SUMMARY
The biomedical utility of induced pluripotent stem cells (iPSCs) will be diminished if most iPSC lines harbor deleterious genetic mutations. Recent microarray studies have shown that human iPSCs carry elevated levels of DNA copy number variation compared to embryonic stem cells, suggesting that these and other classes of genomic structural variation (SV) including inversions, smaller duplications and deletions, complex rearrangements and retroelement transpositions may frequently arise as a consequence of reprogramming. Here we employ whole genome paired-end DNA sequencing and sensitive mapping algorithms to identify all classes of SV in several fully pluripotent mouse iPSC lines. Despite the improved scope and resolution of this study, we find few spontaneous mutations per line (1–2) and no evidence for endogenous retroelement transposition. These results show that genome stability can persist throughout reprogramming, and argue that it is possible to generate iPSCs lacking gene disrupting mutations using current reprogramming methods.
INTRODUCTION
The process of direct reprogramming transforms differentiated somatic cells into induced pluripotent stem cell (iPSC) lines that possess the capacity to generate all cell types in an organism. While iPSCs are functionally similar to embryonic stem cells (ESCs), several aspects of iPSC production suggest that these cells may harbor increased numbers of mutations relative to ESCs. First, reprogramming involves expression of known oncogenes such as c-Myc and Klf4, and is enhanced by downregulating genes that promote genome stability such as p53; reviewed in (Deng and Xu, 2009). Second, reprogramming involves global epigenetic remodeling including histone alteration, genome-wide demethylation and de novo DNA methylation, which may be mutagenic or lead to activation of endogenous retroelements (Koche et al., 2011; Lister et al., 2011). Third, ESCs employ less error prone DNA repair mechanisms than do somatic cells, and failure to reset these during reprogramming could contribute mutations to iPSCs (Fan et al., 2011; Momcilovic et al., 2011). Finally, iPSCs are derived from differentiated cell types instead of early embryos, suggesting that somatic mutations in donor cells may contribute genetic diversity to these cell lines, which could be deleterious.
A mutational class of particular concern is genomic structural variation (SV). SVs include duplications, deletions, insertions, inversions, translocations and complex rearrangements. Since SVs can affect gene copy number and/or structure and arise at high rates in unstable genomic regions (Zhang et al., 2009), they are most likely to have a functional impact on iPSCs or their derivatives. Moreover, a highly mutagenic source of SV is transposition of endogenous retroelements, such as LINEs, which have recently been shown to cause unanticipatedly high levels of genome diversity in germ line cells (Akagi et al., 2008; Beck et al., 2010; Iskow et al., 2010; Quinlan et al., 2010; Xing et al., 2009), tumors (Iskow et al., 2010) and some somatic lineages (Coufal et al., 2009; Garcia-Perez et al., 2007; Muotri et al., 2005). Whether retroelements become active during or after reprogramming is not known.
Recent genome-wide surveys have reported that human iPSC lines harbor high levels of de novo SV. One study (Mayshar et al., 2010) used RNA expression analysis to indirectly assess aneuploidy and large copy number variants (CNVs) at low resolution (~10 mb), and additional studies (Hussein et al., 2011; Laurent et al., 2011; Martins-Taylor et al., 2011) used array-based methods to map smaller CNVs. All report a highly significant excess of CNVs in iPSCs relative to ESCs and fibroblasts. However, these studies were blind to smaller (<10 kb) variants that comprise the vast majority of CNVs (Mills et al., 2011), and could not detect balanced rearrangements or transposon insertions, both of which are common in mammalian genomes (Zhang et al., 2009). In this context the high mutational burden reported by these studies is alarming, and raises the question of whether the reprogramming process is inherently mutagenic.
This study aims to determine whether reprogramming to pluripotency involves inherently mutagenic steps independent of the effects of somatic development, extensive passaging and incomplete reprogramming. Therefore, we sought to measure levels of de novo SV in early passage iPSCs derived from low passage mouse embryonic fibroblasts (MEFs), using methods that distinguish between mutations inherited from donor cells and those acquired during reprogramming. We also controlled for incomplete reprogramming by profiling fully pluripotent iPSCs that generate viable mice derived entirely from iPSCs (Boland et al., 2009).
We analyzed the genomes of three iPSC lines using whole genome paired-end DNA sequencing and highly sensitive SV detection algorithms, yet we observed strikingly few mutations (1-2 per line) and found no evidence for retroelement activation. These results argue that it is possible to identify iPSCs lacking deleterious genomic changes using current reprogramming methods.
RESULTS
Mapping structural variation
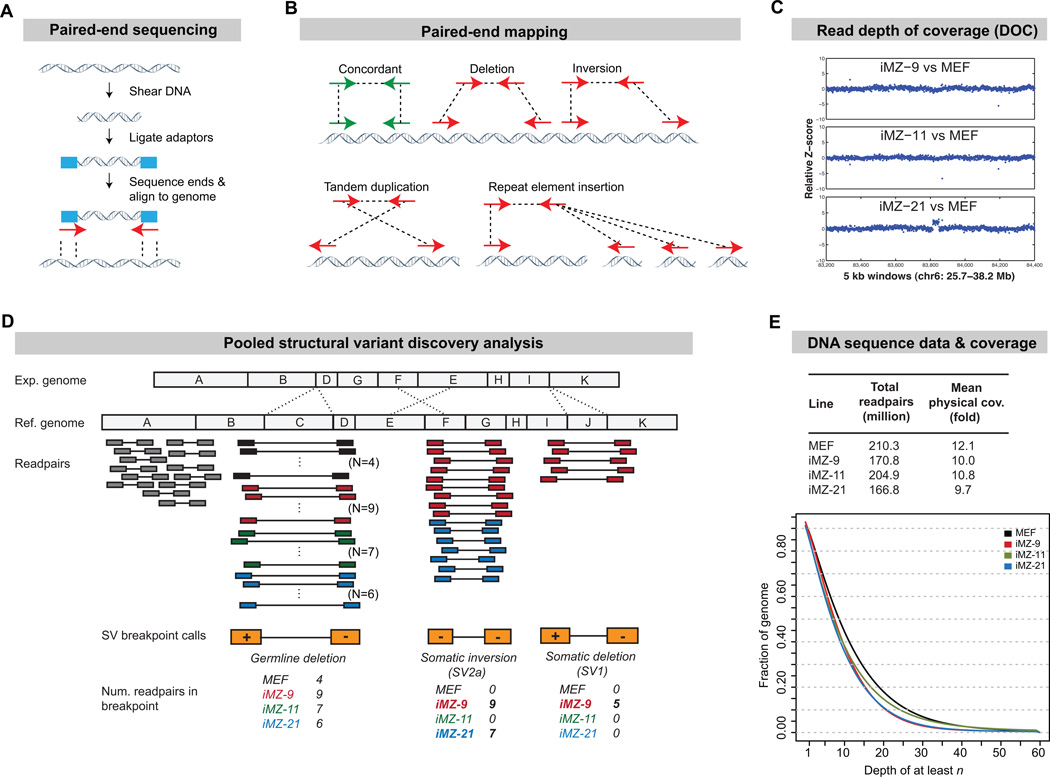
The most common methods for identifying CNVs are array comparative genomic hybridization (array-CGH) and SNP genotyping arrays which have limited resolution and cannot detect balanced rearrangements or transposon insertions. To overcome these limitations we examined iPSC genomes using Illumina DNA sequencing and improved SV detection algorithms (Figure 1A). We obtained 170-220 million paired-end sequence reads (readpairs) from three iPSC lines and their parent fibroblasts, representing 10-12X physical coverage (Figure 1E), and used two complementary approaches to identify SVs: paired end mapping (PEM) and read depth of coverage analysis (DOC) (Figure 1B, C).
Figure 1. Structural variant detection.
(A) Schematic of Illumina paired-end DNA sequencing.
(B) Breakpoint detection by PEM. Most readpairs are concordant (green) and map to the reference genome with the expected size and orientation (arrows), but readpairs that span SV breakpoints map in "discordant" fashion (red). Each breakpoint class yields a distinctive pattern.
(C) CNV detection by read depth of coverage analysis (DOC). DOC uses local read depth to measure DNA copy number in a manner that is analogous to array-CGH. Shown is 12.5 mb region that harbors the lone de novo CNV identified by DOC. Each data point is a 5 kb window, shown in genome order (X-axis), and DNA copy number is expressed as the Z-score (Y-axis) of the indicated iPSC line relative to the donor MEF sample. Note that iMZ-21 clearly shows a 358 kb duplication (SV3) relative to the MEF sample
(D) A multi-sample PEM method using pooled data. A hypothetical region from an experimental genome (Exp.) is shown above the reference genome (Ref.). In this schematic segment C is deleted, E and F are inverted and J is deleted. HYDRA screens for clusters of discordant readpairs (colored) that support the same breakpoint. Germline SVs will be present in all four lines. De novo SVs will be present in a subset of iPSC lines. With our method, the 4 samples are combined into a single HYDRA analysis and breakpoint “genotypes” are inferred from the number of readpairs contributed by each. The origin of each readpair is indicated by its color, which corresponds to the colors of the labeled samples below. Shown are SV1, SV2a and one germline variant.
(E) The total number of readpairs and genome coverage collected for each strain (top) and a plot showing the fraction of the genome in each line having greater than or equal to various levels of physical coverage.
PEM involves clustering readpairs that span SV breakpoints and can, in principle, identify all forms of SV. We used an improved version of HYDRA, an algorithm we developed (Quinlan et al., 2010). Importantly, HYDRA incorporates alternate mappings for readpairs derived from repetitive elements (Figure 1B), which allows for identification of breakpoints involving transposons and segmental duplications, which are among the most mutable genomic elements. Our iPSC lines were derived from a mixed strain mouse, and are thus expected to differ from the C57BL/6J reference genome by thousands of inherited SVs (Quinlan et al., 2010). Distinguishing de novo SV from inherited SV in this context is a difficult and unsolved technical problem. We therefore developed a new method to identify breakpoints from pooled multi-sample data (Figure 1D; http://code.google.com/p/hydra-sv/), which greatly increases the accuracy of determining whether a given SV is a true de novo variant. Here we achieved ~300 bp resolution, which is at least 30-fold greater than the highest resolution iPSC genome surveys to date (Hussein et al., 2011; Laurent et al., 2011).
DOC analysis relies on the observation that the local read depth is directly related to DNA copy number, and is conceptually similar to array-CGH, yet more sensitive. We performed DOC analysis with a custom algorithm (Quinlan et al., 2010) that provides ~15 kb resolution (Figure 1C).
iPSC derivation and lineage analysis
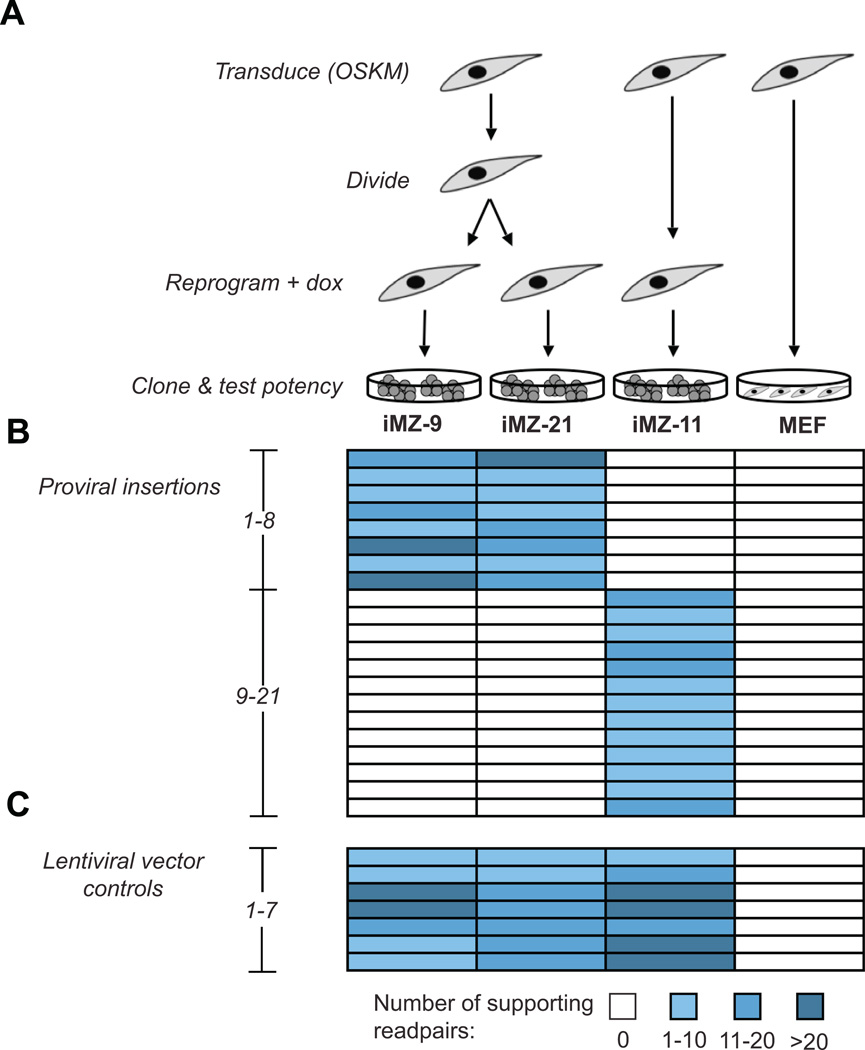
To assess the inherent mutagenicity of reprogramming we wished to examine fully reprogrammed iPSCs that had not been subjected to extensive passaging or clonal selection. Here, we sequenced DNA from low passage iPSC lines that generate viable mice in tetraploid embryo complementation (TEC) assays, demonstrating that they have completed reprogramming (Boland et al., 2009). We produced these lines by transducing mouse embryonic fibroblasts (MEFs) with lentiviruses containing Oct4, Klf4, Sox2 and c-Myc under the control of a doxycycline (dox)-inducible promoter. Following viral transduction, the MEFs were allowed to divide one or two times before initiating reprogramming by adding dox. This protocol allows us to identify pairs of “sister” iPSC lines that arise from the same donor cell (Figure 2A). Sister colonies arising at separate locations will have identical patterns of proviral insertions but will have undergone distinct reprogramming events. Mutations common to both sister lines, yet absent from other iPSCs are likely to be SVs inherited from a donor cell, while mutations unique to a single sister line must have arisen during or after reprogramming. In this study we sequenced a pair of sister iPSCs (iMZ-9 and 21) and a line that arose from a different donor cell (iMZ-11)(Boland et al., 2009).
Figure 2. iPSC lineages.
(A) iPSC lineages. MEFs were transduced with five lentiviruses encoding the four reprogramming factors and a drug inducible transcriptional activator (rTTAM2.2). After viral transduction MEFs were split and allowed to divide one time prior to induction of reprogramming. This scheme produces clonally transduced fibroblasts that undergo different reprogramming events and produce distinct iPSC lines.
(B) The patterns of proviral integration events identified by HYDRA demonstrate that iMZ-9 and iMZ-21 have identical proviral insertions while iMZ-11 is distinct. Thus, iMZ-9 and iMZ-21 are derived from the same original fibroblast cell. Genomic differences between these lines represent post-transduction changes while shared SVs likely represent somatic mutations present in the donor cell.
(C) Positive control “variant” calls resulting from the structure of the lentiviral vectors. The vectors contain the 4 reprogramming genes. The junctions between vector and transgene sequences manifest as 4 SVs. In addition, 3 of the 4 transgenes lack introns relative to the their copies in the reference genome, which produces 3 control variants.
To confirm the lineage of sister iPSC lines and establish the sensitivity of HYDRA we mapped proviral insertion sites. We aligned readpairs to both the reference genome and the lentiviral gene sequences and identified HYDRA breakpoint calls consistent with proviral integration events. This confirmed the clonal origin of the iPSCs and accurately identified 21 proviral insertions (Figure 2B), which is three more than could be clearly distinguished in Southern blots (Boland et al., 2009). We also detected the intron-less reprogramming genes in the lentiviral vectors, as expected (Figure 2C). These data provide an initial estimate of the validity and sensitivity of our methods.
The extent and origin of structural variation in iPSCs
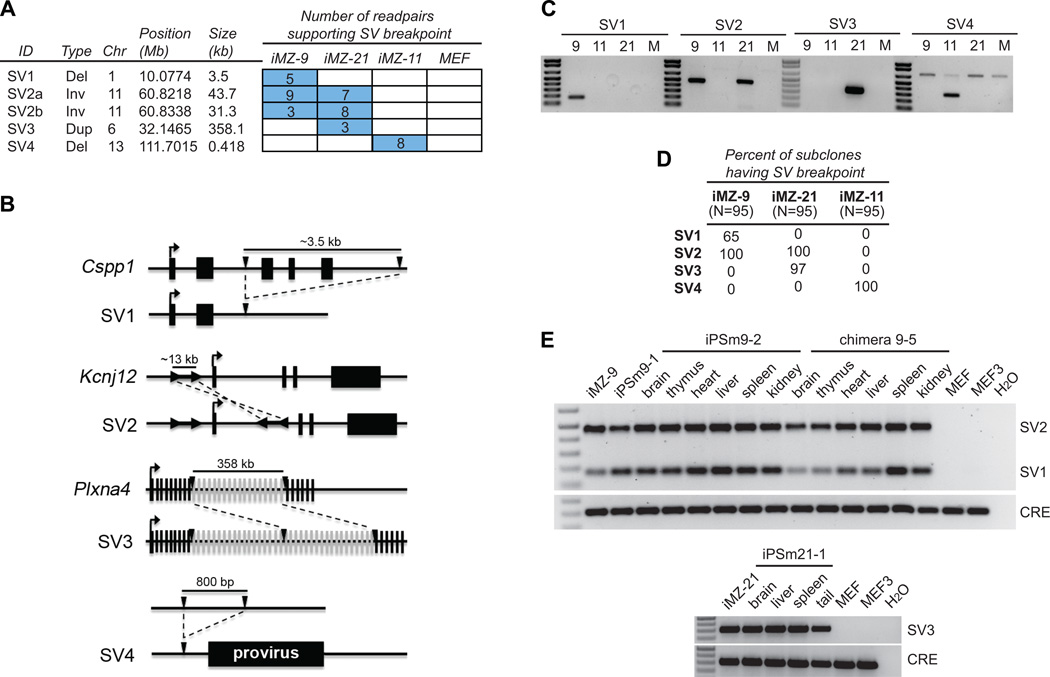
We applied DOC and PEM analyses to identify candidate de novo SVs that are present in one or more iPSC lines but not in the parental MEFs. Surprisingly, DOC analysis identified only one de novo CNV (SV3), a ~358 kb duplication in PlxnA4 that was also detected by PEM and was present only in iMZ-21 (Figure 1C, Figure 3A,B).
Figure 3. SVs arise prior to and during reprogramming and contribute to tissues.
(A) The number of supporting readpairs per breakpoint call are indicated by the numbers in the boxes. The table at left describes the type, size and location of each SV and associated breakpoint.
(B) Schematic diagrams of SV1-4. Three of four SVs interrupt genic regions. Black lines denote non-exonic DNA and black or gray boxes represent exons or the proviral insertion near SV4. Transcription start sites are denoted by arrows above exons. Breakpoints are denoted by inverted black triangles for SV1,3 and 4 and by the black arrows flanking the inverted duplication in SV2. The schematics are not to scale but the size of each region is shown. Dashed lines indicate the change between the wild type and mutated chromosome.
(C) PCR confirmation of 3 SVs. Primers were designed to amplify breakpoint-spanning PCR products that produce a unique band in the line(s) harboring the SV.
(D) The percentage of 95 iPSC subclones for each line that are positive for a given SV by PCR assays.
(E) Tissues from iPSC mice (iPSm 9-1, 9-2 and iPSm 21-1) and a chimeric mouse with iMZ-9 contribution were examined for the presence of SV1-2 (upper panel) and SV3 (lower panel). All SVs are present in all tested tissues, but not the parental MEFs (MEF) or those harvested at the same time from a sibling embryo (MEF3). PCR for the Cre recombinase gene (CRE) present in the iMZ iPSCs serves as a control for iPSC contribution.
More sensitive PEM analyses using the HYDRA algorithm identified 16,579 high-confidence SV breakpoints. Of these, 13099 (79%) were detected in the parental MEF sample and are thus, by definition, inherited variants. The remaining 3,480 breakpoints are candidate de novo mutations. This number of candidates is expected to occur by chance given the abundance of SV between mouse strains (Quinlan et al., 2010) and the moderate physical coverage of our datasets. While HYDRA achieves presence/absence breakpoint “genotyping” at ~89-94% accuracy in the 4 cell lines, the large number of inherited SVs produces false mutation calls at breakpoints that, by chance, lack sequence coverage in one or more samples. We addressed this by using multinomial sampling to prioritize candidate mutations whose readpair distribution among the iPSC lines was unlikely to occur by chance. We used PCR to validate the 182 candidates that exceeded this threshold (Figure S1, Table S1). Of these, 101 were present in all samples and thus represent germline variants; 18 candidates failed two independent PCR attempts, either because they were false positive calls or due to primer failure; and 64 breakpoints were validated as de novo mutations (Table S3). These represent 45 distinct SVs (Table S2).
Structural variants include multiple mutational classes. Of the 45 de novo SVs we identified, only four fell into the “canonical” class defined as deletions, duplications and inversions (Figure 3A-C). The remaining 41 were insertions of an exogenous retroelement, which we discuss later. The sister lines (iMZ-9 and 21) shared one mutation (SV2) not found in iMZ-11 or the MEFs, suggesting that SV2 originated as a somatic mutation in the donor MEF. SV2 is a complex rearrangement on chromosome 11 marked by two large (31.3 kb and 43.7 kb) overlapping inversion breakpoint calls (Figure 3B). The simplest explanation for this breakpoint pattern is a ~12 kb inverted duplication in which the duplicated segments are separated by ~31 kb of non-duplicated sequence. DOC analysis supports this interpretation. The duplicated segment lies between two alternatively spliced first exons of Kcnj12, an inwardly-rectifying potassium channel expressed in the brain, heart and other tissues (Oyamada et al., 2005). Each line also contained one additional line-specific SV. The iMZ-21 line carried SV3, a 358 kb multi-exon duplication in PlxnA4, which is a cell surface signaling protein expressed in multiple tissues including the brain, blood and heart (Suto et al., 2003) Line iMZ-9 carried SV1, a ~3.5 kb deletion that removes 3 exons of Cspp1, a widely expressed gene involved in cytokinesis and cell cycle (Asiedu et al., 2009). Strikingly, line iMZ-11 carried only a single 400 bp deletion in a non-genic region (SV4).
To infer the origin of each putative de novo SV, we generated 95 subclones of each iPSC line and performed PCR (Figure 3D). This showed that SV2, as expected, is present in all subclones while SV1 and SV3 are mosaic (65% and 96% respectively), consistent with their arising early in reprogramming or providing a selective advantage to the iPSCs. SV4 was present in all subclones, consistent with a somatic donor origin. However, SV4 is located only 524 bp from a proviral integration site, suggesting an alternative scenario in which it arose concomitantly with viral insertion (Figure 3B). The mechanism for such an event is unclear, but we note that adjacent deletions are associated with a subset of SINE retroelement insertions in the human genome (Xing et al., 2009).
To gain insight into the mechanisms responsible for these rearrangements, we sequenced each SV breakpoint (Figure S2). None exhibit more than 5 bp of homology and SV3 contains a 12 bp insertion. This indicates that the SVs did not arise through non-allelic homologous recombination (NAHR) but instead through non-homologous end-joining (NHEJ) or microhomology-mediated break-induced replication (MMBIR)(Hastings et al., 2009). The complex multi-breakpoint structure of SV2 and the insertion in SV3 are more characteristic of MMBIR.
SVs contribute to tissues of chimeric and iPSC mice
To address the functional relevance of these variants, we analyzed tissues from iPSC mice and from chimeric mice with iPSC contribution. Previous studies indicate that mice generated by these methods derive from at most three pluripotent cells (Wang and Jaenisch, 2004). Thus, selection against SV1 in the early embryo (65% of iMZ-9 cells) could result in its absence from tissues of iPSC mice, while selection against SV2 (100% of iMZ-9 cells) could exclude this SV from tissues in chimeric mice. However, genomic PCR showed that SVs could be detected in tissues of chimeric or TEC mice (Figure 3E, iMZ-11 data not shown). In addition, RT-qPCR analyses of the genes affected by SV1, SV2 and SV3 indicate that their expression is reduced in iPSCs and/or relevant tissues (Figure S2).
Retroelement silencing is maintained in iPSCs
The mouse genome contains numerous endogenous retroelements that are repressed in somatic cells but active in germ cells, early embryonic lineages and in cells with epigenetic perturbations (Maksakova et al., 2008). The epigenetic remodeling that occurs during reprogramming could activate these normally repressed retroelements, which would be highly deleterious. HYDRA allows us to address this important unanswered question at the whole genome level.
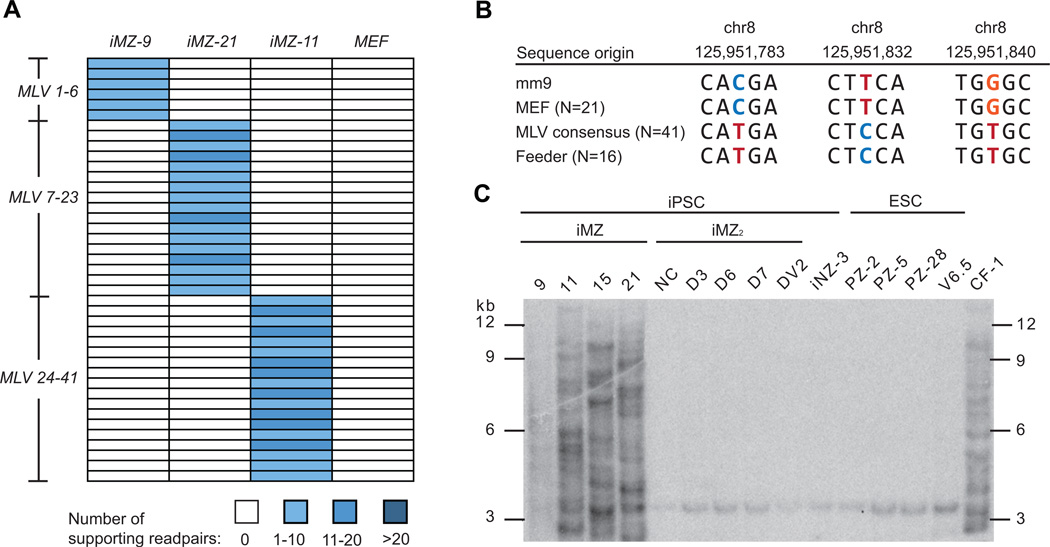
We identified 41 retroelement insertion events in the three iPSC lines. Strikingly, all 41 were endogenous retrovirus elements from the mouse leukemia virus (MLV) family. Each iPSC line displayed a distinct insertional pattern, indicating that transposition occurred during or after reprogramming (Figure 4A). However, closer inspection of the MLV sequence showed that is was not an endogenous element because it contained 58 single nucleotide polymorphisms (SNPs) that were not present in the most similar MLV sequence in the donor MEFs (Figure S4). SNP genotyping confirmed that the mutagenic MLV element matched an MLV found in CF-1 MEFs that were used as feeder cells, suggesting that these feeders transmitted an activated MLV to the iPSCs (Figure 4B). This effect seems to be batch specific because other similarly derived fully pluripotent iPSC lines that were expanded on a different batch of CF-1 feeders lack insertions (Figure 4C) and (KKB, unpublished). This underscores the precautions that must be taken when working with feeder cells.
Figure 4. Analysis of repetitive element insertions.
(A) No endogenous transposon insertions were detected, however, multiple MLV insertions were apparent in each iPSC line. The 41 MLV insertions are shown in a heatmap, following the conventions outlined in Figure 1. Since each MLV insertion is private to a given cell line, they occurred during or after reprogramming.
(B) A portion of the MLV element was amplified by PCR and individual clones were sequenced and analyzed for diagnostic SNPs. Non-reference genome (mm9) alleles are found in the MLV consensus sequence, which was assembled from 41 MLV copies present in the iPSC lines, and in the CF-1 feeder cells (“feeder”), which demonstrates that the additional iPSC MLV copies originate from the feeders.
(C) MLV Southern blot analysis of iPSCs and ESCs. A PCR fragment was used to probe DNA isolated from iMZ iPSCs and iPSCs derived by the same method on different lots of feeders (iMZ2 and iMZ3) as well as ESCs derived on CF-1 feeders. The CF-1 primary MEFs used to generate feeders contain multiple copies of the MLV element as do the iMZ iPSCs, but the other iPSCs and ESCs possess only the chr8 band.
Importantly, we did not detect a single de novo endogenous retroelement transposition among the iPSC lines, despite the demonstrated sensitivity of our detection methods. This suggests that reprogramming using the canonical “Yamanaka” factors can preserve retroelement silencing throughout the dramatic epigenetic changes required to produce fully pluripotent iPSC lines.
Estimating the false negative rate
Given the paucity of new mutations discovered in this study a key question is how many we may have missed. To address this we used two independent methods to estimate the false negative rate (FNR) for SV detection. First, we evaluated HYDRA’s ability to detect the lentiviral insertions used for reprogramming. Using a combination of Southern blots (Boland et al., 2009), PEM, manual inspection of raw sequence data and PCR validation, we identified 24 proviral insertions representing 48 breakpoints. This is 3 more insertions than were detected by HYDRA (Figure 2) and 6 more than Southern blots. We then measured the fraction of breakpoints that were detected by HYDRA at sufficient confidence to be selected as candidate mutations for experimental validation. This resulted in a per variant FNR of 4-47%, depending on the iPSC line and SV class (i.e., one or two breakpoint SVs). Second, we assessed detection of 2284 inherited SVs caused by segregating variation among mouse strains (Quinlan et al., 2010). This predicted a per variant FNR of 22-53%. Therefore, in the worst case we have missed roughly half of the SVs that exist in these genomes, which means that the iPSCs harbor 2-4 total mutations. These calculations strongly support our conclusion that very few de novo SVs exist in these iPSC lines.
One caveat is that PEM cannot detect breakpoints formed by NAHR between large repeats, but this should be a minor source of false negatives. NAHR mutations are detectable by DOC analysis, and NAHR is less frequent in somatic cells than in the germline (Hampton et al., 2009; Hillmer et al., 2011), where it accounts for merely 10-22% of inherited SV (Conrad et al., 2010; Kidd et al., 2010; Mills et al., 2011). A second caveat is that current genome-wide methods cannot detect SVs that are present only in small subsets of cells, such as mutations that arise late during cell expansion.
DISCUSSION
We have examined the mutational burden of a set of fully pluripotent mouse iPSCs using whole genome DNA sequencing and comprehensive SV detection algorithms. Despite the resolution and scope of our methods we observed only four SVs among the three iPSC lines, and iMZ-11 had only a single mutation in a non-genic region. Importantly, we did not observe a single new retrotransposon insertion. Our results argue that current reprogramming methods can produce fully pluripotent iPSC lines that lack severe genomic alterations, even in the presence of c-Myc.
Our results contrast with recent microarray-based studies that have reported high levels of CNV in human iPSC lines (Hussein et al., 2011; Laurent et al., 2011; Martins-Taylor et al., 2011; Mayshar et al., 2010). One explanation for our results is that the lines we analyzed are atypical. Two lines of evidence argue against this. First, our lines were not hand-selected for pluripotency but are instead the first three of seven lines generated using a specific protocol that efficiently produces fully pluripotent iPSCs (6/6 lines tested, KKB unpublished). Second, it is unlikely that we have selected mutation-poor lines by chance alone. The highest resolution study to date examined 22 iPSC lines and discovered a mean excess of 97 CNVs in iPSC lines relative to ESC and fibroblast lines (Hussein et al., 2011). If we randomly select three datasets from Hussein et al. the probability of finding fewer than 42 CNVs is 0.05 (100,000 permutations) (Figure S3). Here, we found a total of four variants in three iPSC lines, only one of which is detectable by microarrays. Moreover, the resolution of our analysis is ~30-fold higher than that Hussein et al., and application of HYDRA to three human datasets with similar levels of coverage to our iPSC datasets reveals 46-fold more SV breakpoints than they reported for control fibroblast lines (mean of 2517 variants vs. 55). These results suggest that the iPSC populations surveyed in the two studies truly differ in their SV burden.
The reduced numbers SV in these mouse iPSCs could reflect inherent differences between mouse and human iPSCs. Alternatively, it could be related to unique aspects of our reprogramming methods or be a consequence of more complete reprogramming of the mouse iPSC lines. Additional experiments are needed to resolve this important question.
A noteworthy aspect of our study is that it was designed to establish the likely origin of each mutation. Encouragingly, we identified only two SVs that were associated with reprogramming per se, and one that was likely caused by lentiviral insertion. This suggests that some iPSCs generated by our method may be completely free of de novo SVs, at least prior to passaging or expansion. However, we also identified a rearrangement (SV2) that almost certainly arose in the donor somatic cell. This is surprising given that donor cells were derived from embryonic day 13.5 fibroblasts. In contrast, adult cells have likely undergone orders of magnitude more divisions as well as exposure to mutagens and potential changes in genome stability that arise during aging or differentiation. Thus, many more somatic SVs may be apparent in iPSC lines derived from adult tissues. In support of this, recent exome sequencing-based studies revealed elevated numbers of point mutations in human iPSCs, of which some were found in donor cells (Gore et al., 2011; Howden et al., 2011). While our current datasets cannot determine whether our iPSC lines also harbor fewer point mutations than human lines, we expect that future genome sequencing efforts will resolve this question.
With the rapid adoption of iPSC and reprogramming technologies, the prospect of bringing patient-specific cell replacement therapy to the clinic is becoming increasingly likely. Here we establish a method to survey iPSC genomes for SVs that arise either during somatic development or reprogramming, and we show that it is possible to achieve reprogramming to full pluripotency with a very low level of mutation. These results underscore the importance of using whole genome sequencing to compare the relative mutagenicity of different reprogramming protocols in order to accelerate the production of mutation-free iPSCs for clinical and research applications.
EXPERIMENTAL PROCEDURES
DNA Sequencing of iPSCs
We derived iPSC lines and chimeric and iPSC-derived mice as described (Boland et al., 2009). We removed iPSC colonies from MEF feeders, isolated DNA and constructed paired-end sequencing libraries according to standard protocols (Bentley et al., 2008). We prepared 2-5 libraries per line and sequenced with an Illumina GA2. Read lengths were 42 bp and the median fragment length was ~330 bp. Readpairs were first aligned with BWA (Li and Durbin, 2009), and subsequently realigned with NOVOALIGN to identify concordant readpairs missed by BWA, and to report up to 1100 alignments per readpair.
SV discovery
SV breakpoints were identified using HYDRA (Quinlan et al., 2010). We combined discordant mappings from the four lines into a single input file, and after breakpoint mapping we calculated the number of readpairs contributed by each sample using the hydraFrequency program in the HYDRA suite. There were 67797 breakpoint calls in the raw unfiltered output file, which includes all breakpoints identified by 2 or more readpairs.
To obtain a final set of high-confidence HYDRA calls we used BEDTOOLS (Quinlan and Hall, 2010) to exclude calls whose aligned ends overlapped an annotated simple sequence repeat (SSR) by more than 50%. This is necessary because SSRs are highly repetitive and often poorly assembled in the reference genome, causing numerous false positives (Quinlan et al., 2010). Second, we required that the readpairs comprising each HYDRA call aligned to the reference genome with a mean edit distance <2 on both ends. This step increases accuracy because false calls can result from low quality alignments that occur when readpairs originating from repetitive or misassembled genomic regions are aligned to incorrect genome positions. Third, we required that the mean number of mappings for the readpairs contained in a HYDRA call were less than 1000 when one of the two ends was unique, and less than 100 when both ends were repetitive. These filters reduced the 67797 raw HYDRA breakpoint calls to a final high-confidence set of 16579.
To identify CNVs we analyzed read depth of coverage (corrected for GC-content) in 5 kb windows using a previously described Hidden Markov Model (HMM) based method (Quinlan et al., 2010)
Identification of candidate mutations
Of the 16579 breakpoint calls, 3480 were not found in the MEF donor sample and represent candidate de novo SVs. However, many of these are expected to occur by chance due to mouse strain variation. We used a multinomial sampling approach that accounts for the relative sequence coverage in each strain to rank variants. We performed validation experiments on all 84 candidate mutations that had a probability of occurring by chance of less than 0.001. We also selected an additional 98 candidates with a probability less than 0.01. These 98 include all breakpoints that 1) involved the MLV element; 2) were identified in both the iMZ-9 and iMZ-21 lines, but not iMZ-11 and MEF; 3) were identified in a single iPSC line or 4) were identified by DOC analysis.
False negative rate calculations
To intersect HYDRA calls with “true” breakpoints we used pairToPair in the BEDTOOLs suite, requiring strand-specific overlap between both ends. To acquire the set of 48 “true” proviral integration breakpoints we used results from HYDRA and Southern blots, and we inspected raw sequence data to identify single readpairs that mapped to the reference genome on one end and the lentiviral vector sequence on the other. Since the latter can be caused by artifacts (e.g., chimeras) we performed PCR to ensure their validity (3/7 validated). To acquire the set of 2284 “true” inherited germline breakpoints we intersected the 67797 unfiltered HYDRA calls from this study with 7784 breakpoints previously identified in the DBA/2J strain (Quinlan et al., 2010). For calculations of FNR we assessed the fraction of breakpoints in each set that were identified by HYDRA at sufficient confidence to be put forward for experimental validation, using precisely the same filtering and prioritization approach used to identify candidate mutations. To assess the FNR of presence/absence breakpoint genotyping in each iPSC line, we calculated the fraction of 1854 high confidence germline breakpoints that were not detected by one or more readpairs.
Supplementary Material
HIGHLIGHTS.
Reprogramming can produce iPSCs with few (1-2) de novo structural variants
Structural variants arise in donor somatic cells and during iPSC generation
iPSCs harboring de novo mutations contribute to tissues of iPSC-derived mice.
Endogenous retroelements remain inactive during reprogramming
ACKNOWLEDGEMENTS
We thank M. Lindberg for algorithm development and R. Clark for computational support, and A. Prorock and Y. Bao (UVA Sequencing Core) for DNA sequencing. We acknowledge S. Kupriyanov and G. Martin (TSRI Mouse Genetics Facility) for assistance with ESC derivation, and K. Nazor for assistance with cell culture. Support to ARQ was from an NRSA postdoctoral fellowship (1F32HG005197-01); to KKB and MJB from the California Institute for Regenerative Medicine, a Pew Scholar Award, the Esther B. O’Keeffe Family Foundation and the Shapiro Family Foundation; to IMH from a Burroughs Wellcome Fund Career Award and the NIH Director’s New Innovator Award (DP2OD006493-01).
Footnotes
ACCESSION NUMBERS
Sequence data have been submitted to the Short Read Archive (http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi) under accession no. SRA043600.
SUPPLEMENTAL INFORMATION
Supplemental information includes 4 figures, 3 tables and a detailed version of the experimental procedures, and can be found with this article online.
The authors declare no competing financial interests.
REFERENCES
- Akagi K, Li J, Stephens RM, Volfovsky N, Symer DE. Extensive variation between inbred mouse strains due to endogenous L1 retrotransposition. Genome Res. 2008;18:869–880. doi: 10.1101/gr.075770.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asiedu M, Wu D, Matsumura F, Wei Q. Centrosome/spindle pole-associated protein regulates cytokinesis via promoting the recruitment of MyoGEF to the central spindle. Molecular biology of the cell. 2009;20:1428–1440. doi: 10.1091/mbc.E08-01-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck CR, Collier P, Macfarlane C, Malig M, Kidd JM, Eichler EE, Badge RM, Moran JV. LINE-1 retrotransposition activity in human genomes. Cell. 2010;141:1159–1170. doi: 10.1016/j.cell.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland MJ, Hazen JL, Nazor KL, Rodriguez AR, Gifford W, Martin G, Kupriyanov S, Baldwin KK. Adult mice generated from induced pluripotent stem cells. Nature. 2009;461:91–94. doi: 10.1038/nature08310. [DOI] [PubMed] [Google Scholar]
- Conrad DF, Bird C, Blackburne B, Lindsay S, Mamanova L, Lee C, Turner DJ, Hurles ME. Mutation spectrum revealed by breakpoint sequencing of human germline CNVs. Nat Genet. 2010;42:385–391. doi: 10.1038/ng.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coufal NG, Garcia-Perez JL, Peng GE, Yeo GW, Mu Y, Lovci MT, Morell M, O'Shea KS, Moran JV, Gage FH. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460:1127–1131. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Xu Y. Genome integrity: linking pluripotency and tumorgenicity. Trends in genetics : TIG. 2009;25:425–427. doi: 10.1016/j.tig.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Robert C, Jang YY, Liu H, Sharkis S, Baylin SB, Rassool FV. Human induced pluripotent cells resemble embryonic stem cells demonstrating enhanced levels of DNA repair and efficacy of nonhomologous end-joining. Mutat Res. 2011;713:8–17. doi: 10.1016/j.mrfmmm.2011.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Perez JL, Marchetto MC, Muotri AR, Coufal NG, Gage FH, O'Shea KS, Moran JV. LINE-1 retrotransposition in human embryonic stem cells. Hum Mol Genet. 2007;16:1569–1577. doi: 10.1093/hmg/ddm105. [DOI] [PubMed] [Google Scholar]
- Gore A, Li Z, Fung HL, Young JE, Agarwal S, Antosiewicz-Bourget J, Canto I, Giorgetti A, Israel MA, Kiskinis E, et al. Somatic coding mutations in human induced pluripotent stem cells. Nature. 2011;471:63–67. doi: 10.1038/nature09805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton OA, Den Hollander P, Miller CA, Delgado DA, Li J, Coarfa C, Harris RA, Richards S, Scherer SE, Muzny DM, et al. A sequence-level map of chromosomal breakpoints in the MCF-7 breast cancer cell line yields insights into the evolution of a cancer genome. Genome Res. 2009;19:167–177. doi: 10.1101/gr.080259.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillmer AM, Yao F, Inaki K, Lee WH, Ariyaratne PN, Teo AS, Woo XY, Zhang Z, Zhao H, Ukil L, et al. Comprehensive long-span paired-end-tag mapping reveals characteristic patterns of structural variations in epithelial cancer genomes. Genome Research. 2011;21:665–675. doi: 10.1101/gr.113555.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howden SE, Gore A, Li Z, Fung HL, Nisler BS, Nie J, Chen G, McIntosh BE, Gulbranson DR, Diol NR, et al. Genetic correction and analysis of induced pluripotent stem cells from a patient with gyrate atrophy. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1103388108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussein SM, Batada NN, Vuoristo S, Ching RW, Autio R, Narva E, Ng S, Sourour M, Hamalainen R, Olsson C, et al. Copy number variation and selection during reprogramming to pluripotency. Nature. 2011;471:58–62. doi: 10.1038/nature09871. [DOI] [PubMed] [Google Scholar]
- Iskow RC, McCabe MT, Mills RE, Torene S, Pittard WS, Neuwald AF, Van Meir EG, Vertino PM, Devine SE. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell. 2010;141:1253–1261. doi: 10.1016/j.cell.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd JM, Graves T, Newman TL, Fulton R, Hayden HS, Malig M, Kallicki J, Kaul R, Wilson RK, Eichler EE. A human genome structural variation sequencing resource reveals insights into mutational mechanisms. Cell. 2010;143:837–847. doi: 10.1016/j.cell.2010.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koche RP, Smith ZD, Adli M, Gu H, Ku M, Gnirke A, Bernstein BE, Meissner A. Reprogramming factor expression initiates widespread targeted chromatin remodeling. Cell Stem Cell. 2011;8:96–105. doi: 10.1016/j.stem.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent LC, Ulitsky I, Slavin I, Tran H, Schork A, Morey R, Lynch C, Harness JV, Lee S, Barrero MJ, et al. Dynamic changes in the copy number of pluripotency and cell proliferation genes in human ESCs and iPSCs during reprogramming and time in culture. Cell Stem Cell. 2011;8:106–118. doi: 10.1016/j.stem.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G, Antosiewicz-Bourget J, O'Malley R, Castanon R, Klugman S, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksakova IA, Mager DL, Reiss D. Keeping active endogenous retroviral-like elements in check: the epigenetic perspective. Cell Mol Life Sci. 2008;65:3329–3347. doi: 10.1007/s00018-008-8494-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins-Taylor K, Nisler BS, Taapken SM, Compton T, Crandall L, Montgomery KD, Lalande M, Xu RH. Recurrent copy number variations in human induced pluripotent stem cells. Nature biotechnology. 2011;29:488–491. doi: 10.1038/nbt.1890. [DOI] [PubMed] [Google Scholar]
- Mayshar Y, Ben-David U, Lavon N, Biancotti JC, Yakir B, Clark AT, Plath K, Lowry WE, Benvenisty N. Identification and classification of chromosomal aberrations in human induced pluripotent stem cells. Cell Stem Cell. 2010;7:521–531. doi: 10.1016/j.stem.2010.07.017. [DOI] [PubMed] [Google Scholar]
- Mills RE, Walter K, Stewart C, Handsaker RE, Chen K, Alkan C, Abyzov A, Yoon SC, Ye K, Cheetham RK, et al. Mapping copy number variation by population-scale genome sequencing. Nature. 2011;470:59–65. doi: 10.1038/nature09708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momcilovic O, Navara C, Schatten G. Cell cycle adaptations and maintenance of genomic integrity in embryonic stem cells and induced pluripotent stem cells. Results Probl Cell Differ. 2011;53:415–458. doi: 10.1007/978-3-642-19065-0_18. [DOI] [PubMed] [Google Scholar]
- Muotri AR, Chu VT, Marchetto MC, Deng W, Moran JV, Gage FH. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- Oyamada Y, Yamaguchi K, Murai M, Hakuno H, Ishizaka A. Role of Kir2.2 in hypercapnic ventilatory response during postnatal development of mouse. Respir Physiol Neurobiol. 2005;145:143–151. doi: 10.1016/j.resp.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Clark RA, Sokolova S, Leibowitz ML, Zhang Y, Hurles ME, Mell JC, Hall IM. Genome-wide mapping and assembly of structural variant breakpoints in the mouse genome. Genome Res. 2010;20:623–635. doi: 10.1101/gr.102970.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suto F, Murakami Y, Nakamura F, Goshima Y, Fujisawa H. Identification and characterization of a novel mouse plexin, plexin-A4. Mech Dev. 2003;120:385–396. doi: 10.1016/s0925-4773(02)00421-5. [DOI] [PubMed] [Google Scholar]
- Wang Z, Jaenisch R. At most three ES cells contribute to the somatic lineages of chimeric mice and of mice produced by ES-tetraploid complementation. Dev Biol. 2004;275:192–201. doi: 10.1016/j.ydbio.2004.06.026. [DOI] [PubMed] [Google Scholar]
- Xing J, Zhang Y, Han K, Salem AH, Sen SK, Huff CD, Zhou Q, Kirkness EF, Levy S, Batzer MA, et al. Mobile elements create structural variation: analysis of a complete human genome. Genome Res. 2009;19:1516–1526. doi: 10.1101/gr.091827.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Gu W, Hurles ME, Lupski JR. Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet. 2009;10:451–481. doi: 10.1146/annurev.genom.9.081307.164217. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.