A nanometer thick, micron scale-patterned interface on a polymeric material directs fibroblast proliferation into a highly aligned, confluent cell monolayer. These cells assemble fibronectin extracellular matrix (ECM) fibrils that are aligned with the pattern, and matrix alignment on the synthetic polymer surface is maintained throughout a decellularization process. Biologic relevance of this ECM-synthetic material composite is illustrated by directing oriented neurite outgrowth in register with the aligned matrix fibrils.
Oriented ECM fibers are essential for normal tissue development and homeostasis; they give tissues their characteristic properties. As examples, collagen fibers in tendons and ligaments are aligned in parallel bundles in the direction of major tension,1 and neurite outgrowth occurs along linear paths composed of ECM and cells.2, 3 When tissue is damaged, this critical organization of ECM proteins is lost and is replaced initially with a provisional matrix composed of fibrin and fibronectin that is remodeled over time to form, ultimately, a scar;4 native properties of this ECM are not completely regenerated. ECM fibrils with native tissue anisotropy are crucial for guiding cellular re-growth, such as is needed for oriented regeneration of neuronal, corneal and musculoskeletal tissues. 5 Thus, a major challenge for tissue regeneration is to devise templates that direct the assembly of cells and their ECM into arrangements that possess the unique physical and mechanical properties of the undamaged tissue.
Various methods exist to control cell organization on materials, including using cell adhesive or anti-adhesive patterning and surface topography.6, 7 However, these methods cannot always maintain cell organization over the extended time periods that are required for cells to assemble ECM proteins in spatially defined ways. In vitro approaches to generate aligned matrix fibrils, such as by fabricating purified ECM proteins into fibers,8–10 lack other cell-derived components and the native three-dimensional organization of a cell-assembled matrix. The diverse molecules in a cell-derived matrix, as used here, are likely to provide biochemical signals that synergize with other types of guidance cues. Conceptually, this type of material could be a powerful tool for personalized medicine regimes to allow use of autologous stem cells with simple surface chemistry to generate a naturally assembled ECM for spatially predetermined tissue repair.
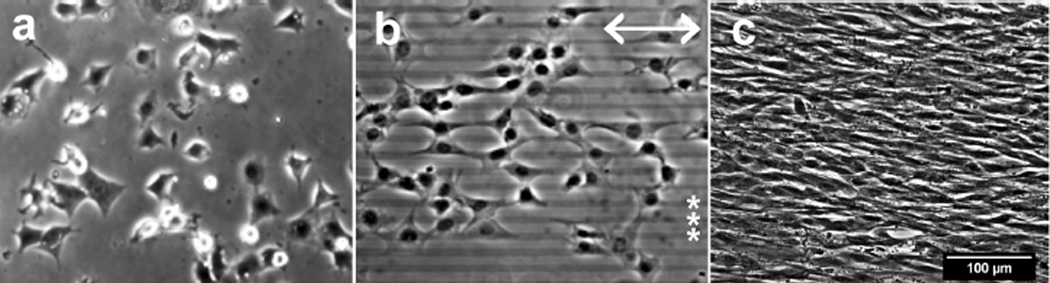
We have reported that surface patterning with a two-component, nanometer thin, micron scale, self-assembled alkylphosphonate monolayer (SAMP) interface synthesized on in situ generated ZrO2 (ZrO2/SAMP) can be used to effect alignment of cell spreading across an entire two-dimensional surface with high fidelity to the pattern.11 These patterned interfaces are prepared using commercially available small molecules through a simple photolithographic process that is readily scalable and that is applicable to a range of polymeric substrates. Unlike physically adsorbed protein patterns, the ZrO2/SAMP coating is robustly bonded to the polymer substrate through coordination complexation, and is prepared in two simple steps. First, volatile zirconium tetra(tert-butoxide) is condensed from the vapor phase onto the patterned substrate; it is then heated to generate cross-linked ZrO2 that is bound to the polymer surface through coordination of its ester oxygen groups to the metal centers, which is substantiated by angle-dependent XPS analysis of the interface (see Supplementary Figure 1). Then, the substrate is dipped into a solution of the phosphonic acid to form the polymer-attached ZrO2/SAMP, which enjoys a long shelf life at ambient temperature. In this way patterns of 10 µm wide stripes of ZrO2/SAMP and 10 µm wide gaps (10 µm × 10 µm pattern) were prepared on poly(ethylene terephthalate) (PET), a polymer that is used in vascular graft biomaterials and is replete with ester carbonyl groups.12 The ZrO2/SAMP-terminated substrate supports cell adhesion and spreading (Figure 1a). NIH 3T3 fibroblasts on the 10 × 10 µm striped pattern spread in alignment with the pattern (Figure 1b). Note that because these cells are larger than 10 µm wide, they tend to spread over two or more stripes,11 but in many cases, one edge of a cell is aligned with the edge of the pattern. The cells remained aligned with the pattern as they grew to confluence (Figure 1c).
Figure 1.
Fibroblast alignment on micro-patterned PET. NIH3T3 cells cultured on unpatterned (a) or a 10 × 10 µm striped pattern (b, c) were visualized by phase contrast microscopy after 4 hours (a, b) and 10 days (c). Scale bars = 100µm. Double arrow indicates the orientation of the ZrO2/SAMP pattern (in b) and the right ends of several pattern stripes are indicated by asterisks (*, in b).
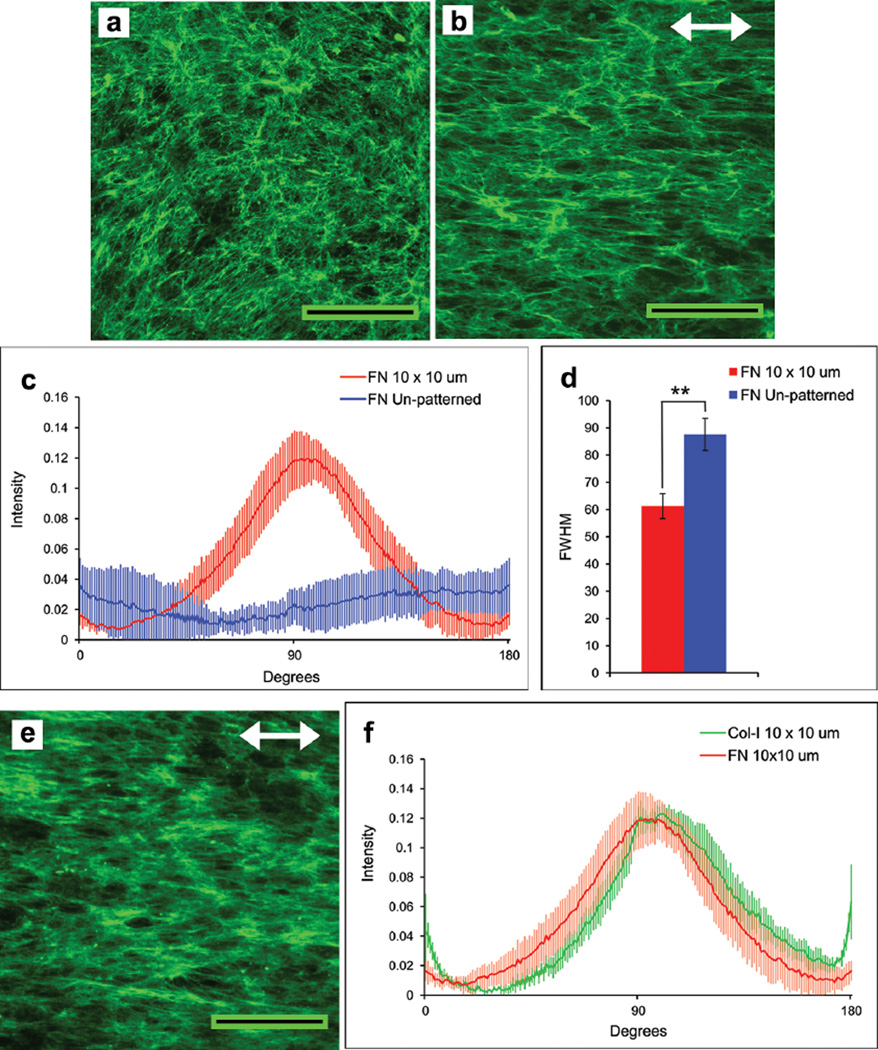
Fibroblasts plated on our patterned substrates assemble a natural fibrillar matrix composed primarily of fibronectin, which is an essential ECM component that interacts directly with integrins and other cell receptors to organize the actin cytoskeleton.13, 14 Fibronectin has binding domains for a variety of ECM molecules including collagens, proteoglycans, and other glycoproteins; in this capacity, fibronectin is a foundational matrix that is required for deposition of type I collagen and other ECM proteins.14, 15 During growth of NIH 3T3 fibroblasts for 10 days on 10 µm × 10 µm stripe-patterned PET, a robust ECM was assembled. Immunofluorescence detection with anti-fibronectin antibodies showed matrix fibrils that were oriented with the pattern (Figure 2b) in contrast to the matrix on the unpatterned surface (Figure 2a). The degree of matrix alignment was quantified by performing two-dimensional fast-Fourier transform analysis (FFT) of the images. The radial summation of pixel intensity in the FFT output for each angle was plotted between 0 to 180°. A peak centered at 90° for the patterned substrates indicated alignment of matrix fibrils parallel to underlying patterned stripes (Figure 2c). No peak was obvious for matrix on an unpatterned substrate. The variation in height and shape of the major peak is represented quantitatively by full width-half maximum (FWHM) values. A significantly lower FWHM for the patterned sample indicates higher alignment of fibrils compared to the random fibrils on the unpatterned substrates (Figure 2d). In addition to fibronectin, the matrix also contained type I collagen fibrils (Figure 2e), which were co-aligned with the fibronectin fibrils and the pattern (Figure 2f).
Figure 2.
Oriented assembly of a matrix composed of fibronectin and collagen fibrils. NIH 3T3 fibroblasts were grown to confluence on unpatterned (a) or 10 × 10 µm patterned (b, e) ZrO2/SAMP-modified PET substrates. The matrix was detected on day 10 by immunostaining with anti-fibronectin antiserum (a, b) or anti-type I collagen antibodies (e). Scale bars = 100 µm. Double arrows indicate pattern orientation. Images were processed by two-dimensional FFT, and the radial summation of pixel distributions around the origin in the FFT outputs was plotted between 0° and 180° (c, f). The FWHM of each pixel intensity curve in (c) was calculated and is plotted in (d). Col-I: type I collagen; FN: fibronectin. ** p < 0.0005.
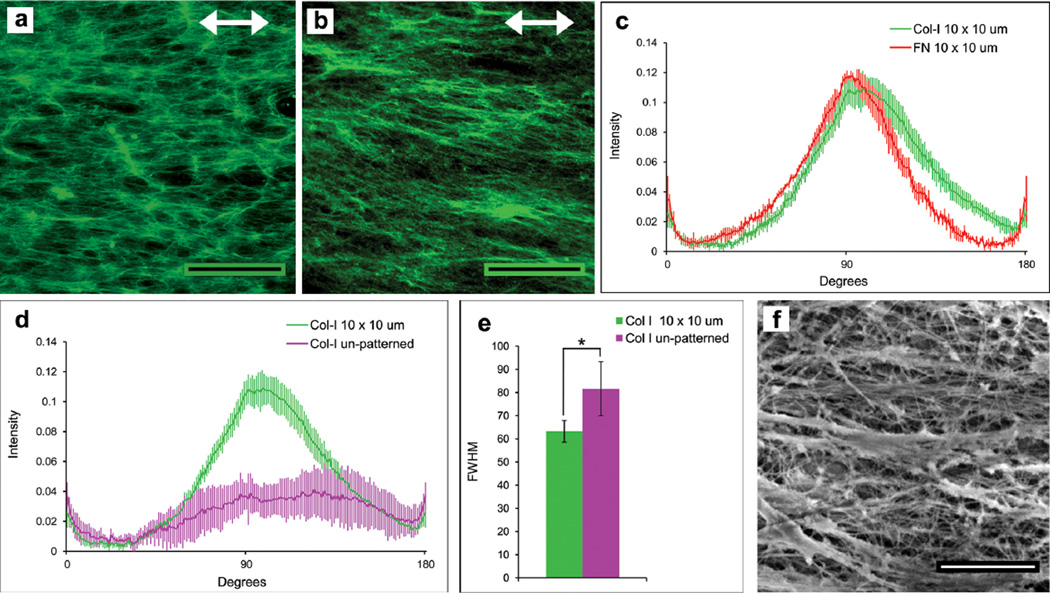
A mild cell-lysis protocol was used to decellularize the matrices that were cell-assembled on PET. We showed previously that this procedure maintains the organization and dimensionality of the matrix.16 Compared to cells on a two-dimensional fibronectin-coated surface, cell activities such as adhesion, migration, and matrix assembly are enhanced when cells are grown on a decellularized three-dimensional ECM (as used here).16, 17 Immunofluorescence and FFT analyses of fibronectin and type I collagen matrix fibrils after decellularization confirmed that the matrix remained attached to the PET substrate and that fibril alignment was maintained with the pattern (Figure 3a–c). Quantification showed significant alignment of type I collagen fibrils in decellularized matrix on patterned PET compared to unpatterned surfaces (Figure 3d, e). Analysis of the decellularized matrix by scanning electron microscopy (SEM) highlights the density of fibrils and the three-dimensionality of the matrix (Figure 3f).
Figure 3.
Characterization of decellularized matrices. After decellularization, matrices were stained with anti-fibronectin (a) or anti-collagen type I antibodies (b) and the FFT pixel intensity distribution plots for fibronectin (FN) and type I collagen (Col-I) in this matrix are shown in (c). (d, e) Images of type I collagen fibrils in decellularized matrix on unpatterned or 10 × 10 µm ZrO2/SAMP patterned PET were used for FFT analysis (d) and the FWHM of these curves were determined (e). (f) SEM image of decellularized matrix. Scale bars = 100 µm (a, b) and 2 µm (f). * p < 0.05.
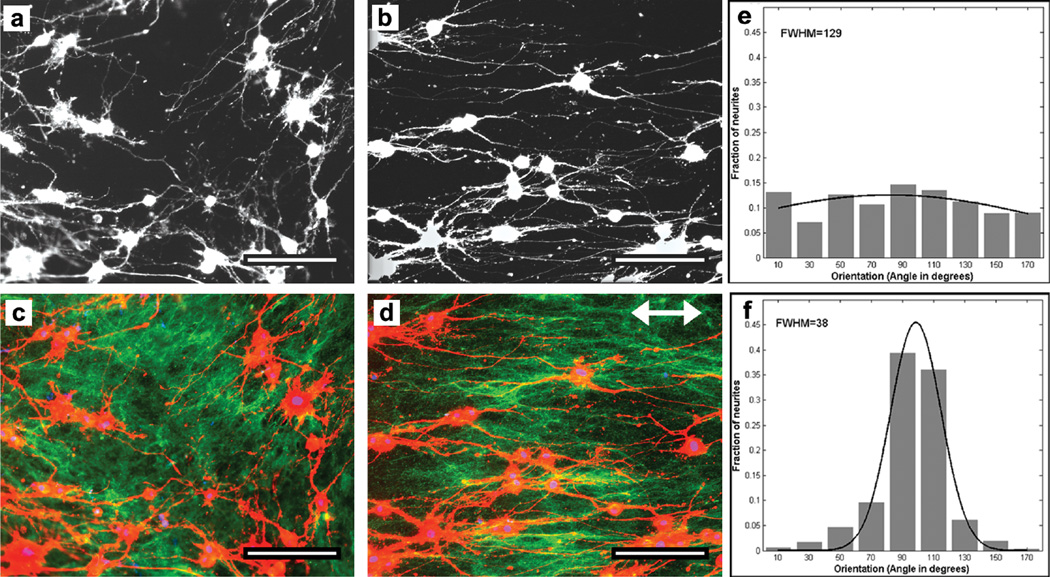
Oriented cell growth is essential in nerve regeneration, which depends on linear extension of axons. Many scaffold devices use conduits fabricated from type I collagen, chitosan, porcine small intestinal submucosa, or poly-ε-caprolactone (PCL),18 or materials micro-patterned with an applied coating of purified ECM proteins;6, 19 others modulate surface topology by microgrooving or use electrospun materials as substrates.10, 20 These devices do not recreate the microarchitecture or composition of native ECM. Given that both fibronectin and type I collagen have been shown to guide neurite outgrowth,6, 21–23 and as proof of function for our decellularized, aligned matrix, we tested the response to it of PC12 cells, which are a model system for neural differentiation.24, 25 PC12 cells were plated on this aligned ECM/PET material and were stimulated for neurite outgrowth, which was quantitatively evaluated through directional analysis (Figure 4). The cells cultured on patterned or unpatterned, decellularized matrix extended neurites, which were visualized by staining the actin cytoskeleton (Figure 4a, b). Co-staining of the matrix fibrils indicated correspondence between the direction of neurite outgrowth and the orientation of fibrils (compare Figure 4c and 4d). Measurements of neurite projection angles relative to the patterned matrix showed the majority of neurites in line with the 10 µm × 10 µm pattern (90° peak, Figure 4f) compared to the lack of a single peak in neurite angles on the unpatterned matrix (Figure 4e). The aligned orientation of neurites is also reflected in the three-fold decrease in FWHM value compared to the unpatterned substrate (Figure 4e, f). In this way, we showed that our cell-assembled aligned ECM supports neurite outgrowth that is oriented in register with the underlying ZrO2/SAMP-patterned surface. This system therefore provides a platform for studying the effects of alignment in devices used for nerve reconstruction.
Figure 4.
Directional neurite outgrowth on decellularized matrix. PC12 cells were induced to extend neurites with NGF for 72 hr on unpatterned (a, c) or 10 × 10 µm patterned (b, d) decellularized matrix PET substrates. Cells were visualized by staining the actin cytoskeleton with rhodamine-phalloidin (a–d; red in c, d). Matrix fibrils were immunostained with anticollagen type I antibodies (c, d; green). Double arrow indicates pattern orientation. Scale bars = 100 µm. (e, f) Angles of neurites were measured relative to an arbitrary orientation for unpatterned (e) or to the pattern stripes (f) and binned in ranges of 20° centered on 90° which is parallel to the pattern. Each histogram of the angular distributions of neurites was fitted to a Gaussian to calculate the FWHM values.
In conclusion, we have shown that cells assemble their ECM in alignment with the ZrO2/SAMP pattern and that ECM alignment is maintained following decellularization. Neural cells adhere to the decellularized matrix and extend their neurites oriented in line with the ECM fibrils. Several cell types, including human mesenchymal stem cells, proliferate in register with our ZrO2/SAMP-terminated surface patterns.11 Like fibroblasts, mesenchymal stem cells assemble a fibronectin-rich matrix (unpublished observations) that will be suitable for decellularization studies. Together, our results support the contention that our technology will be applicable to spatially direct the assembly of a broad range of tissue-specific ECMs by using the appropriate matrix-forming cells. With such tissue-specific matrices in hand, two methods might then be envisaged to address tissue repair and regeneration scenarios: One would involve ex situ repopulation of an aligned matrix with tissue-specific cells prior to being placed in a wound bed; another would apply the cell-assembled, aligned matrix directly to a wound in which it would then serve in situ as a “smart band-aid” to template tissue regeneration.
Supplementary Material
Acknowledgments
We thank Prof. Lynn Enquist and Dr. Tal Kramer, both of the Department of Molecular Biology, Princeton University, for supplying the PC12 cells and for technical assistance. This research was supported by grants from the NIH (CA160611 to JES), the NSF (CHE-0924104 to JS), the National Football League Charities, and the New Jersey Center for Biomaterials NIBIB Post-doctoral Training Program (T32EB005583 to SS).
Contributor Information
Shivani Singh, Department of Molecular Biology, Princeton University, Princeton, NJ 08544-1014 (USA).
Stephen B. Bandini, Department of Chemistry, Princeton University, Princeton, NJ 08544-1009 (USA)
Patrick E. Donnelly, Department of Chemistry, Princeton University, Princeton, NJ 08544-1009 (USA)
Jeffrey Schwartz, Department of Chemistry, Princeton University, Princeton, NJ 08544-1009 (USA).
Jean E. Schwarzbauer, Department of Molecular Biology, Princeton University, Princeton, NJ 08544-1014 (USA)
References
- 1.Birk DE, Brückner P. In: The Extracellular Matrix: an Overview. Mecham RP, editor. Berlin Heidelberg: Springer; 2011. pp. 77–115. [Google Scholar]
- 2.Huber AB, Kolodkin AL, Ginty DD, Cloutier JF. Annu. Rev. Neurosci. 2003;26:509–563. doi: 10.1146/annurev.neuro.26.010302.081139. [DOI] [PubMed] [Google Scholar]
- 3.Lowery LA, Van Vactor D. Nat. Rev. Mol. Cell Biol. 2009;10:332–343. doi: 10.1038/nrm2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clark RA, Ghosh K, Tonnesen MG. J. Invest. Dermatol. 2007;127:1018–1029. doi: 10.1038/sj.jid.5700715. [DOI] [PubMed] [Google Scholar]
- 5.Silvestri A, Boffito M, Sartori S, Ciardelli G. Macromol. Biosci. 2013;13:984–1019. doi: 10.1002/mabi.201200483. [DOI] [PubMed] [Google Scholar]
- 6.Meng F, Hlady V, Tresco PA. Biomaterials. 2012;33:1323–1335. doi: 10.1016/j.biomaterials.2011.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whitesides GM, Ostuni E, Takayama S, Jiang X, Ingber DE. Annu. Rev. Biomed. Eng. 2001;3 doi: 10.1146/annurev.bioeng.3.1.335. [DOI] [PubMed] [Google Scholar]
- 8.Burck J, Heissler S, Geckle U, Ardakani MF, Schneider R, Ulrich AS, Kazanci M. Langmuir. 2013;29:1562–1572. doi: 10.1021/la3033258. [DOI] [PubMed] [Google Scholar]
- 9.Cisneros DA, Friedrichs J, Taubenberger A, Franz CM, Muller DJ. Small. 2007;3:956–963. doi: 10.1002/smll.200600598. [DOI] [PubMed] [Google Scholar]
- 10.Neal RA, Tholpady SS, Foley PL, Swami N, Ogle RC, Botchwey EA. J. Biomed. Mat. Res. A. 2011;100A:406–423. doi: 10.1002/jbm.a.33204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donnelly PE, Jones CM, Bandini SB, Singh S, Schwartz J, Schwarzbauer JE. J. Mater. Chem. B. 2013;1:3553–3561. doi: 10.1039/C3TB20565G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Devine C, McCollum C. J. Vasc. Surg. 2004;40:924–931. doi: 10.1016/j.jvs.2004.08.033. [DOI] [PubMed] [Google Scholar]
- 13.Schwarzbauer JE, Desimone DW. Cold Spring Harb. Perspect. Biol. 2011;3 doi: 10.1101/cshperspect.a005041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singh P, Carraher C, Schwarzbauer JE. Annu. Rev. Cell Dev. Biol. 2010;26:397–419. doi: 10.1146/annurev-cellbio-100109-104020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sottile J, Shi F, Rublyevska I, Chiang HY, Lust J, Chandler J. Am. J. Physiol. Cell Physiol. 2007;293:C1934–C1946. doi: 10.1152/ajpcell.00130.2007. [DOI] [PubMed] [Google Scholar]
- 16.Mao Y, Schwarzbauer JE. J. Cell Sci. 2005;118:4427–4436. doi: 10.1242/jcs.02566. [DOI] [PubMed] [Google Scholar]
- 17.Cukierman E, Pankov R, Stevens DR, Yamada KM. Science. 2001;294:1708–1712. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- 18.Bell JHA, Haycock JW. Tissue Eng. Part B Rev. 2012;18:116–128. doi: 10.1089/ten.TEB.2011.0498. [DOI] [PubMed] [Google Scholar]
- 19.Williams C, Tsuda Y, Isenberg BC, Yamato M, Shimizu T, Okano T, Wong JY. Adv. Mater. 2009;21:2161–2164. [Google Scholar]
- 20.Spivey EC, Khaing ZZ, Shear JB, Schmidt CE. Biomaterials. 2012;33:4264–4276. doi: 10.1016/j.biomaterials.2012.02.043. [DOI] [PubMed] [Google Scholar]
- 21.Abu-Rub MT, Billiar KL, van Es MH, Knight A, Rodriguez BJ, Zeugolis DI, McMahon S, Windebank AJ, Pandit A. Soft Matter. 2011;7:2770–2781. [Google Scholar]
- 22.Volkenstein S, Kirkwood J, Lai E, Dazert S, Fuller G, Heller S. Eur. Arch. Otorhinolaryngol. 2012;269:1111–1116. doi: 10.1007/s00405-011-1775-8. [DOI] [PubMed] [Google Scholar]
- 23.Mukhatyar VJ, Salmeron-Sanchez M, Rudra S, Mukhopadaya S, Barker TH, Garcia AJ, Bellamkonda RV. Biomaterials. 2011;32:3958–3968. doi: 10.1016/j.biomaterials.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fujita F, Lazarovici P, Guroff G. Environ. Health Perspect. 1989;80:127–142. doi: 10.1289/ehp.8980127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vogelezang M, Forster U, Han J, Ginsberg M, ffrench-Constant C. BMC Neuroscience. 2007;8:44. doi: 10.1186/1471-2202-8-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.