Abstract
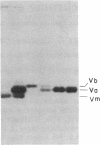
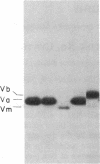
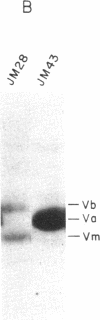
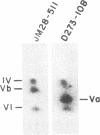
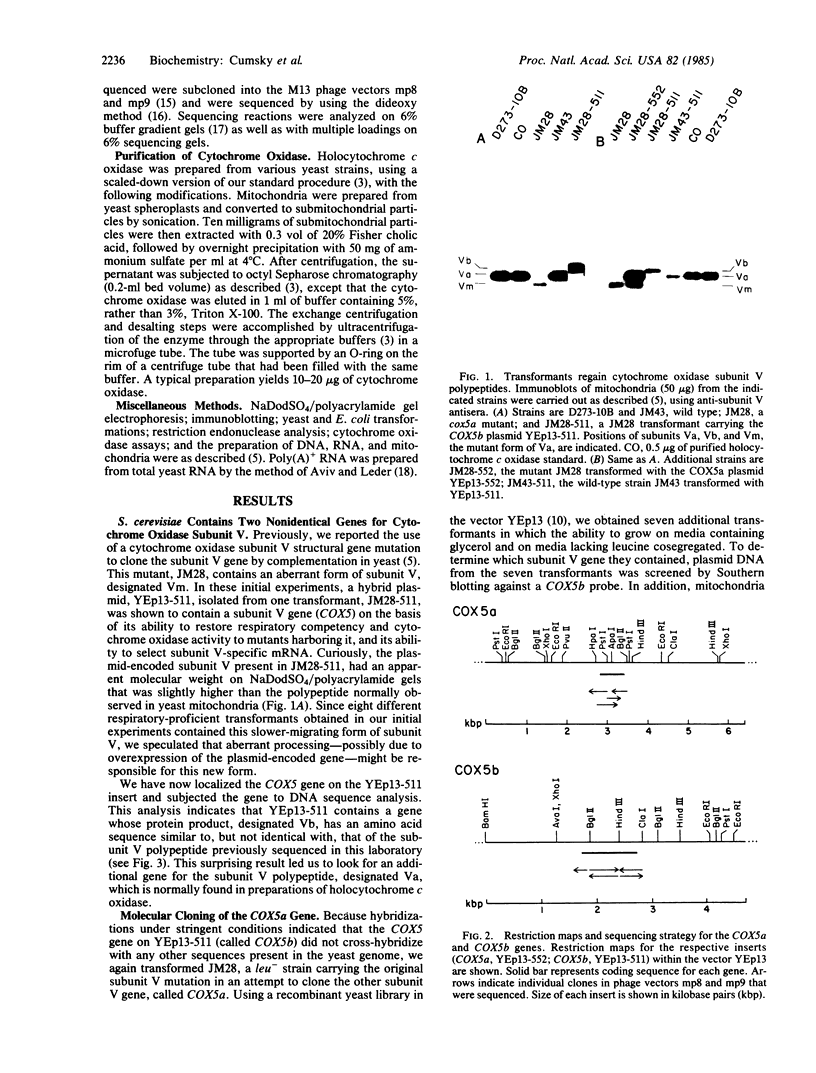
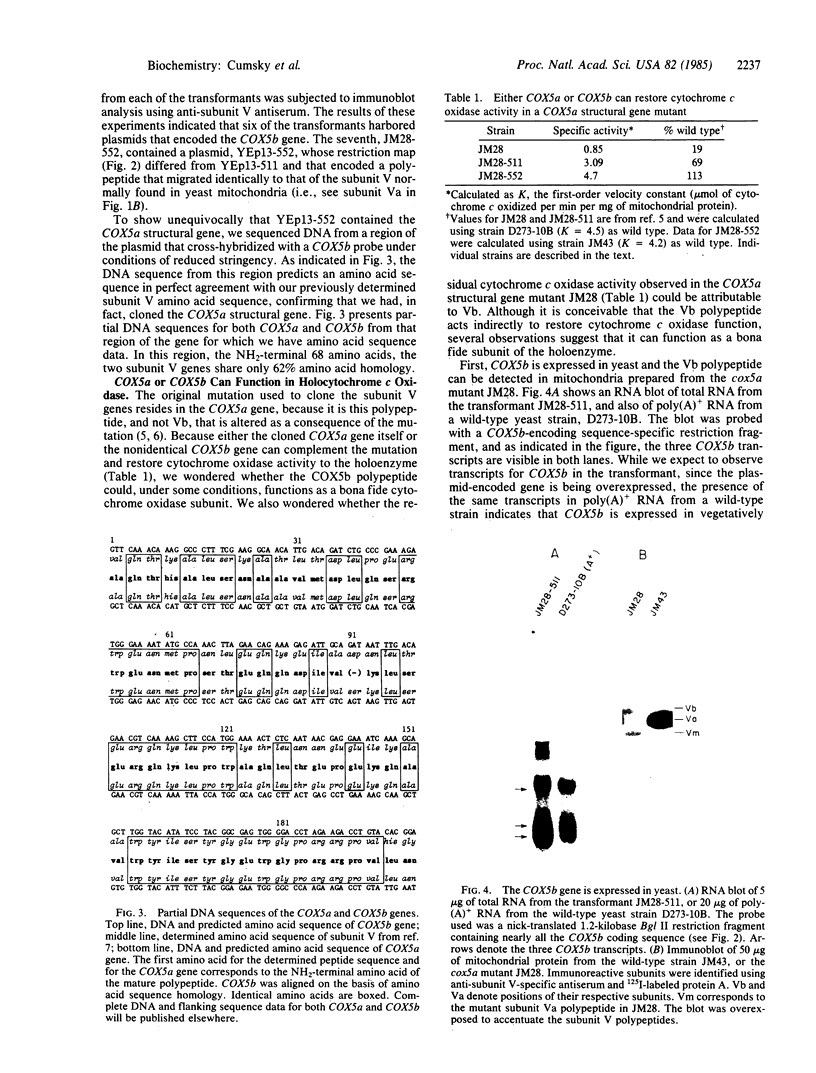
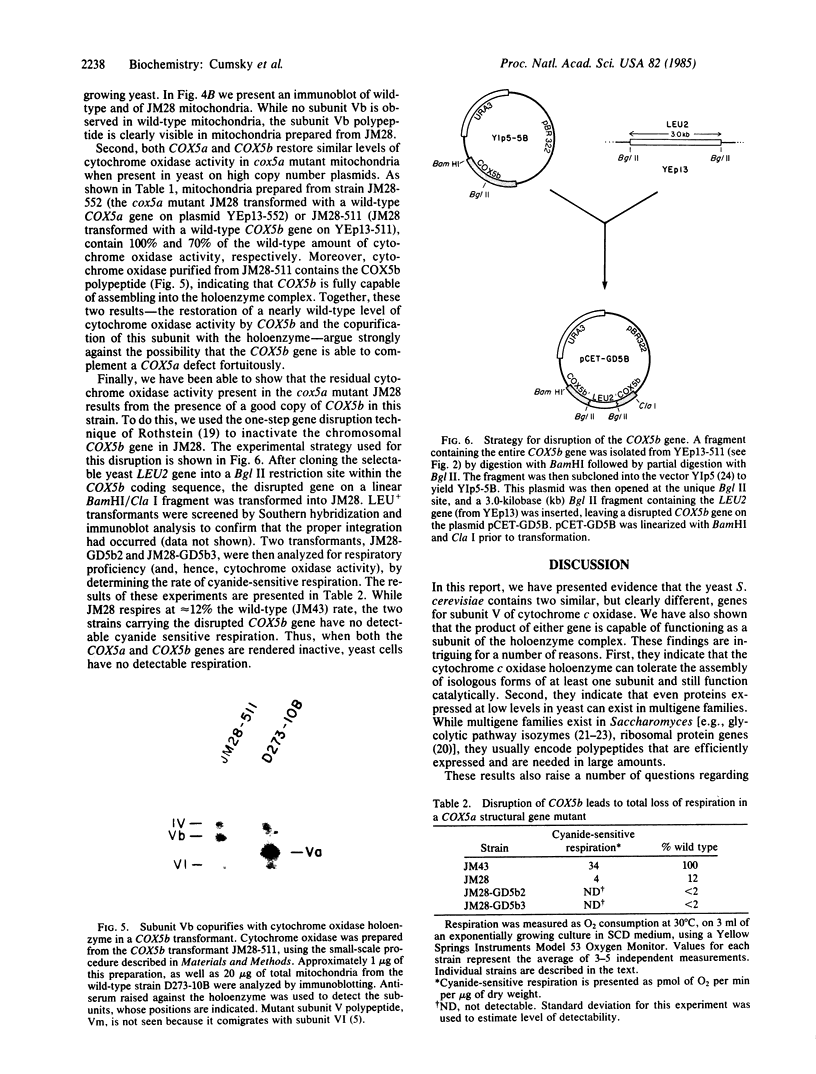
In Saccharomyces cerevisiae, the inner mitochondrial membrane protein cytochrome c oxidase is composed of nine polypeptide subunits. Six of these subunits (IV, V, VI, VII, VIIa, VIII) are encoded by the nuclear genome, and the remaining three (I, II, III) are encoded by mitochondrial DNA. We report here the existence of two nonidentical subunit V polypeptides, which are encoded by separate genes within the yeast genome. One gene, COX5a, encodes the polypeptide Va, normally found in preparations of holocytochrome c oxidase. The other gene, COX5b, encodes the polypeptide Vb, which cross-reacts with anti-subunit Va antiserum and restores respiratory competency and cytochrome oxidase activity in transformants of cox5a structural gene mutants. This polypeptide also copurifies with the holoenzyme prepared from these transformants. We have found that COX5b is expressed in vegetatively growing yeast cells, and that the Vb polypeptide can be detected in mitochondria from strain JM28, a cox5a mutant. This mutant has 15%-20% residual cytochrome oxidase activity, and it respires at 10%-15% the wild-type rate. By disrupting the COX5b gene in this strain, we show that this residual activity is directly attributable to the presence of a chromosomal copy of the COX5b gene. Taken together, these results suggest that Va or Vb can function as cytochrome oxidase subunits in yeast and that Vb may be used under some specific, as yet undefined, physiological conditions.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abovich N., Rosbash M. Two genes for ribosomal protein 51 of Saccharomyces cerevisiae complement and contribute to the ribosomes. Mol Cell Biol. 1984 Sep;4(9):1871–1879. doi: 10.1128/mcb.4.9.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein D., Falco S. C., Stewart S. E., Brennan M., Scherer S., Stinchcomb D. T., Struhl K., Davis R. W. Sterile host yeasts (SHY): a eukaryotic system of biological containment for recombinant DNA experiments. Gene. 1979 Dec;8(1):17–24. doi: 10.1016/0378-1119(79)90004-0. [DOI] [PubMed] [Google Scholar]
- Broach J. R., Strathern J. N., Hicks J. B. Transformation in yeast: development of a hybrid cloning vector and isolation of the CAN1 gene. Gene. 1979 Dec;8(1):121–133. doi: 10.1016/0378-1119(79)90012-x. [DOI] [PubMed] [Google Scholar]
- Cumsky M. G., McEwen J. E., Ko C., Poyton R. O. Nuclear genes for mitochondrial proteins. Identification and isolation of a structural gene for subunit V of yeast cytochrome c oxidase. J Biol Chem. 1983 Nov 25;258(22):13418–13421. [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Holland J. P., Labieniec L., Swimmer C., Holland M. J. Homologous nucleotide sequences at the 5' termini of messenger RNAs synthesized from the yeast enolase and glyceraldehyde-3-phosphate dehydrogenase gene families. The primary structure of a third yeast glyceraldehyde-3-phosphate dehydrogenase gene. J Biol Chem. 1983 Apr 25;258(8):5291–5299. [PubMed] [Google Scholar]
- Holland M. J., Holland J. P., Thill G. P., Jackson K. A. The primary structures of two yeast enolase genes. Homology between the 5' noncoding flanking regions of yeast enolase and glyceraldehyde-3-phosphate dehydrogenase genes. J Biol Chem. 1981 Feb 10;256(3):1385–1395. [PubMed] [Google Scholar]
- Jarausch J., Kadenbach B. Tissue-specificity overrides species-specificity in cytoplasmic cytochrome c oxidase polypeptides. Hoppe Seylers Z Physiol Chem. 1982 Sep;363(9):1133–1140. doi: 10.1515/bchm2.1982.363.2.1133. [DOI] [PubMed] [Google Scholar]
- Kadenbach B., Hartmann R., Glanville R., Buse G. Tissue-specific genes code for polypeptide VIa of bovine liver and heart cytochrome c oxidase. FEBS Lett. 1982 Feb 22;138(2):236–238. doi: 10.1016/0014-5793(82)80450-x. [DOI] [PubMed] [Google Scholar]
- McEwen J. E., Cumsky M. G., Ko C., Power S. D., Poyton R. O. Mitochondrial membrane biogenesis: characterization and use of pet mutants to clone the nuclear gene coding for subunit V of yeast cytochrome c oxidase. J Cell Biochem. 1984;24(3):229–242. doi: 10.1002/jcb.240240305. [DOI] [PubMed] [Google Scholar]
- Messing J., Crea R., Seeburg P. H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Montgomery D. L., Leung D. W., Smith M., Shalit P., Faye G., Hall B. D. Isolation and sequence of the gene for iso-2-cytochrome c in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1980 Jan;77(1):541–545. doi: 10.1073/pnas.77.1.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. A., Tatchell K. The structure of transposable yeast mating type loci. Cell. 1980 Mar;19(3):753–764. doi: 10.1016/s0092-8674(80)80051-1. [DOI] [PubMed] [Google Scholar]
- Power S. D., Lochrie M. A., Poyton R. O. The nuclear-coded subunits of yeast cytochrome c oxidase. III. Identification of homologous subunits in yeast, bovine heart, and Neurospora crassa cytochrome c oxidases. J Biol Chem. 1984 May 25;259(10):6575–6578. [PubMed] [Google Scholar]
- Power S. D., Lochrie M. A., Sevarino K. A., Patterson T. E., Poyton R. O. The nuclear-coded subunits of yeast cytochrome c oxidase. I. Fractionation of the holoenzyme into chemically pure polypeptides and the identification of two new subunits using solvent extraction and reversed phase high performance liquid chromatography. J Biol Chem. 1984 May 25;259(10):6564–6570. [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright R. M., Ko C., Cumsky M. G., Poyton R. O. Isolation and sequence of the structural gene for cytochrome c oxidase subunit VI from Saccharomyces cerevisiae. J Biol Chem. 1984 Dec 25;259(24):15401–15407. [PubMed] [Google Scholar]