CD55-Phe154Ala developed elongated forms (multiplying within vacuoles), upregulated CD55–MT complexes, acquired CD55 coat, and escaped phagolysosomal fusion.
Conclusions. E. coli target complement-protective CD55 epitopes for invasion and exploit CD55–MT complexes to escape phagolysosomal fusion, leading to a nondestructive parasitism that allows bacteria to persist intracellularly.
Keywords: Dr+ E. coli, CD55, invasion, intracellular persistence, microtubules
Escherichia coli–bearing Dr-adhesins (Dr+ E. coli), the first CD55-recognizing pathogen described, can cause chronic/recurrent infections. Dr+ E. coli are reportedly associated with a risk of chronic pyelonephritis in pregnant women, recurrent cystitis, and chronic diarrhea in children [1–4]. Dr+ E. coli use human CD55/decay accelerating factor as the receptor to invade epithelial cells [1]. CD55 is a cell surface protein that protects host cells from lysis by autologous complement [5–7]. CD55 is also a receptor for multiple enteroviruses and Helicobacter pylori, all of which have the capacity to establish chronic infections [8–14]. This poses the question of how CD55, which is normally a host-protective protein, promotes chronic/recurrent infections.
The attachment of Dr-adhesin to CD55 is a crucial step that precedes E. coli invasion [15, 16]. Using a recombinant model of the CD55–E. coli–Dr-adhesin interaction [17] and a 3-dimensional (3D) model of human CD55-SCR3 [18], the Dr-adhesin binding site was narrowed down to the SCR3 domain of CD55. A substitution of Ser155Ala and Ser165Leu (a rare polymorphism of human CD55 designated Dr[a-]) abolished binding of Dr+ E. coli [17, 19]. CD55 residues Phe123, Phe148, Phe154, and Ser165 confer complement protective function but not bacterial attachment [18].
Because the molecular mechanism of Dr+ E. coli invasion remains unresolved, we evaluated a series of CD55 mutants for their ability to promote the rate of invasion of and/or the fate of Dr+ E. coli once internalized.
METHODS
Materials
Primary antibodies for CD55 (sc9156), β-actin (ab8227), and tubulin (ab7291) were procured from Santa-Cruz Biotechnologies and Abcam (ab8227 and ab7291), respectively. Alexa Fluor 488/546 secondary antibodies were obtained from Invitrogen.
Plasmids
The EGFP-LC3 construct was a kind gift from Dr Waldemar Popik, Meharry Medical College, Nashville, TN.
CD55 SCR3 Mutants
The construction, characterization, and transfection of human CD55 SCR2/SCR3 mutants in CHO cells has been described elsewhere [18].
Bacterial Strains, Cell Lines, and Growth Conditions
Dr-fimbriated pyelonephritis strain IH11128 (O75:K5:H−) was grown on Luria–Bertani (LB) agar at 37°C. E. coli EC901 strain BN406, containing plasmid pBJN406 (expressing Dr-adhesin [Dr+], and its transposon mutant [BN 17] with a mutation in draE adhesin subunit gene [Dr−]), was grown on LB agar containing chloramphenicol (25 μg/mL) [1]. The expression of Dr fimbrae by E. coli was evaluated by determining the minimal hemagglutination titer as detailed in the Supplementary Methods. The maintenance of Chinese hamster ovary (CHO) K-1 (American Type Culture Collection, CCL61) cells has been described elsewhere [18].
Binding and Invasion Assays
The assays are detailed in the Supplementary Materials. Briefly, for the invasion assay, CHO cells expressing wild-type (WT) or mutated CD55 were infected with bacteria for 3 hours at 37°C. Extracellular bacteria were killed with gentamicin. Cells were then washed and lysed and the lysate plated on L-agar plates and incubated at 37°C. The colonies were counted and results expressed as colony-forming units per well.
For the modified invasion assay, monolayers were incubated with bacterial suspension for time periods ranging from 15 minutes to 3 hours. Monolayers were then processed as was done for the invasion assay.
Intracellular Survival Assay
CHO cell monolayers were incubated with bacterial suspensions for 1 hour at 37°C to allow for invasion. Monolayers were washed with F-12 plus gentamicin (200 mg/mL) and incubated in the same medium for 1–2 hours. Cells were then washed, lysed, and plated, as was done for the invasion assay. Since extracellular bacteria were killed, recovered colonies represented bacteria that survived.
Labeling of Bacteria
The bacteria were fluorescently labeled using Alexa Fluor 488 (AF488) or 633 (AF633) protein labeling kits (Invitrogen). AF488-labeled bacteria were used in experiments for labeling only microtubules (MT); AF633-labeled bacteria were used for CD55–MT costaining experiments.
Immunofluorescence Microscopy
CHO cells were cultured on glass coverslips in 24-well plates for 24 hours, rinsed with phosphate-buffered saline (PBS), and fixed in 4% paraformaldehyde for 20 minutes at room temperature. Thereafter, cells were washed with PBS (×3) and permeabilized with ice-cold 100% methanol for 10 minutes at −20°C. The cells were washed with PBS (×3), blocked in 1:1 blocking buffer (LI-COR Biosciences) and PBS, and incubated with primary antibodies (CD55/MT at 1:200) overnight at 4°C in blocking buffer with 0.1% Tween-20. Slides were then washed 5 times with PBST (PBS with 0.1% Tween-20) followed by a 1-hour incubation with the Alexa Fluor secondary antibody. The slides were washed with PBST (×5), mounted in SlowFade Gold antifade reagent (Invitrogen), and examined using a confocal laser scanning microscope (Nikon A1R).
Proximity Ligation Assay
The proximity ligation assay (PLA) was performed to detect CD55–MT interactions in situ. The assay was performed using a Duolink II fluorescence kit (Olink Biosciences). Details are included in the Supplementary Materials.
RESULTS
Dr+ E. coli Recognize CD55 Ser165Ala Invasion Epitope
The CD55 mutants that were used involved primarily alanine substitutions of selected amino acids in the structurally conserved SCR2/SCR3 regions of CD55. SCR mutants were analyzed to identify the hypothetical internalization domain that allowed Dr+ E. coli to invade epithelial cells. For this purpose, a standard 3-hour gentamicin protection/invasion assay was performed. All experiments were done using a clinical isolate of Dr+ E. coli (IH11128) and a control Dr− E. coli Dr14 (IH11128ΔDr) as described [20] and of recombinant Dr+ and Dr− E. coli as shown in Supplementary Figure 1. CD55 mutants were classified into 5 groups based on their E. coli binding and invasion characteristics (Table 1); 5 alanine mutants from this list were used in this study. In each case, their properties were compared with WT-CD55 or vector-transfected (CD55−) CHO cells. They included S165A (normal binding characteristics but a significantly lower invasion rate), G159A (reduced E. coli binding and invasion rates), F123A (normal binding and invasion rates), and 2 group V mutants (F148A and F154A) that have normal binding characteristics along with a significantly elevated rate of invasion. The positions of mutated key AA residues on the CD55 SCR2/SCR3 region that were used are depicted in Figure 1A. The levels of CD55 expression on selected mutants were similar, as evaluated by flow cytometry/Western blotting (Supplementary Table 1; Supplementary Figure 1).
Table 1.
Effect of CD55 Mutations on Attachment and Invasion of Dr+ Escherichia coli
| Group | CD55+ and Mutants | % Binding vs CD55+ | % Invasion vs CD55+ |
|---|---|---|---|
| CD55+ | 100 | 100 | |
| Group I | (CD55−) | (0*) | (10*) |
| S155A | 0* | 1.6* | |
| C156A | 0* | 2.0* | |
| S165L | 0* | 0.4* | |
| Group II | G159A | 40* | 77 |
| Y160A | 25* | 35* | |
| L162A | 25* | 34* | |
| Group III | S165A | 100 | 28* |
| Group IV | F163A | 75 | 100 |
| L171A | 80 | 74 | |
| F123A | 95 | 84 | |
| Group V | F148A | 100 | 142* |
| F154A | 100 | 201* |
Asterisk (*) indicates statistical significance (P ≤ .05) compared with WT-CD55+.
Figure 1.
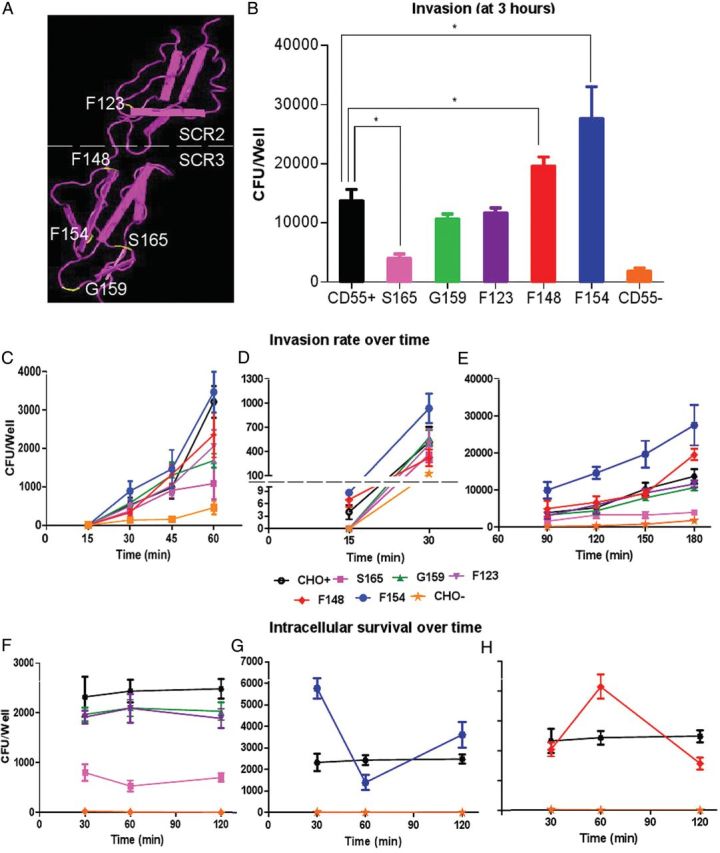
CD55 SCR3 mutants differ in Escherichia coli invasion and survival capacities. A, Location of mutated amino acids on CD55's SCR2 and SCR3 domains. This figure was obtained using Cn3D (PubMed structure). The yellow labels represent amino acids mutated for the study. B, Results from a 3-hour Dr+ E. coli invasion assay using Chinese hamster ovary (CHO) cells expressing wild-type (WT) and mutated human CD55; the CD55(−) control represents group I mutants with no binding or invasion characteristics. Other mutant group representatives (all selected mutants are alanine substitutions) include group II (G159), group III (S165), group IV (F123), and group V (F148 and F154). Results are shown as means ± standard deviation (SD; n = 3). Asterisk (*) indicates significance (P ≤ .05) vs CD55(+) WT control. C, Results from temporal invasion assays with invasion stopped at earlier time points (15, 30, 45, or 60 minutes). D, Results from invasion assays stopped at 15 or 30 minutes; data are reported using expanded scales for both time points. E, Invasion results from assays stopped at 90 minutes and 180 minutes. F, Survival assay comparing WT, mutant, or CD55(−) CHO control cells that had no significant change in the number of bacteria within CHO cells between 30 and 120 minutes. G, Mutant F154A survival assay data supporting killing followed by multiplication of bacteria over time. H, Mutant F148A survival assay data supporting multiplication followed by killing of bacteria over time. Results shown are mean ± SD for 3 independent experiments.
Figure 1B shows the results of an E. coli invasion assay using CHO cells expressing WT-CD55(+), CD55 mutants, or CD55(−) control. There was a significant reduction in the number of colony-forming units per well in the invasion-deficient S165A mutant and modest to no changes in invasion with G159A and F123A mutations, but a significantly higher number of E. coli were found intracellularly when either mutant F148A or F154A was used.
Ser165Ala Decelerates While Phe154Ala Accelerates Initial Phase of Invasion
In order to investigate the mechanism involved in the reduced ability of E. coli to invade S165A, we focused on the rate of entry at early stages of invasion when both intracellular multiplication and killing would have no, or minimal, effect. The overall time-dependent invasion rates (between 15 and 60 minutes) for S165A and other mutants are shown in Figure 1C. It took at least 15 minutes to observe a demonstrable invasion of E. coli in various groups (Figure 1D). Therefore, this may represent time required for the initial binding, signaling, and cytoskeleton-related processes to occur that result in invagination and internalization. The most rapid increase in internalization of E. coli occurred within 15–30 minutes. Clearly, after 90–180 minutes, CD55(−) and S165A had the fewest colony-forming units, while F148A and F154A had the most (Figure 1E).
With this extended period of incubation, however, the question arose as to whether or not these findings included a component of intracellular multiplication or destruction of bacteria.
Ser165Ala Promotes Dormant Persistence While Phe154Ala Promotes Intracellular Multiplication
We then focused on the survival patterns of bacteria using an intracellular survival assay (Figure 1F–H). CHO cells containing WT-CD55 and mutants S165A, G159A, and F123A appeared to stay dormant with a steady number of colony-forming units (Figure 1F). A parallel evaluation of AF488 showed a lack of any dividing E. coli or stained bacterial debris, suggesting the absence of intracellular multiplication and/or killing (see below). Unexpectedly, E. coli mutant F154A showed a dramatic decrease in colony-forming units (to less than half) at 60 minutes; this increased again at 120 minutes, indicating an alternating cycle of killing and multiplication (Figure 1G). In contrast, the number of colony-forming units in mutant F148A doubled at 60 minutes compared with the number at 30 minutes and then decreased rapidly to less than half at 120 minutes (Figure 1H). This led us to consider that once internalized, CD55 mutants either led to a seemingly dormant intracellular persistence of E. coli (G159A and F123A) or were subjected to processes of destruction and/or duplication (F154A and F148A).
To support or refute these suppositions, we used transmission electron microscopy (TEM) and fluorescence confocal microscopy analyses as described below.
TEM Analysis of CD55 SCR3 Mutants
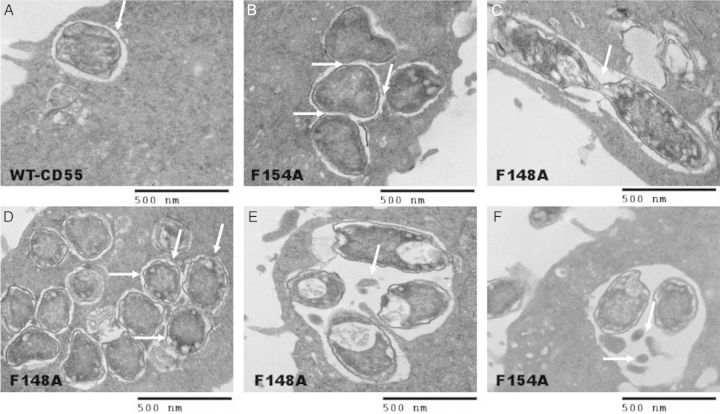
After a 2-hour invasion, TEM analysis of WT-CD55 identified intracellular vacuoles, each containing a single bacterium without evidence of multiplication (Figure 2A). Similar observations were made for S165A (data not shown). In contrast, several other types of vacuoles were observed in mutants F148A and F154A (Figure 2B–F). Bacteria occurred in clusters in these mutants and formed microcolonies. In F154A, 4 bacteria were found in a vacuole with tightly apposing, limiting membranes; this suggests that this vacuole was formed as a result of fusion of the 4 individual vacuoles to form a microcolony (Figure 2B). Nevertheless, some microcolonies in mutants F148A/F154A appeared to be formed by E. coli multiplication within a tight vacuole (Figure 2C). Other vacuoles contained multiple bacteria or intact E. coli along with bacterial debris (Figure 2E and 2F). Overall, unlike Shigella, where bacteria escape to the cytoplasm to multiply, Dr+ E. coli appear to be able to multiply within vacuoles [21–23].
Figure 2.
Transmission electron microscopy analysis of Escherichia coli invasion in wild-type (WT)-CD55 and mutants F148A and F154A. A, Dr+ E. coli 2-hour invasion results of Chinese hamster ovary (CHO) cells containing WT-CD55 showing a tight vacuole containing a single bacterium. B, Dr+ E. coli 2-hour invasion results of CHO cells containing CD55 mutant F154A showing a microcolony containing 4 bacteria in a tight vacuole. The arrows indicate the tightly apposing limiting membranes of 4 individual vacuoles fusing to form the big vacuole. C, Following a 2-hour invasion assay, a CD55 mutant F148A example showing a dividing bacterium within a single vacuole. D, Mutant F148A showing a larger colony with several bacteria, each within its own vacuole (arrows point to individual bacteria within their own vacuoles). E, Mutant F148A showing a loose vacuole containing 4 bacteria and some debris after a 2-hour invasion. F, Mutant F154A showing a loose vacuole containing 2 bacteria and some bacterial debris after a 2-hour invasion (arrows point to the bacterial debris). Bar = 500 nm.
Fluorescence confocal microscopy findings were consistent with TEM results. Confocal microscopy showed single 1- to 3-µm dormant intracellular E. coli without evidence of division and/or elongation in WT-CD55 and S165A, while F148A and F154A contained elongated (20-µm, see below) and dividing bacteria, with clusters of microcolonies localized below the cell membrane.
CD55 SCR3 Mutations and E. coli–MT Interaction as Evidenced by Confocal Fluorescence Microscopy
Our previous studies showed that depolymerization of MT with nocodazole abolished invasion [20]. Here we investigated whether mutations in CD55 affected bacterial interactions with the MT network and therefore their disposition and fate within cells. WT and mutant CD55-expressing CHO cells were infected with AF488-labeled Dr+ E. coli for 30 minutes, 90 minutes, or 3 hours. Figure 3A shows a 3D reconstruction of the confocal microscopy images, indicating the interaction between bacteria (green) and MT (red). E. coli appear tethered to MT in both controls and mutants, as shown in the insets in the 3-hour images (Figure 3A). To show their close association, the average Pearson correlation coefficient values for WT-CD55 ranged from 0.592 to 0.624 at 30 and 90 minutes and at 3 hours. Compared with WT-CD55, the level of association with MT was far more pronounced in F148A and F154A, but significantly reduced in S165A, and absent in CD55(−) cells (20 random cells were evaluated for each; Supplementary Figure 2). Likely the heightened invasion kinetics in CD55 mutants F148A and F154A as well as the propensity to form microcolonies once internalized (arrows in F148A and F154A) produced some of the observed intense MT staining patterns. We found that F148A had an 86% ± 5% and F154A had a 91% ± 4% greater propensity to form microcolonies compared with WT-CD55 cells. We also quantitated MT staining intensity and performed Western blot analysis (not shown) and found that mutants F154A and F148A showed 2- and 2.5-fold increases, while mutant S165A showed about a 2-fold decrease in MT fluorescence intensity compared with WT-CD55 after a 3-hour invasion (Figure 3B). This increased and/or decreased MT fluorescence intensity corresponded to the E. coli invasion assay results at 3 hours (Figure 1B), which in turn supports the concept that MT expression and the level of bacteria internalization are correlated. MT in CD55(−) CHO cells remained unaffected, presumably due to the lack of CD55 receptors and therefore the lack of significant bacterial attachment or invasion.
Figure 3.
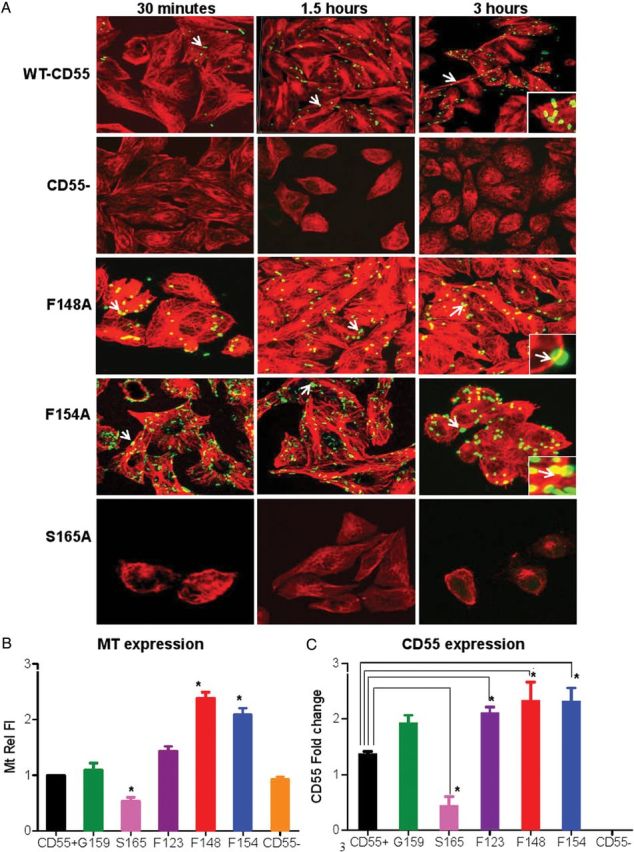
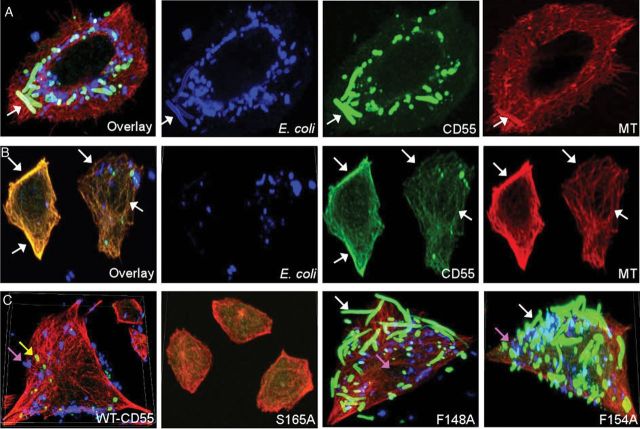
Dr+ Escherichia coli interact with microtubules (MT) and MT and CD55 follow the same expression patterns in wild-type (WT) and mutant CD55 expressing Chinese hamster ovary (CHO) cells. A, Immunofluorescence confocal microscopy images showing invasion of Dr+ E. coli in CHO cells expressing WT-CD55, mutated CD55, or cells lacking CD55(−). Red = MT and green = bacteria. The panels indicate projected images of internal Z stacks (60 × magnification) of 0.5 μm each. Mutants F148A and F154A showed an increased invasion capacity compared with WT-CD55, while mutant S165A behaved like the negative control CD55(−). White arrows in WT-CD55 panels indicate a single bacterium. Arrows in mutants F148A and F154A indicate larger microcolonies of bacteria. Insets in 3-hour images show bacteria apparently tethered to, or in very close association with MT. B, Quantitation of MT fluorescence intensity (MtRelFl) at 3 hours for WT-CD55, mutants, and negative controls. Values represent the mean ± standard deviation (SD) obtained from at least 20 cells. All images were taken at the same intensity (set at 5) and gain (set at 3) with a pixel dwell time of 1.1 μs/pixel. C, Graphic representation of the densitometric analysis of immunoblots showing changes in CD55 protein expression upon infection of WT and mutant CD55 expressing CHO cells with Dr+ E. coli. Quantitation and analysis of bands were performed using Odyssey software. β-actin was used as a normalization control, and the results are expressed relative to the level of β-actin. Asterisk (*) indicates significance, P ≤ .05, vs CD55+ controls. Values are mean ± SD (n = 3).
As the invasion of E. coli is also dependent upon the density of CD55, we determined whether changes in MT density correlated with changes in CD55 protein expression. After infection with E. coli, expression of CD55 in F148A and F154A increased approximately 2-fold, while S165A had about half as much expression as WT-CD55, as shown by Western blot analysis (Figure 3C) and immunostaining (data not shown). Therefore, the changes in CD55 density corresponded to the changes in MT immunofluorescence intensity. We also considered that CD55 and MT were physically associated. Thus, we performed confocal microscopy on WT-CD55 cells infected with labeled bacteria (AF633, blue) for 3 hours and stained for both CD55 (green) and MT (red) (Figure 4A) after removing extracellular bacteria with gentamicin.
Figure 4.
Dr+ Escherichia coli bind to a complex including microtubules (MT) and CD55 and develop a CD55-mutant–dependent morphological plasticity. A, Wild-type (WT)-CD55–containing Chinese hamster ovary (CHO) cell assays only. Taken from a 3-hour E. coli invasion assay. Examples show a colocalization overlay, followed by E. coli (blue), CD55 (green), and MT (red) immunofluorescences alone. Arrows indicate the position of a single E. coli that is colocalized with CD55 and MT, as an example. B, WT-CD55–containing CHO cell assays only. From a 3-hour invasion assay that shows colocalization of CD55 and MT yielding a yellow overlay. Arrows pinpoint various spots and indicate colocalization of CD55 and MT. C, Four comparative overlays. A comparison of E. coli 3-hour invasion assays using WT-CD55 or selected CD55 mutants (S165A, F148A, and F154A). Individual figures show the differences in the number of bacteria invading the cells and also the different forms of E. coli with respect to size and extent of CD55 coating (white arrows, bacteria that were heavily coated with CD55; yellow arrows, bacteria that were lightly coated with CD55; and fuchsia arrows, bacteria that had an undetectable CD55 coating). Alexa Fluor 633–labeled bacteria (blue) were used to infect cells in these experiments. A–C are projected images of Z stacks (60 × magnification), 0.5 μm each.
We noted that E. coli were bound to CD55 molecules that either colocalized with MT or with CD55–MT “complexes” within CHO cells (note the arrow in Figure 4A points to a single bacterium at the base of each panel). We also observed that MT were decorated with CD55 molecules, as shown by the overlay, thereby producing a yellow color (arrows in Figure 4B). The heavy staining of the bacteria-associated CD55 does not allow for the visualization of MT-associated CD55 in panel A of Figure 4.
Finally, we identified the following 3 patterns of E. coli staining within cells (Figure 4C): (1) bacteria that were heavily coated with CD55 (F148A/F154A, white arrows), (2) bacteria that were lightly coated with CD55 (WT, yellow arrow), and (3) bacteria that had an undetectable CD55 coating (fuchsia arrows). In addition, elongated bacteria were always heavily coated with CD55 but they had a 50%–80% reduction in blue staining compared with other bacteria in WT cells, which is consistent with an interpretation that the amount of staining per unit of E. coli length was reduced due to their intracellular elongation. We also noted that while WT-CD55 contained few to no elongated/filamentous E. coli, mutants F148A and F154A contained 3–4 times more filamentous bacteria (Figure 4C), indicating that the formation of elongated forms may be dependent upon the CD55 genotype.
CD55 and MT Form Molecular Complexes
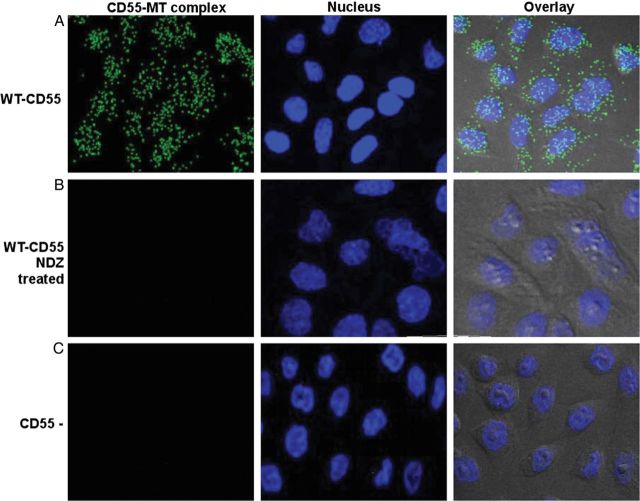
To determine if costaining of CD55 and MT resulted only from coincidental/functional coexpression or represented a close molecular association, we studied the proximity between CD55 and tubulin proteins using a PCR-based PLA. Using WT-CD55 (Figure 5A), the PLA showed fluorescent green spots, indicating that CD55/MT were within ≤40 nm of each other [24]. When the primary antibodies were omitted in the assay (data not shown) or when the CD55(−) controls were used, these green spots were not produced, indicating the specificity of results (Figure 5B). The number of CD55 and tubulin molecules that were in close association (55–90 dots/cell) decreased upon treatment of the cells with the MT depolymerizing agent nocodazole (NDZ) at 5 µg/µL for 1 hour (few or no dots; Figure 5C). Overall, these results demonstrate that CD55 and MT are physically associated and that NDZ dissociates these CD55–MT complexes.
Figure 5.
CD55 and microtubules (MT) form molecular complexes. A, In situ proximity ligation assay (PLA) showing green spots (from fluorescein isothiocyanate–labeled probe) indicate the close association of CD55 and MT as seen by immunofluorescence (60 × magnification) in wild-type (WT)-CD55 cells. B, PLA showing WT-CD55 cells after nocodazole (NDZ) treatment (5 µg/µL for 1 hour). C, CD55 (−) Chinese hamster ovary cell patterns. Diamidino-2-phenylindole (blue) was used to stain the nuclei.
Dr+ E. coli Escape Phagolysosomal Fusion
Our EM data indicate that E. coli persist within tight vacuoles, suggesting they escape phagolysosomal fusion. To investigate this further, we transfected WT-CD55 CHO cells with the autophagosome marker EGFP-LC3. These cells were infected with AF633-labeled E. coli (red) for 3 hours, fixed after the removal of extracellular bacteria, and examined by confocal microscopy. As shown in Figure 6A, bacteria did not colocalize with EGFP-LC3, suggesting that Dr+ E. coli escaped phagolysosomal degradation.
Figure 6.
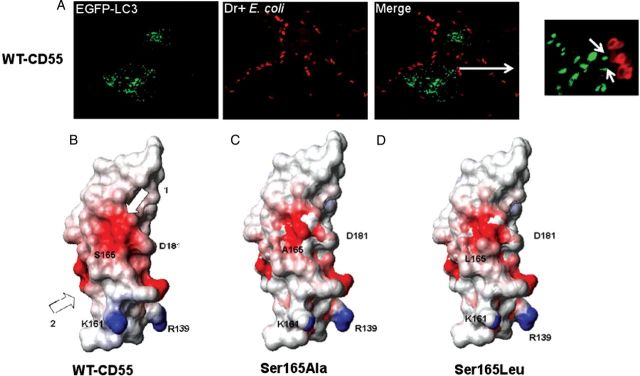
Dr+ Escherichia coli escape phagolysosomal fusion in WT-CD55 Chinese hamster ovary (CHO) cells; modeled structures of CD55 SCR3 generated using MPACK showing CD55 SCR3 and Ser165 mutants. A, Wild-type (WT)-CD55 expressing CHO cells transfected with EGFP-LC3 were infected with Alexa Fluor 633–labeled Dr+ E. coli (red) for 3 hours. White arrows in the merged inset image show that bacteria do not colocalize with LC3. The panels are projected images of Z stacks (60 × magnification) 0.5 μm each. B, The negatively charged surface in Ser165 and Asp181 of WT-CD55 is indicated by arrow 1, and the surface of Dr-adhesin binding site is indicated by arrow 2. This figure was generated using MOLecule analysis and MOLecule display (MOLMOL). Other key residues (D181, K161, and R139) are also noted. Areas in red have a negative potential of −0.5 units and those in blue have a positive potential of +0.5 units. C, A modeled structure of a CD55 Ser165Ala mutant that binds to Dr+ E. coli as well as WT-CD55 but has a significantly reduced capability of promoting the invasion of bacteria. D, A modeled structure of a CD55 Ser165Leu mutant that neither binds to Dr+ E. coli nor promotes its invasion capabilities.
Molecular Models of CD55 SCR3 and Ser165 Mutants Defective in E. coli Invasion
Our calculations with GETAREA [25–27], using an experimental structure of CD55 SCR3, showed that Ser165 is located on the exposed surface of the molecule. The models for WT vs the Ser165 mutant showed a small backbone root mean square deviation of 0.28Å. Calculated electrostatic potentials, using MOLMOL, were mapped onto the surface of the models of mutant and WT-CD55 SCR3 (Figure 6B). The red patch indicates the presence of a negative potential developed by the presence of polar or charged atoms. WT-CD55 SCR3 shows a large polarized area (red) in the region surrounding Ser165. There was a significant change in this distribution in point mutants Ser165Ala and Ser165Leu (Figure 6C and 6D) compared with WT. A hypothetical possibility exists that the presence of charged or polar atoms contributes to the CD55–MT association.
DISCUSSION
Dr+ E. coli can cause chronic/recurrent infections [2–4, 28–33]. Previously, we developed an experimental animal model of chronic pyelonephritis with Dr+ E. coli in which infection was abolished by mutation of Dr-adhesin, which inactivated invasion [34]. In the current study, we describe how Dr+ E. coli use CD55 receptors to invade and persist within the intracellular environment.
CD55 SCR3 single-point mutants were analyzed to identify the internalization domain for Dr+ E. coli. While Ser165Ala decreased invasion, Phe154Ala significantly increased invasion. As Ser165 and Phe154 are important for the protective function of CD55 against complement autolysis and at the same time influence invasion, one may speculate that Dr+ E. coli exploit pathways established for host cells to escape complement injury in order to persist intracellularly [18, 20, 34]. Surprisingly, while S155A abolished binding, the adjacent F154A did not impact binding but enhanced invasion. This observation is consistent with the apparent strategy of E. coli to separate binding and invasion into 2 closely spaced but distinct events involving separate single CD55 AA residues [35]. One may consider that the Ser155-binding domain allows colonization by multiple E. coli, while Ser165 permits only a few bacteria to invade, leading to an intracellular colonization that may serve as a reservoir for chronic/recurrent infection. Alternatively, invasion via the hypothetical single-attachment invasion epitope (Ser155-Phe154Ala) could allow bacteria to achieve an overwhelming level of invasion that, instead of a chronic subclinical process, turns into an acute infection that could damage both host and pathogen.
To further define the mechanistic role of Ser165 and Phe154, we analyzed temporary invasion patterns of E. coli and discovered that Ser165Ala decelerated, while Phe154Ala accelerated bacterial invasion at an early entry stage. The acceleration/deceleration pattern was associated with the pathogen's capacity to upregulate/downregulate CD55 in a corresponding fashion. We propose that the resulting crosstalk between Dr-adhesin and CD55 invasive epitopes defines the quantitative and/or qualitative status of MT, which likely serve as an E. coli translocation vehicle while passing through the cell membrane.
We also found that these CD55 mutants represent separate survival classes that respond differently to infection by Dr+ E. coli. WT-CD55, G159A, F123A, and S165A had flat survival curves (a constant number of bacteria), which supports the proposed mechanism of a dormant, nonproliferative tissue reservoir that permits chronic/recurrent infections. Unexpectedly, E. coli in F154A and F148A showed a pattern of rapid bacterial multiplication alternating with a limited rate of killing. To our knowledge, this study provides the first experimental evidence that indicates that a single-point mutation in a host receptor may change a dormant intracellular microorganism into a hypervirulent pathogen. This implies that an evolution of the host may have reshaped an aggressive host–pathogen interaction to a less violent coexistence strategy.
The modeling study revealed changes in the distribution of negative potential in Ser165 mutants. These changes could impact Dr-adhesin–CD55-receptor interactions, leading to a much lower invasion rate. Although the role of charge distribution in pathogen–host receptor entry is not well understood, it is recognized that residues that contribute to a negative surface can help determine ligand selectivity [36].
After mapping the invasion epitope to Ser165, we traced the location of intracellular E. coli and found that it colocalized with MT. Further mutant analysis revealed that invasion rates, MT density, and CD55 expression in the CD55 mutants followed the same patterns. Confocal microscopy and PLA proved that CD55 and MT were close enough to form a molecular complex. Although the role of CD55 as a regulator of complement activation has been investigated for years, the literature does not show that the complement system and CD55, in particular, may involve the cytoskeleton and MT. We postulate that the CD55–MT complex plays a previously unrecognized function in regulating pathogen survival.
An intriguing finding was that CD55-coated intracellular E. coli in WT-CD55 and selected CD55 mutants displayed plasticity. Horvath and others from Hultgren group suggested that uropathogenic E. coli use morphological plasticity (elongated forms) as a way to subvert host immunity [37]. We therefore considered that bacteria coated with WT-CD55/MT are recognized by host cells as a self-antigen and hence they evade the elimination process. In contrast, E. coli, sensing CD55-F154A/MT complexes, form destruction-resistant, highly virulent elongated forms.
To elaborate how E. coli survive intracellularly, we looked at phagolysosomal degradation, which is a prominent player in the elimination of intracellular pathogens. We postulated that Dr+ E. coli may have evolved like some intracellular bacteria, for example, Legionella, Brucella, and Mycobacterium tuberculosis, to escape phagolysosomal fusion in order to survive [22–23]. Our data also support our hypothesis that E. coli developed an alternate, as-yet-unknown strategy that involves CD55/MT complexes to achieve this survival.
While further studies to define the signals/molecular mechanisms involved in the subversion of autophagy by Dr+ E. coli are underway, the findings presented here add to our understanding of the cellular mechanisms that allow CD55-mediated intracellular survival of these bacteria. Understanding Dr+ E. coli host–cell interactions may allow us to identify new targets (like CD55–MT complexes) and design strategies to treat and/or prevent chronic/recurrent urinary tract infections.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We thank Dr Diana Marver for her thoughtful comments regarding the manuscript.
Financial support. This work was supported by Public Health Service grant HD055648 (to S. N.), HD041687 (to B. J. N. and S. N.), U54RR026140 from National Institutes on Minority Health and Health Disparities-NIMHD (to S. N.), and a CTSA (Clinical and Translational Science Awards) award (UL1TR000445). The Morphology Core at Meharry is supported by U54MD007593, U54CA091408, and S10RR025970. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Nowicki B, Truong L, Moulds J, Hull R. Presence of the Dr receptor in normal human tissues and its possible role in the pathogenesis of ascending urinary tract infection. Am J Pathol. 1988;133:1–4. [PMC free article] [PubMed] [Google Scholar]
- 2.Hart A, Nowicki BJ, Reisner B, et al. Ampicillin-resistant Escherichia coli in gestational pyelonephritis: increased occurrence and association with the colonization factor Dr adhesin. J Infect Dis. 2001;183:1526–9. doi: 10.1086/320196. [DOI] [PubMed] [Google Scholar]
- 3.Greer LG, Roberts SW, Sheffield JS, et al. Ampicillin resistance and outcome differences in acute antepartum pyelonephritis. Infect Dis Obs Gyn. 2008 doi: 10.1155/2008/891426. doi:10.1155/2008/891426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rizvi M, Khan F, Shukla I, Malik A, Shaheen Rising prevalence of antimicrobial resistance in urinary tract infections during pregnancy: necessity for exploring newer treatment options. J Lab Phys. 2011;3:98–103. doi: 10.4103/0974-2727.86842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hourcade D, Holers VM, Atkinson JP. The regulators of complement activation (RCA) gene cluster. Adv Immunol. 1989;45:381–416. doi: 10.1016/s0065-2776(08)60697-5. [DOI] [PubMed] [Google Scholar]
- 6.Osuka F, Endo Y, Higuchi M, et al. Molecular cloning and characterization of novel splicing variants of human decay-accelerating factor. Genomics. 2006;88:316–22. doi: 10.1016/j.ygeno.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 7.Wu J, Li H, Zhang S. Regulator of complement activation (RCA) group 2 gene cluster in zebrafish: identification, expression, and evolution. Funct Integr Gen. 2012;12:367–77. doi: 10.1007/s10142-012-0262-7. [DOI] [PubMed] [Google Scholar]
- 8.Hafenstein S, Bowman VD, Chipman PR, et al. Interaction of decay-accelerating factor with coxsackievirus B3. J Virol. 2007;81:12927–35. doi: 10.1128/JVI.00931-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Brien DP, Romero-Gallo J, Schneider BG, et al. Regulation of the Helicobacter pylori cellular receptor decay-accelerating factor. J Biol Chem. 2008;283:23922–30. doi: 10.1074/jbc.M801144200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Plevka P, Hafenstein S, Harris KG, et al. Interaction of decay-accelerating factor with echovirus 7. J Virol. 2010;84:12665–74. doi: 10.1128/JVI.00837-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Renois F, Hong SS, Le Naour R, et al. Development of a recombinant CHO cell model for the investigation of CAR and DAF role during early steps of echovirus 6 infection. Virus Res. 2011;158:46–54. doi: 10.1016/j.virusres.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 12.Shafren DR, Bates RC, Agrez MV, Herd RL, Burns GF, Barry RD. Coxsackieviruses B1, B3, and B5 use decay accelerating factor as a receptor for cell attachment. J Virol. 1995;69:3873–7. doi: 10.1128/jvi.69.6.3873-3877.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sobo K, Rubbia-Brandt L, Brown TD, Stuart AD, McKee TA. Decay-accelerating factor binding determines the entry route of echovirus 11 in polarized epithelial cells. J Virol. 2011;85:12376–86. doi: 10.1128/JVI.00016-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williams DT, Chaudhry Y, Goodfellow IG, Lea S, Evans DJ. Interactions of decay-accelerating factor (DAF) with haemagglutinating human enteroviruses: utilizing variation in primate DAF to map virus binding sites. J Gen Virol. 2004;85:731–8. doi: 10.1099/vir.0.19674-0. [DOI] [PubMed] [Google Scholar]
- 15.Selvarangan R, Goluszko P, Popov V, et al. Role of decay-accelerating factor domains and anchorage in internalization of Dr-fimbriated Escherichia coli. Infect Immun. 2000;68:1391–9. doi: 10.1128/iai.68.3.1391-1399.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoepelman AI, Tuomanen EI. Consequences of microbial attachment: directing host cell functions with adhesins. Infect Immun. 1992;60:1729–33. doi: 10.1128/iai.60.5.1729-1733.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nowicki B, Hart A, Coyne KE, Lublin DM, Nowicki S. Short consensus repeat-3 domain of recombinant decay-accelerating factor is recognized by Escherichia coli recombinant Dr adhesin in a model of a cell-cell interaction. J Exp Med. 1993;178:2115–21. doi: 10.1084/jem.178.6.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hasan RJ, Pawelczyk E, Urvil PT, et al. Structure-function analysis of decay-accelerating factor: identification of residues important for binding of the Escherichia coli Dr adhesin and complement regulation. Infect Immun. 2002;70:4485–93. doi: 10.1128/IAI.70.8.4485-4493.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lublin DM, Thompson ES, Green AM, Levene C, Telen MJ. Dr(a-) polymorphism of decay accelerating factor. Biochemical, functional, and molecular characterization and production of allele-specific transfectants. J Clin Invest. 1991;87:1945–52. doi: 10.1172/JCI115220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goluszko P, Popov V, Selvarangan R, Nowicki S, Pham T, Nowicki BJ. Dr fimbriae operon of uropathogenic Escherichia coli mediate microtubule- dependent invasion to the HeLa epithelial cell line. J Infect Dis. 1997;176:158–67. doi: 10.1086/514018. [DOI] [PubMed] [Google Scholar]
- 21.Cersini A, Salvia AM, Bernardini ML. Intracellular multiplication and virulence of Shigella flexneri auxotrophic mutants. Infect Immun. 1998;66:549–57. doi: 10.1128/iai.66.2.549-557.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cossart P, Sansonetti PJ. Bacterial invasion: the paradigms of enteroinvasive pathogens. Science. 2004;304:242–8. doi: 10.1126/science.1090124. [DOI] [PubMed] [Google Scholar]
- 23.Ray K, Marteyn B, Sansonetti PJ, Tang CM. Life on the inside: the intracellular lifestyle of cytosolic bacteria. Nat Rev Microbiol. 2009;7:333–40. doi: 10.1038/nrmicro2112. [DOI] [PubMed] [Google Scholar]
- 24.Gustafsdottir SM, Schallmeiner E, Fredriksson S, et al. Proximity ligation assays for sensitive and specific protein analyses. Anal Biochem. 2005;345:2–9. doi: 10.1016/j.ab.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 25.Barlow PN, Steinkasserer A, Norman DG, et al. Solution structure of a pair of complement modules by nuclear magnetic resonance. J Mol Biol. 1993;232:268–84. doi: 10.1006/jmbi.1993.1381. [DOI] [PubMed] [Google Scholar]
- 26.Fraczkiewicz R, Braun W. Exact and efficient analytical calculation of the accessible surface areas and their gradients for macromolecules. J Comp Chem. 1998;19:319–33. [Google Scholar]
- 27.Soman KV, Schein CH, Zhu H, Braun W. Homology modeling and simulations of nuclease structures. Methods Mol Biol. 2001;160:263–86. doi: 10.1385/1-59259-233-3:263. [DOI] [PubMed] [Google Scholar]
- 28.Albert MJ, Faruque AS, Faruque SM, Sack RB, Mahalanabis D. Case-control study of enteropathogens associated with childhood diarrhea in Dhaka, Bangladesh. J Clin Microbiol. 1999;37:3458–64. doi: 10.1128/jcm.37.11.3458-3464.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Girón JA, Jones T, Millán-Velasco F, et al. Diffuse-adhering Escherichia coli (DAEC) as a putative cause of diarrhea in Mayan children in Mexico. J Infect Dis. 1991;163:507–13. doi: 10.1093/infdis/163.3.507. [DOI] [PubMed] [Google Scholar]
- 30.Ishitoya S, Yamamoto S, Kanamaru S. Distribution of afaE adhesins in Escherichia coli isolated from Japanese patients with urinary tract infection. J Urol. 2003;169:1758–61. doi: 10.1097/01.ju.0000057968.53213.b1. [DOI] [PubMed] [Google Scholar]
- 31.Johnson JR, Owens K, Gajewski A, Kuskowski MA. Bacterial characteristics in relation to clinical source of Escherichia coli isolates from women with acute cystitis or pyelonephritis and uninfected women. J Clin Microbiol. 2005;43:6064–72. doi: 10.1128/JCM.43.12.6064-6072.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kachroo BB. Association between antibiotic resistance and the expression of Dr adhesin among uropathogenic Escherichia coli. Chemotherapy. 2001;47:97–103. doi: 10.1159/000048507. [DOI] [PubMed] [Google Scholar]
- 33.Nowicki B, Svanborg-Edén C, Hull R, Hull S. Molecular analysis and epidemiology of the Dr hemagglutinin of uropathogenic Escherichia coli. Infect Immun. 1989;57:446–51. doi: 10.1128/iai.57.2.446-451.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goluszko P, Moseley S, Truong LD, Kaul A, Nowicki S, Nowicki B. Development of experimental model of chronic pyelonephritis with Escherichia coli O75:K5:H- bearing Dr fimbriae: mutation in the dra region prevented tubulointerstitial nephritis. J Clin Invest. 1997;99:1–11. doi: 10.1172/JCI119329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Das M, Hart-Van Tassell A, Urvil PT, et al. Hydrophilic domain II of Escherichia coli Dr fimbriae facilitates cell invasion. Infect Immun. 2005;73:6119–26. doi: 10.1128/IAI.73.9.6119-6126.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Albuquerque EX, Pereira EFR, Alkondon M, Rogers SW. Mammalian nicotinic acetylcholine receptors: from structure to function. Physiol Rev. 2009;89:73–120. doi: 10.1152/physrev.00015.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Horvath DJ, Jr, Li B, Casper T, et al. Morphological plasticity promotes resistance to phagocyte killing of uropathogenic Escherichia coli. Microbes Infect. 2011;13:426–37. doi: 10.1016/j.micinf.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.