Abstract
We used single cell genomic approaches to map DNA copy number variation (CNV) in neurons obtained from human induced pluripotent stem cell (hiPSC) lines and post-mortem human brains. We identified aneuploid neurons as well as numerous subchromosomal CNVs in euploid neurons. Neurotypic hiPSC-derived neurons had larger CNVs than fibroblasts, and several large deletions were found in hiPSC-derived neurons but not in matched neural progenitor cells. Single cell sequencing of endogenous human frontal cortex neurons revealed that 13%-41% of neurons have at least one megabase-scale de novo CNV, that deletions are twice as common as duplications, and that a subset of neurons have highly aberrant genomes marked by multiple alterations. Our results show that mosaic copy number variation is abundant in human neurons.
Neuronal genomes exhibit elevated levels of aneuploidy (1-3) and retrotransposition (4-6) relative to other cell types; this finding has fueled speculation that somatic genome variation may contribute to functional diversity in the human brain (7-10). The prevalence of CNVs has been difficult to assess given the limited ability of conventional genome-wide methods to detect CNVs that are rare within a population of cells, as most somatic mutations are expected to be. Recently, two methods have been developed to map large-scale CNVs in single cells: microarray analysis of multiple displacement amplification (MDA) products (11), and single cell sequencing (12). Here, we applied both of these approaches to single human neurons.
We examined human neurons from two neurotypic sources (Fig. S1A): 1) human induced pluripotent stem cells [i.e., hiPSC-derived neurons (Fig. S2)] and 2) human post-mortem frontal cortex (FCTX) neurons (Fig. S3). We employed fluorescence activated cell sorting (FACS) to obtain neurons from neuronogenic hiPSC cultures based on synapsin::GFP expression and from post-mortem tissue based on NeuN immunostaining (13). After multiple displacement amplification (MDA) (14), we hybridized single hiPSC-derived neuronal genomes to Affymetrix 250K SNP arrays (as in (11)). We subjected single neurons from post-mortem tissue to Illumina DNA sequencing using a custom version of the single cell sequencing protocol developed by Navin et al. (12), which combines the GenomePlex whole-genome amplification method with Nextera-based library preparation (15). We developed stringent quality control measures to ensure that only the highest quality amplification reactions and datasets were included in downstream analyses (see Methods).
To detect CNVs, we first aggregated raw copy number measurements over very large genomic intervals. We then selected interval sizes that were 1 - 2 orders of magnitude larger than the local amplification biases reported for single cell DNA amplification (16, 17). For SNP array data, we calculated the median copy number in 100-probe bins, which corresponds to a mean genomic interval of 666 Kb; for sequencing data, we measured read-depth in bins composed of 500 Kb of uniquely mappable sequence (mean size of 687 Kb). CNVs were identified using circular binary segmentation (18) combined with strict filtering based on the number of consecutive bins identified by segmentation and the amplitude of CNV predictions relative to the noise (median absolute deviation) of each dataset. These methods and filtering criteria resulted in a mean CNV size detection limit of 6.7 Mb for SNP array data and 3.4 Mb for sequencing data. A subset (n = 7) of the MDA-amplified hiPSC-derived neurons, analyzed by both SNP array and sequencing, showed high concordance (Fig. S1B and Fig. S4). Sub-chromosomal deletions (Fig. 1A, C) and duplications (Fig. 1B, D) were identified in both groups of neurons.
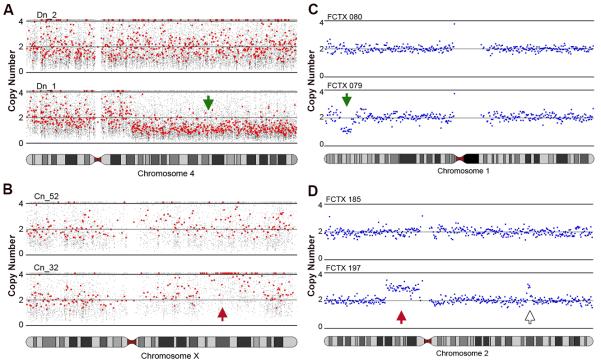
Figure 1. Mosaic copy number variation (CNV) is detected in human neurons.
(A, B) Subchromosomal deletions (green arrow) and duplications (red arrow) are observed in hiPSC-derived neurons. (A) Neuron Dn_1 has a deletion on chromosome (chr) 4q (lower panel); neuron Dn_2 has no CNV on Chr4 (upper panel). Small gray dots show the predicted copy number at individual SNPs; red dots show every 30th SNP. (B) Neuron Cn_32 has a duplication on ChrXq (lower panel); neuron Cn_2 does not (upper panel). (C, D) Single cell sequencing reveals subchromosomal deletions (green arrow) and duplications (red arrow) in FCTX neurons. (C) FCTX079 has a deletion on Chr1p (lower panel); FCTX080 does not (upper panel). Blue dots show raw copy number predictions obtained by read-depth analysis (mean window size ~687 Kb; see methods) (D) Neuron FCTX197 has a duplication on Chr2p (lower panel) whereas FCTX185 does not (upper panel). Another likely duplication on Chr2q in FCTX197 (open arrow) was comprised of only four consecutive bins and therefore failed our five-bin confidence threshold.
We examined neurons from three hiPSC lines, referred to as C, D, and E, that were generated from three different individuals as neurotypic controls for a hiPSC-based disease model (19). Analysis of bulk DNA from C and D line donor fibroblasts or hiPSC-derived neuronal progenitor cells (NPCs) revealed no clonal genomic aberrations. Of 40 single neurons analyzed [C(n = 21), D(n = 6), E(n = 13)], 27 had copy number profiles consistent with bulk DNA, but 13 had unique genomes. In total, we identified seven whole chromosome gains, four whole chromosome losses, and 12 sub-chromosomal CNVs (range: 7.0 Mb – 156 Mb) in 13 hiPSC-derived neurons (Fig. 2A, Fig. S5, and Table S1). Each CNV was identified in merely one neuron, suggesting that the CNVs are not early clonal events but rather are unique to single cells or distinct lineages.
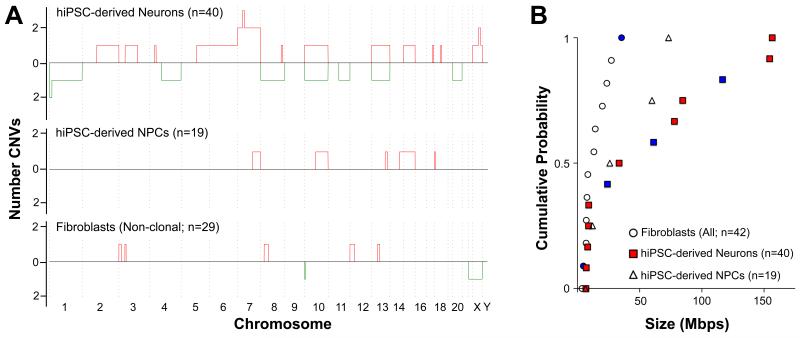
Figure 2. Large CNVs are found in hiPSC-derived neurons.
(A) Whole and subchromosomal duplications (red) and deletions (green) are summarized for 40 hiPSC-derived neurons (upper panel). The y-axis value represents the number of times each genomic interval was deleted (below in green) or duplicated (above in red). CNVs were detected in 9 of 21 C neurons (Cn), 2/6 D neurons (Dn) and 2/13 E neurons (En). In donor hiPSC-derived NPC populations (middle panel), CNVs were detected in 1/10 D NPCs (Dp) and 3/9 C NPCs (Cp). In donor fibroblast populations (lower panel), CNVs were detected in 7/20 D fibroblasts (Df) and 0/9 C fibroblasts (Cf). Note that chromosomes are not plotted to scale because data is summarized in 100-SNP bins. (B) Subchromosomal CNVs in fibroblasts were significantly smaller than in hiPSC-derived neurons (KS test, P < .001). No deletions were observed in NPCs. Deletions are denoted with blue markers; all other markers indicate duplications. Aneuploidies are not included in this plot. For completeness, subchromosomal CNVs from clonal fibroblasts (Fig. 3) were included in this plot, bringing the total n to 42 fibroblasts.
The CNVs detected in C and D line hiPSC-derived neurons were distinct from those seen in either C or D line fibroblasts or NPCs (Fig. 2). Of 29 fibroblasts, six had single CNVs (range: 5.2 - 27.7 Mb) and one was aneuploid (−22, −X) (Fig. 2A). Among 19 hiPSC-derived NPCs, only six duplications were observed (Fig. 2A). Technical replicates of five fibroblasts and three hiPSC-derived neurons showed high concordance, and principal component analysis also showed that replicates from each individual neuron clustered distinctly from both the fibroblasts and the other two neurons (Fig. S2E). Comparison of CNVs in the three cell types (Fig. 2B) showed that neurons have significantly larger CNVs than fibroblasts (KS test, P < .001). In addition, we found deletions only in hiPSC-derived neurons and not in hiPSC-derived NPCs.
We performed two additional experiments to confirm that low-level aneuploidy and CNVs occur in single fibroblasts. First, we obtained single cell clones by limiting dilution. Each single fibroblast was expanded to ~20 sister cells over seven days; then we obtained individual sister fibroblasts from three different clonal expansions. In one of these clones, chromosome missegregation was observed as a gain of Chr2 in one cell and a loss of Chr2 in a sister cell (Fig. 3A). Non-clonal CNVs were also detected, so we performed a second experiment using fluorescence in situ hybridization (FISH) for a common hiPSC CNV on Chr20 (20) and for ChrX. Consistent with genomic analysis of bulk DNA, 20 metaphase spreads from this population karyotyped as euploid, but 13/200 were aneuploid for ChrX (Fig. 3B) and 26/200 nuclei had a Chr20 CNV (Fig. 3C). These data show that two distinct approaches (SNP array and FISH) detect large non-clonal CNVs that arise in single human cells in culture.
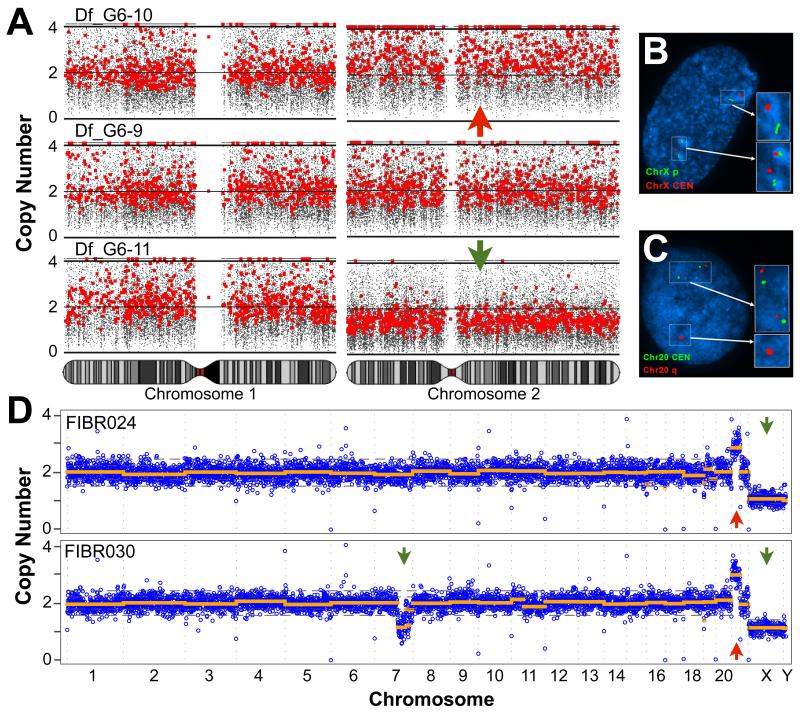
Figure 3. Large CNVs are found in cultured fibroblasts.
(A) Single fibroblasts obtained by limiting dilution were expanded to a population of ~20 clonal fibroblasts after seven days in vitro (DIV). In one clonal population, a reciprocal chromosome missegregation event was detected. One fibroblast was trisomic for Chr2 (upper panel) and a sister was monosomic for Chr2 (lower panel). Chromosomes 1 - 3 are shown alongside the third euploid cell. (B, C) Two groups of Df (passage 7 and 8) were summarized in (Fig 2A); a parallel culture of the p7 group was sent for karyotyping and FISH. 20/20 metaphase chromosome spreads were euploid. (B) FISH was performed for a ChrX p arm telomere (green) and ChrX centromere (red). 13/200 nuclei were aneuploid. (C) FISH was performed for the Chr20 centromere (green) and Chr20 CNV (red). 26/200 nuclei had the CNV. (D) Single cell sequencing of 2 male fibroblasts with karyotypically defined trisomy 21. Genome-wide copy number profiles show that, in both cells, most of the genome is present at 2 copies, Chr21 is present at 3 copies, and ChrX is present at 1 copy. In addition, we identified a large deletion on Chr7q in FIBR030. DNA copy number (y-axis) was calculated by read-depth analysis of variably sized genomic windows containing 500 Kb of uniquely mappable sequence (blue), and CNVs were detected by circular binary segmentation (orange). Green and red arrows denote deletions and duplications, respectively, that were identified by segmentation and passed filtering criteria. Reported CNVs comprise five or more consecutive bins and exceed two MADs. Dotted gray lines show 1 and 2 median absolute deviations (MADs) from the median copy number of each dataset. See Figs. S6 and S7 for plots of additional cells.
We next sought to determine if mosaic CNVs were also present in FCTX neurons from postmortem human brains. For these experiments we used the single cell sequencing method (12), which offers superior sensitivity to microarray approaches due to the digital nature of DNA sequence data (12, 21). After benchmarking the sequencing approach with trisomic, male fibroblasts where we identified 100% trisomy 21 and monosomy X (Fig. 3D, Fig. S6, and Table S2), we sequenced 110 FCTX neurons from three different individuals [a 24-year-old female (NICHD Brain Bank ID#5125; n = 19), a 26-year-old male (ID#1583; n = 41), and a 20-year-old female (ID#1846; n = 50)] and used strict filtering criteria to identify high confidence CNVs (see Methods) composed of five or more consecutive bins. We identified 100% monosomy X and Y in the 41 male neurons (Fig. 4A, Fig. S7, and Table S3) as expected, and simulation experiments indicate that our methods detected CNVs at high sensitivity and specificity, with a predicted mean false negative rate of 17% and a predicted mean false discovery rate of 0.6% (Fig S8; see Methods).
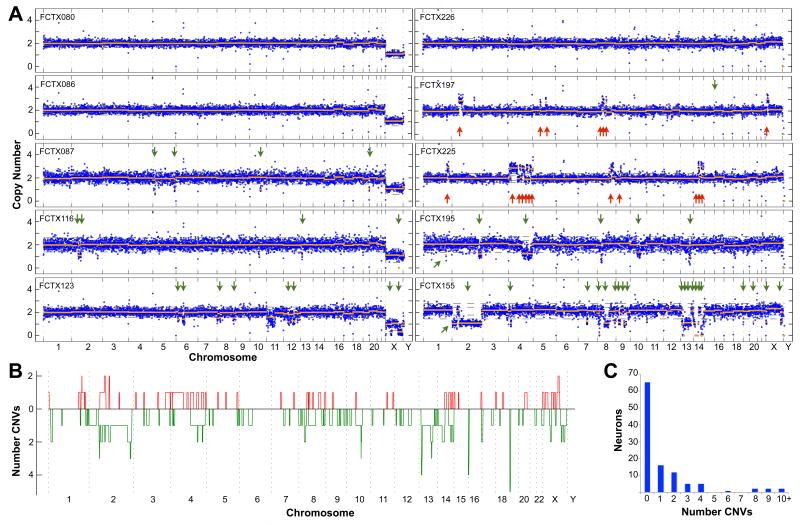
Figure 4. Identification of CNVs in post-mortem neurons using single cell sequencing.
(A) Genome-wide copy number profiles of five male (left panels) and five female (right) neurons from two individuals, #1583 and #1846, respectively. Data are plotted exactly as in Fig. 3D. Arrows denote deletions (green) and duplications (red) that were identified by copy number segmentation and passed filtering criteria. Note that single copy “losses” of ChrX in cells from male individual #1583 are not indicated by arrows, but were identified in 100% of cells. See Fig. S7 for plots of all cells. (B) Whole and subchromosomal duplications (red) and deletions (green) are summarized for the 110 FCTX neurons as in Fig. 2A. (C) Bar chart showing the number of individual neurons (y-axis) that exhibited a given number of CNVs (X-axis). See Fig. S11 for results at different CNV detection stringency thresholds.
We identified one or more somatic CNVs in 45 of the 110 (41%) FCTX neurons analyzed (Fig. 4, Fig. S7, and Table S2). The vast majority of somatic CNVs were subchromosomal alterations ranging in size from 2.9 to 75 Mb, although we also identified one putative chromosome gain and two losses where CNV calls affected >50% of the chromosome (e.g., FCTX155, Fig. 4A). Subchromosomal CNVs were distributed throughout the genome, and in only one case did two independent CNVs share the same breakpoints (a 3 Mb subtelomeric deletion on Chr16 in FCTX198 and FCTX224 (Fig. S7, and Table S2). However, a number of loci were affected by multiple “small” CNVs less than 20Mb in size (N=133), and small CNVs were preferentially found at telomeres (Fig. 4B), with 23.3% extending to the chromosome end (2067-fold enrichment by Monte-Carlo, see Methods). Small CNVs are not enriched with features known to affect genome stability such as transposons, segmental duplications or fragile sites; neither are they enriched with germline CNVs or known genes (Fig. S9). Subchromosomal deletions were prevalent in each of the three individuals and were twice as common as duplications, on average, which might be explained by a bias towards DNA loss in non-dividing post-mitotic neurons; however, the third individual (#1846) was unique in also showing abundant duplications (Fig. S3D - G. These results demonstrate that somatic CNVs are a common feature of neuronal genomes and suggest that the relative abundance of different CNV classes may vary among individuals.
The overall high mutational load that we report in neurons is predominantly due to a small number of cells with highly aberrant genomes. Whereas the majority of FCTX neurons exhibited 0 (59%) or 1-2 CNVs (25%), 17 cells (15%) accounted for 108 of the 148 CNV calls (73%) and seven cells accounted for nearly half (49%) of all calls (Fig. 4C). Aberrant cells are marked by multiple copy number switches on distinct chromosomes, with interdigitated altered and unaltered segments that adhere well to the expectation of integer-like copy number states measured by digital DNA sequencing technology. Similar, if less dramatic, examples of this phenomenon were apparent in hiPSC neurons, where several cells harbored multiple alterations. For example, hiPSC-derived neuron Cn_32 had five events: loss of Chr13, three duplications, and one deletion (Fig. S10). Similarly, two FCTX neurons had more than 10 events. One of these, FCTX 155, was aneuploid for most of Chr2 and had 18 deletions and one duplication (Fig. 4A). We did not observe similarly aberrant copy number profiles among the 16 control fibroblasts analyzed by sequencing (Fig. S6) or among the 42 fibroblasts or 19 NPCs analyzed by SNP array (Fig. S5). Taken together, these results suggest that a subset of neurons is especially prone to large-scale genome alterations.
Single cell genome analysis is inherently challenging because all existing approaches require amplification of the genome prior to measurement; thus, validation is impossible because one cannot know the state of a single cell’s genome before it was amplified. However, several lines of evidence argue that the vast majority of events we report are true CNVs. First, we used methods that were previously validated on clonally related cell populations, including tumors (12) and eight-cell embryos (11). Second, we report megabase-scale CNVs that are orders of magnitude larger than the amplicons generated by whole genome amplification. Indeed, previous studies have noted that amplification artifacts tended to be small (<10kb) and distributed relatively uniformly across the genome (11, 12); therefore, simple amplification effects cannot readily explain the large-scale deviations in copy number that we observe. It is also difficult to explain how such effects could cause both gains and losses of DNA that produce integral copy number values by sequencing. Third, the post-mortem interval is unlikely to contribute significantly to our results because DNA degradation cannot generate duplications and because we observed large deletions in both FCTX and hiPSC-derived neurons. Fourth, Monte-Carlo simulation experiments showed that our CNV detection methods identify hemizygous gains and losses at high sensitivity and are not affected by random fluctuations in sequence coverage. Fifth, we have employed strict quality control measures to exclude datasets with uneven or noisy amplification or that (in the case of sequence data) do not exhibit expected integer-like copy number profiles (see Methods). Finally, and perhaps most importantly, many of our CNV calls appear to be extremely high quality based on their size, amplitude and integer-like properties (see Fig. 4A; Fig. S6; Fig. S7), and a subset (30-56%) is robust to a series of increasingly strict CNV detection parameters (Fig. S11). At increased stringency, the overall number of CNVs diminishes but the core results do not change: CNVs are apparent in a significant fraction of neurons (13-24%), there is a predominance of deletions relative to duplications (Fig. S11A), and we observe a subset of neurons with highly aberrant genomes marked by multiple copy number oscillations (Fig. S11D). Therefore, although we cannot definitively exclude the possibility of as-yet-undescribed single cell amplification artifacts, the above observations strongly argue that the central results and conclusions of our study are not attributable to technical factors.
Using three completely independent single cell approaches (SNP array, sequencing, and FISH), we find that a subset of cultured fibroblasts has megabase-scale CNVs. Recently, small CNVs (<1Mb) have been estimated to occur in skin fibroblasts at a frequency of perhaps 30%; however, no large CNVs were reported in this study (25). In order to study single somatic cells, Abyzov, et al. reprogrammed fibroblasts and performed deep whole genome sequencing on the hiPSC cell lines that emerged. In contrast, we analyzed single cultured fibroblasts directly using lower resolution methods that cannot resolve small CNVs (<1Mb). Given that many large CNVs are expected to be deleterious and may adversely affect reprogramming or clonal expansion in culture, we believe that the two findings are not inconsistent.
Our single cell genomic analysis of human neurons extends the observation of somatic mosaicism in the nervous system to the single cell level. Several studies using bulk DNA from somatic tissues, including brain, have found CNVs among monozygotic twins (22) and in different organs or brain regions from the same individual (23, 24). These studies were only able to detect CNVs present in >10% of the cells in the bulk sample and thus have only provided a coarse assessment of somatic mosaicism. We have shown that mosaic copy number variation is abundant in human neurons. Additional work will be required to fully address the full spectrum of somatic mutation in neurons and other cell lineages; however, it is possible that some neuronal lineages acquire genomic instability during development, leading to subsequent diversification of neuronal genomes, or that individual neurons become prone to large-scale mutational events due to widespread DNA damage. A recent study has implicated electrophysiological activity as a source of double-strand DNA breaks in neurons (28), and small circular DNAs caused by excision have been reported in multiple somatic cell types, including neurons (26, 27). Additionally, retrotransposon activity is known to cause sub-chromosomal deletions and other rearrangements in human cells (29-32); thus, higher levels of retrotransposon activity during human neurogenesis (5, 33) may also contribute to the prevalence of CNVs in neuronal genomes.
The effect of somatic genome diversification on neuronal function remains unknown. One straightforward hypothesis is that neurons with different genomes will have distinct molecular phenotypes due to altered transcriptional or epigenetic landscapes. We expect that ongoing development of single cell technologies will allow for this hypothesis to be tested by measuring multiple states of the same neuron (e.g., the genome and the epigenome/transcriptome/proteome). We have shown that hiPSC-derived neurons recapitulate somatic variation, as observed in endogenous human neurons; thus hiPSCs may offer a tractable system for applying single cell approaches to understanding the consequences of somatic mosaicism. In the future, the ability to manipulate and measure genomic diversity in human neural circuits in vitro may help to reveal the consequences of somatic mosaicism in the brain.
Supplementary Material
Acknowledgments
We thank D. Husband (Salk), L. Moore (Salk), S. Jackmaert (KU Leuven), R. Layer (UVA) and R. Clark (UVA) for technical assistance, A. Prorock and Y. Bao (UVA Sequencing Core) for DNA sequencing, and all members of the Gage laboratory for critical feedback on the project. We thank M.L. Gage for editorial comments. FHG thanks the Center for Academic Research and Training in Anthropogeny (CARTA) for support and perspective. This work was supported by a Crick-Jacobs Junior Fellowship to MJM; a Mather’s Family Foundation Grant, a NIH TR01 (R01 MH095741), the JPB Foundation and a Helmsley Foundation grant to FHG; and an NIH New Innovator Award (DP20D006493-01) and Burroughs Wellcome Fund Career Award to IMH. Human tissue was obtained from the NICHD Brain and Tissue Bank for Developmental Disorders at the University of Maryland, Baltimore, MD, contract HHSN2752009000011C, Ref. No. N01-HD-9-011. The hiPSC lines used in this study are available from the Coriell Cell Repository. Microarray data have been deposited in the NCBI Gene Expression Omnibus (pending), and DNA sequence data have been deposited in the NCBI Short Read Archive (SRP030642).
References
- 1.Rehen SK, et al. Chromosomal variation in neurons of the developing and adult mammalian nervous system. Proceedings of the National Academy of Sciences of the United States of America. 2001 Nov 6;98:13361. doi: 10.1073/pnas.231487398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rehen SK, et al. Constitutional aneuploidy in the normal human brain. J Neurosci. 2005 Mar 2;25:2176. doi: 10.1523/JNEUROSCI.4560-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yurov YB, et al. Aneuploidy and confined chromosomal mosaicism in the developing human brain. PloS one. 2007;2:e558. doi: 10.1371/journal.pone.0000558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muotri AR, et al. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005 Jun 16;435:903. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- 5.Baillie JK, et al. Somatic retrotransposition alters the genetic landscape of the human brain. Nature. 2011 Nov 24;479:534. doi: 10.1038/nature10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evrony GD, et al. Single-neuron sequencing analysis of L1 retrotransposition and somatic mutation in the human brain. Cell. 2012 Oct 26;151:483. doi: 10.1016/j.cell.2012.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ostertag EM, Kazazian HH. Genetics: LINEs in mind. Nature. 2005 Jun 16;435:890. doi: 10.1038/435890a. [DOI] [PubMed] [Google Scholar]
- 8.Singer T, McConnell MJ, Marchetto MC, Coufal NG, Gage FH. LINE-1 retrotransposons: mediators of somatic variation in neuronal genomes? Trends Neurosci. 2010 May 12; doi: 10.1016/j.tins.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin SL. Developmental biology: Jumping-gene roulette. Nature. 2009 Aug 27;460:1087. doi: 10.1038/4601087a. [DOI] [PubMed] [Google Scholar]
- 10.Bushman DM, Chun J. The genomically mosaic brain: Aneuploidy and more in neural diversity and disease. Semin Cell Dev Biol. 2013 Apr;24:357. doi: 10.1016/j.semcdb.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vanneste E, et al. Chromosome instability is common in human cleavage-stage embryos. Nat Med. 2009 May;15:577. doi: 10.1038/nm.1924. [DOI] [PubMed] [Google Scholar]
- 12.Navin N, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011 Apr 7;472:90. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spalding KL, Bhardwaj RD, Buchholz BA, Druid H, Frisen J. Retrospective birth dating of cells in humans. Cell. 2005 Jul 15;122:133. doi: 10.1016/j.cell.2005.04.028. [DOI] [PubMed] [Google Scholar]
- 14.Dean FB, et al. Comprehensive human genome amplification using multiple displacement amplification. Proceedings of the National Academy of Sciences of the United States of America. 2002 Apr 16;99:5261. doi: 10.1073/pnas.082089499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adey A, et al. Rapid, low-input, low-bias construction of shotgun fragment libraries by high-density in vitro transposition. Genome Biol. 2010;11:R119. doi: 10.1186/gb-2010-11-12-r119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lasken RS. Genomic DNA amplification by the multiple displacement amplification (MDA) method. Biochem Soc Trans. 2009 Apr;37:450. doi: 10.1042/BST0370450. [DOI] [PubMed] [Google Scholar]
- 17.Lasken RS, Stockwell TB. Mechanism of chimera formation during the Multiple Displacement Amplification reaction. BMC Biotechnol. 2007;7:19. doi: 10.1186/1472-6750-7-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004 Oct;5:557. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 19.Brennand KJ, et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011 May 12;473:221. doi: 10.1038/nature09915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laurent LC, et al. Dynamic changes in the copy number of pluripotency and cell proliferation genes in human ESCs and iPSCs during reprogramming and time in culture. Cell stem cell. 2011 Jan 7;8:106. doi: 10.1016/j.stem.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baslan T, et al. Genome-wide copy number analysis of single cells. Nature protocols. 2012 Jun;7:1024. doi: 10.1038/nprot.2012.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bruder CE, et al. Phenotypically concordant and discordant monozygotic twins display different DNA copy-number-variation profiles. American journal of human genetics. 2008 Mar;82:763. doi: 10.1016/j.ajhg.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Huallachain M, Karczewski KJ, Weissman SM, Urban AE, Snyder MP. Extensive genetic variation in somatic human tissues. Proceedings of the National Academy of Sciences of the United States of America. 2012 Oct 30;109:18018. doi: 10.1073/pnas.1213736109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Piotrowski A, et al. Somatic mosaicism for copy number variation in differentiated human tissues. Hum Mutat. 2008 Sep;29:1118. doi: 10.1002/humu.20815. [DOI] [PubMed] [Google Scholar]
- 25.Abyzov A, et al. Somatic copy number mosaicism in human skin revealed by induced pluripotent stem cells. Nature. 2012 Dec 20;492:438. doi: 10.1038/nature11629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maeda T, et al. Somatic DNA recombination yielding circular DNA and deletion of a genomic region in embryonic brain. Biochem Biophys Res Commun. 2004 Jul 9;319:1117. doi: 10.1016/j.bbrc.2004.05.093. [DOI] [PubMed] [Google Scholar]
- 27.Shibata Y, et al. Extrachromosomal microDNAs and chromosomal microdeletions in normal tissues. Science. 2012 Apr 6;336:82. doi: 10.1126/science.1213307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suberbielle E, et al. Physiologic brain activity causes DNA double-strand breaks in neurons, with exacerbation by amyloid-beta. Nat Neurosci. 2013 May;16:613. doi: 10.1038/nn.3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gilbert N, Lutz-Prigge S, Moran JV. Genomic deletions created upon LINE-1 retrotransposition. Cell. 2002 Aug 9;110:315. doi: 10.1016/s0092-8674(02)00828-0. [DOI] [PubMed] [Google Scholar]
- 30.Callinan PA, et al. Alu retrotransposition-mediated deletion. J Mol Biol. 2005 May 13;348:791. doi: 10.1016/j.jmb.2005.02.043. [DOI] [PubMed] [Google Scholar]
- 31.Gilbert N, Lutz S, Morrish TA, Moran JV. Multiple fates of L1 retrotransposition intermediates in cultured human cells. Mol Cell Biol. 2005 Sep;25:7780. doi: 10.1128/MCB.25.17.7780-7795.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Symer DE, et al. Human l1 retrotransposition is associated with genetic instability in vivo. Cell. 2002 Aug 9;110:327. doi: 10.1016/s0092-8674(02)00839-5. [DOI] [PubMed] [Google Scholar]
- 33.Coufal NG, et al. L1 retrotransposition in human neural progenitor cells. Nature. 2009 Aug 27;460:1127. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.