Abstract
Long non-coding RNA HOTAIR exerts regulatory functions in various biological processes in cancer cells, such as proliferation, apoptosis, mobility, and invasion. We previously found that HOX transcript antisense RNA (HOTAIR) is a negative prognostic factor and exhibits oncogenic activity in hepatocellular carcinoma (HCC). In this study, we aimed to investigate the role and molecular mechanism of HOTAIR in promoting HCC cell migration and invasion. Firstly, we profiled its gene expression pattern by microarray analysis of HOTAIR loss in Bel-7402 HCC cell line. The results showed that 129 genes were significantly down-regulated, while 167 genes were significantly up-regulated (fold change >2, p < 0.05). Bioinformatics analysis indicated that RNA binding proteins were involved in this biological process. HOTAIR suppression using RNAi strategy with HepG2 and Bel-7402 cells increased the mRNA and protein expression levels of RNA binding motif protein 38 (RBM38). Moreover, the expression levels of RBM38 in HCC specimens were significantly lower than paired adjacent noncancerous tissues. In addition, knockdown of HOTAIR resulted in a decrease of cell migration and invasion, which could be specifically rescued by down-regulation of RBM38. Taken together, HOTAIR could promote migration and invasion of HCC cells by inhibiting RBM38, which indicated critical roles of HOTAIR and RBM38 in HCC progression.
Keywords: RNA binding motif protein 38, HOX transcript antisense RNA, hepatocellular carcinoma, long non-coding RNA
1. Introduction
Hepatocellular carcinoma (HCC) is the second leading cause of cancer related deaths worldwide [1]. China accounts for 55% of the world’s cases each year, due to high incidence of chronic hepatitis B infection and liver cirrhosis [2]. Although great advances in surgical technique and medical care have been achieved over the last two decades, long-term survival of HCC is still low due to the high rate of recurrence and metastasis [3,4]. Moreover, the majority of HCC patients present with advanced metastatic disease at the initial diagnosis. It has been identified that the development and progression of HCC are promoted by multiple abnormal biological processes, such as gene mutations [5], epigenetic alterations [6], and dysregulation of both coding and non-coding genes [7]. For accurate diagnosis and adequate treatment of HCC, there is an urgent need to understand the precise molecular mechanisms underlying HCC pathogenesis and progression.
According to the size, non-coding RNAs (ncRNAs) are subdivided into two major classes: small ncRNAs (<200 nt) and long ncRNAs (lncRNAs, >200 nt) [8]. In recent years, microRNAs have been characterized as oncogenes or tumor suppressor genes to influence biological function of cancer cells through post-transcriptional regulation of protein expression [9]. In contrast, lncRNAs were once considered to be transcriptional noise. However, it has become increasingly clear that lncRNAs execute important functions at various levels, including X chromosomal inactivation, chromatin remodeling, and transcriptional repression [10–12].
HOX transcript antisense RNA (HOTAIR), as a lncRNA, can regulate gene expression through changes in chromatin states and epigenetic modifications [13,14]. HOTAIR, located in the HOXC cluster, trimethylates histone H3 lysine-27 (H3K27me3) of the HOXD locus with the polycomb-repressive complex 2 (PRC2), and subsequently inhibits HOXD gene expression [15]. Recently, the up-regulation of HOTAIR was observed in several cancers, including breast cancer [14,16,17], hepatocellular carcinoma [18], colorectal cancer (CRC) [19], pancreatic cancer [20], non-small cell lung cancer (NSCLC) [21], and esophageal squamous cell carcinoma (ESCC) [22,23]. Furthermore, HOTAIR promoted migration and invasion of breast carcinoma cells [14], CRC cells [19], pancreatic cancer cells [20], NSCLC cells [21], and ESCC cells [22,23]. Our previous studies [18] have shown that HOTAIR is overexpressed in HCC and serves as an independent prognostic factor for recurrence related survival. However, the role and molecular mechanism of HOTAIR in promoting HCC cell migration and invasion remain to be elucidated.
In this study, we profiled its gene expression pattern by microarray analysis of HOTAIR loss in Bel-7402 HCC cell line. Microarray analysis revealed that the expression of QKI, CD82, and RBM38 increased in siHOTAIR groups compared with control groups, which were validated by qRT-PCR and Western blot. Moreover, the expression levels of RBM38, not QKI or CD82, were significantly lower in HCC tissues than paired adjacent noncancerous tissues. In addition, the effects of HOTAIR knockdown on cell migration and invasion could be specifically rescued by down-regulation of RBM38. These results suggest that HOTAIR promotes cell migration and invasion via suppressing RBM38, which indicated critical roles of HOTAIR and RBM38 in HCC progression.
2. Results and Discussion
2.1. Results
2.1.1. Knockdown of HOTAIR Altered Global Gene Expression Patterns in HCC Cells
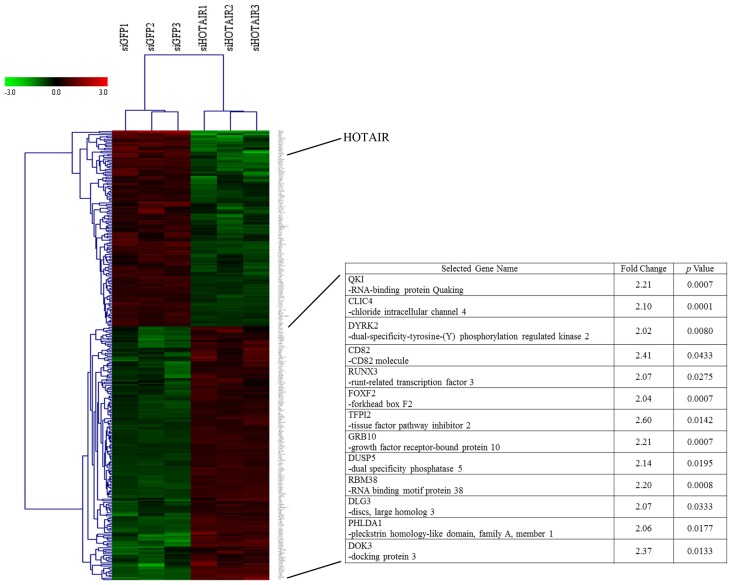
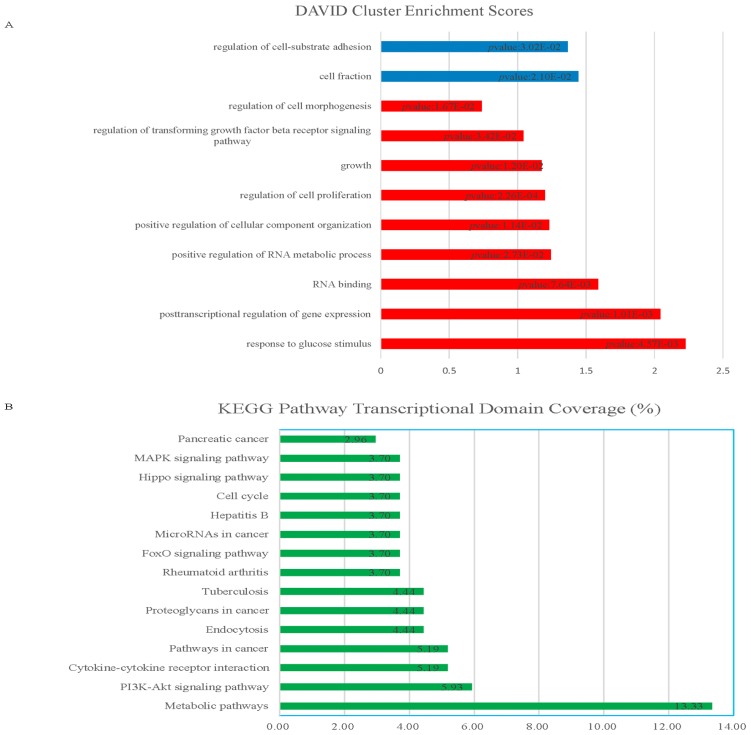
To study the molecular mechanism of HOTAIR knockdown, we profiled its gene expression pattern by microarray analysis. Affymetrix u133 pluss 2.0 was applied to screen for global transcriptional changes in Bel-7402 cells 48 h after siRNA treatment. Overall, 296 genes were differentially expressed in HOTAIR knockdown cells (including 167 up-regulated and 129 down-regulated genes, fold change >2.0 and p value <0.05) (Figure 1). Gene ontology (GO) analysis showed that many differentially expressed genes are involved in biological processes relevant to cancer pathogenesis, such as metabolic process, posttranscriptional regulation of gene expression, RNA binding, cell proliferation, growth, transforming growth factor beta (TGF-β) signaling, and so on (Figure 2A). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis indicated that metabolic pathways, PI3K-AKT signaling pathway, cytokine receptor interaction are top canonical pathways identified in this process (Figure 2B). Based on the predicted interactions of key molecules with their associated pathway, the mRNA-signaling pathway integrated network was built (Figure 2C and Supplementary Figure S1). Notably, TGF-β signaling pathway played an important role in this network.
Figure 1.
Global transcriptional changes by microarray analysis of HOTAIR loss in Bel-7402 HCC cell line. (Left), Heat map depicting transcript profiling of independent biologic replicate Bel-7402 cells with siRNA-mediated HOTAIR depletion (siHOTAIR) versus control siRNA (siGFP). Red represents upregulated genes while green represents downregulated genes; (Right), selected genes for further validation.
Figure 2.
Bioinformatics analysis of HOTAIR loss in Bel-7402 HCC cell line. (A) Gene ontology analysis of HOTAIR knockdown microarray data using the DAVID program. Red bars represent the top hits for upregulated genes. Blue bars represent the top hits for downregulated genes. DAVID enrichment scores are represented with Benjamini-Hochberg-adjusted p values. All error bars in this figure are mean ± S.E.M.; (B) KEGG pathway analysis of HOTAIR knockdown microarray data. Green bars represent the percentage of transcriptional domain coverage; and (C) mRNA-signaling pathway integrated network based on the predicted interactions (partial).
Based on the results of bioinformatics analysis and the fact that overexpression of HOTAIR in cancer cells suppresses downstream gene expression, and increases cancer invasiveness and metastasis [14], we gave a special attention to those genes which were significantly upregulated in the microarray data and had been reported as potential tumor suppressors. After a screening of 167 genes, 13 genes of them met the above requirements. These genes were QKI, CLIC4, DYRK2, CD82, RUNX3, FOXF2, TFPI2, GRB10, DUSP5, RBM38, DLG3, PHLDA1, and DOK3 (Figure 1 right).
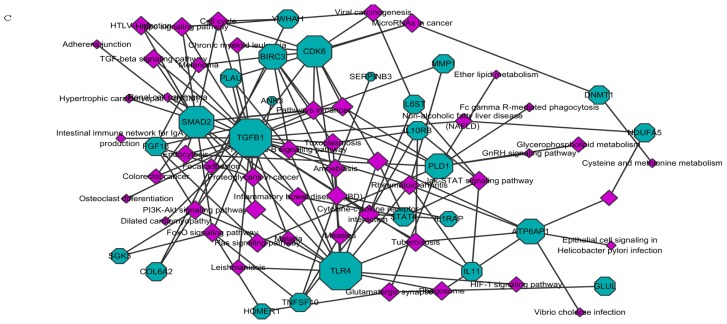
2.1.2. Upregulation of QKI, CD82, and RBM38 Were Validated after Knockdown of HOTAIR
Thirteen upregulated genes were selected for further analysis. Previous studies showed that all of these genes had potential tumor suppressor roles in tumor development and progression. HOTAIR siRNA treatment was introduced to Bel-7402 and HepG2 cells. Then mRNA levels of these genes were assessed after 48 h transfection. Finally, we found three genes (QKI, CD82, and RBM38) significantly upregulated after HOTAIR knockdown in Bel-7402 cells (Figure 3A) and HepG2 cells (Figure 3B). Moreover, the protein levels of QKI, CD82, and RBM38 were increased in siHOTAIR group compared with siGFP group by Western blot (Figure 3C). These results suggest that downregulation of HOTAIR can increase QKI, CD82, and RBM38 expression both on mRNA and protein levels.
Figure 3.
Upregulation of QKI, CD82, and RBM38 were validated after knockdown of HOTAIR. (A) Bel-7402 cells; (B) HepG2 cells were transiently transfected with HOTAIR siRNA for 48 h to detect QKI, CD82, and RBM38 expression by qRT-PCR compared with the siGFP group (* indicated p < 0.05); and (C) showed the protein level of QKI, CD82, and RBM38 after HOTAIR knockdown in Bel-7402 cells by Western blot. All experiments were performed in triplicates.
2.1.3. QKI, CD82, and RBM38 Knockdown by the Corresponding siRNAs Did not Alter Cell Proliferation
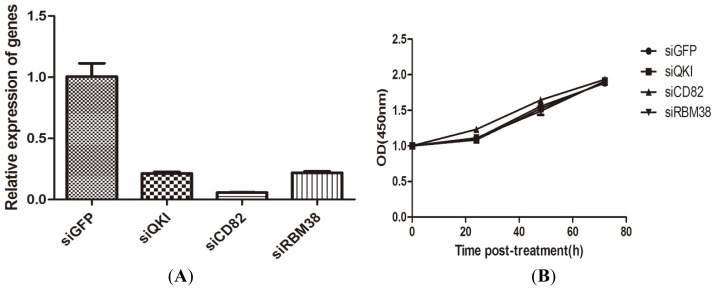
To selectively suppress the expression of QKI, CD82, and RBM38, we used the gene-specific siRNAs (QKI-siRNA, CD82-siRNA and RBM38-siRNA) against QKI, CD82, and RBM38 in Bel-7402 cells. qRT-PCR results showed that QKI, CD82 and RBM38 mRNA levels were significantly reduced by the corresponding siRNAs treatment compared with siGFP groups (Figure 4A). Next we analyzed the effects of silencing of QKI, CD82, and RBM38 on HCC cell proliferation. CCK-8 assays showed that knockdown of these genes did not alter proliferation of Bel-7402 cells compared with siGFP groups (Figure 4B). These results indicated that these genes have little effect on HCC cell proliferation.
Figure 4.
Effects of QKI, CD82, and RBM38 knockdown on cell proliferation. (A) effective silencing of QKI, CD82, and RBM38 expression in Bel-7402 cells after 48 h siRNA treatment according to qRT-PCR analysis; and (B) Viable cell numbers were detected by CCK-8 assay after transferred to 96-wells plates for 24, 48, and 72 h.
2.1.4. The Expression Levels of RBM38 Were Decreased in HCC Samples
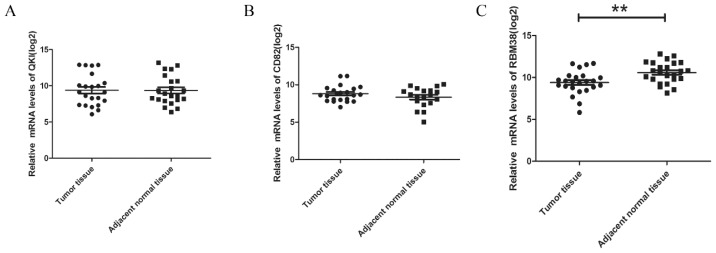
We detected the mRNA levels of QKI, CD82, and RBM38 in 53 pairs of HCC resection specimens by qRT-PCR. Each HCC pair contained the cancer and the corresponding adjacent normal tissues. No significant change for QKI and CD82 were observed in 53 paired HCC samples (Figure 5A,B). However, the relative expression levels of RBM38 were significantly downregulated in HCC tissues compared with paired adjacent noncancerous tissues (p = 0.0043) (Figure 5C).
Figure 5.
The expression levels of RBM38 were downregulated in HCC resection specimens. The expression levels of QKI (A); CD82 (B) and RBM38 (C) in HCC and paired adjacent noncancerous tissues were measured by qRT-PCR and normalized to GAPDH. The significant differences between samples were analyzed using the Student’s t test. (log 2, ** p < 0.01).
To explore whether RBM38 could be an important biomarker for HCC patients, we evaluated the relationship between RBM38 expression and clinicopathological parameters. Thirty-nine of these cases (73.6%) exhibited lower expression levels of RBM38. The cutoff value for high and low expression of RBM38 was the mean value of RBM38 mRNA level. Reduced expression of RBM38 negatively correlated with serum AFP level (p = 0.017). There was no significant correlation between RBM38 expression and other clinicopathological features (Table 1).
Table 1.
Relationship between RBM38 expression and clinicopathological features of 53 hepatocellular carcinoma (HCC) patients.
| Variables | N = 53 | RBM38 expression | χ2 | p value | |
|---|---|---|---|---|---|
|
|
|||||
| high | low | ||||
| Age (years) | |||||
|
| |||||
| ≤50 | 21 | 6 (11.3%) | 15 (28.3%) | 0.083 | 0.773 |
| >50 | 32 | 8 (15.1%) | 24 (45.3%) | ||
|
| |||||
| Gender | |||||
|
| |||||
| Male | 44 | 13 (24.5%) | 31 (58.5%) | 1.306 | 0.253 |
| Female | 9 | 1 (1.9%) | 8 (15.1%) | ||
|
| |||||
| Size of tumor (cm) | |||||
|
| |||||
| ≤5 | 32 | 7 (13.2%) | 25 (47.2%) | 0.856 | 0.355 |
| >5 | 21 | 7 (13.2%) | 14 (26.4%) | ||
|
| |||||
| Number of tumor | |||||
|
| |||||
| Single | 29 | 8 (15.1%) | 21 (39.6%) | 0.045 | 0.832 |
| Multiple | 24 | 6 (11.3%) | 18 (33.9%) | ||
|
| |||||
| Vascular invasion | |||||
|
| |||||
| Negative | 41 | 11 (20.8%) | 30 (56.6%) | 0.016 | 0.899 |
| Positive | 12 | 3 (5.7%) | 9 (17.0%) | ||
|
| |||||
| Pathological Grading | |||||
|
| |||||
| Well/Moderately | 25 | 5 (9.4%) | 20 (37.7%) | 1.002 | 0.317 |
| Poorly | 28 | 9 (17.0%) | 19 (35.8%) | ||
|
| |||||
| AFP (ng/mL) | |||||
|
| |||||
| ≤400 | 20 | 9 (17.0%) | 11 (20.8%) | 5.708 | 0.017 |
| >400 | 33 | 5 (9.4%) | 28 (52.8%) | ||
|
| |||||
| TBIL (μmol/L) | |||||
|
| |||||
| ≤20 | 33 | 10 (18.9%) | 23 (43.4%) | 0.680 | 0.410 |
| >20 | 20 | 4 (7.5%) | 16 (30.2%) | ||
AFP, alpha-fetoprotein; TBIL, total bilirubin; Statistical analyses were performed with Pearson’s chi-square test.
2.1.5. RBM38 Knockdown Promoted Migration and Invasion of HCC Cells
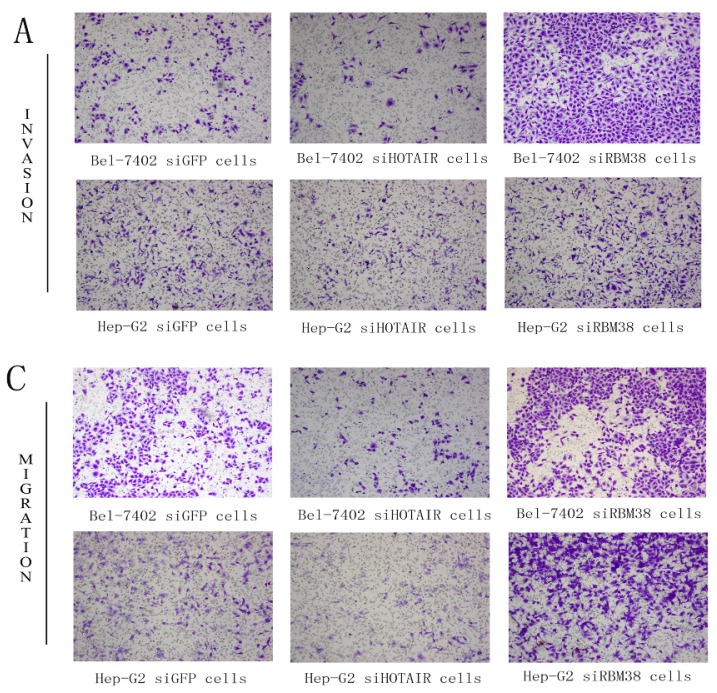
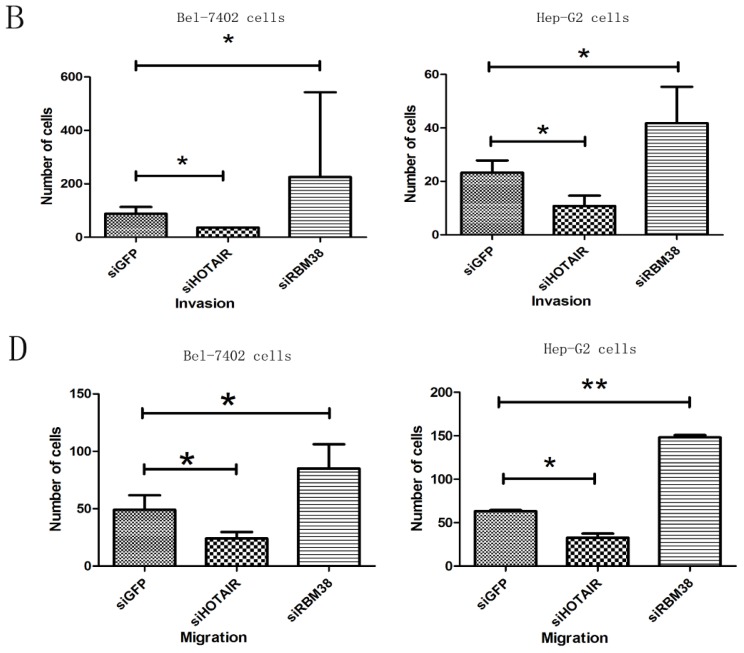
To examine the effect of RBM38 knockdown in liver cancer cells on migration and invasion, we developed a transwell assay with Bel-7402 and HepG2 cells. The siRBM38-transfected cells significantly increased invasion and migration ability compared with the siGFP group in Bel-7402 cells and HepG2 cells (Figure 6A–D). These findings showed that RBM38 could inhibit cell migration and invasion.
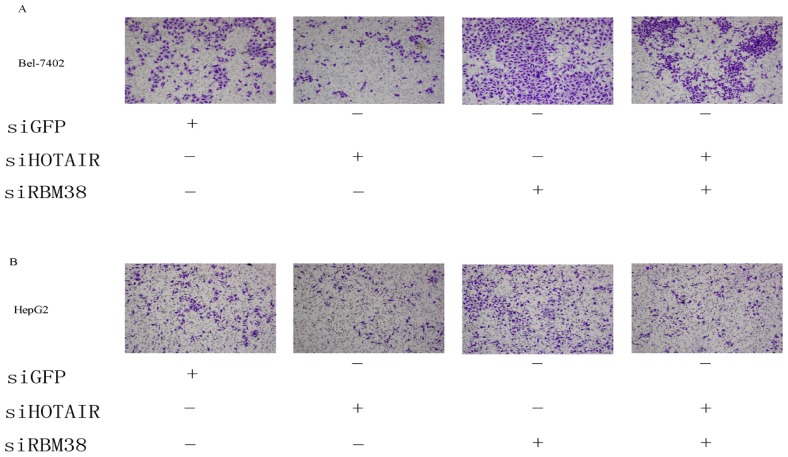
Figure 6.
RBM38 inhibited the invasion and migration of HCC cells. Representative images of invasion (A) and migration (C) of Bel-7402 cells and HepG2 cells transfected with siGFP, siHOTAIR and siRBM38; The number of invaded cells was measured and represented in (B) and (D), respectively. The representative invasion images are shown at 100× magnification.* p < 0.05, ** p < 0.01.
2.1.6. HOTAIR Is Functionally Related to RBM38
Although HOTAIR was shown to negatively regulate RBM38, it is unknown whether a direct or indirect functional relevance to cancer cell invasion exists. In order to determine the relationship between HOTAIR and RBM38, HCC cells were co-transfected with siHOTAIR and siRBM38 and compared with those only transfected with siHOTAIR, siRBM38 or siGFP. The invasion ability of HCC cells transfected with siHOTAIR were decreased compared with siGFP group. On the contrary, the cells transfected with siRBM38 had increased motility. Furthermore, RBM38 knockdown could restore siHOTAIR-mediated inhibition of motility in HCC cells (Figure 7A,B). Thus, these findings indicated that the effect of HOTAIR knockdown on cell motility could be specifically rescued by downregulation of RBM38, thereby, confirming the functional relevance between HOTAIR and RBM38.
Figure 7.
Knockdown of RBM38 restored the HOTAIR siRNA-mediated inhibition of invasion in Bel-7402 cells (A) and HepG2 cells (B). The number of invaded cells was measured. The representative invasion images are shown at 100× magnification.
2.2. Discussion
Long noncoding RNAs represent a novel class of noncoding RNAs that are longer than 200 nucleotides without protein-coding potential [8]. Recently, lncRNAs were found to be dysregulated and involved in various cancer biological processes, such as proliferation, apoptosis, mobility, and invasion [10,24]. But the underlying mechanism remains uncertain. Therefore, understanding the role and precise molecular mechanism of lncRNAs in cancer development and progression are urgently needed.
In our previous study [18], HOTAIR was increased in HCC tissues compared with paired noncancerous tissues. Higher HOTAIR expression level was an independent prognostic marker for HCC recurrence and shorter survival. However, the role and molecular mechanism of HOTAIR in promoting HCC cell migration and invasion are largely limited.
In this study, we profiled its gene expression pattern by microarray analysis of HOTAIR loss in Bel-7402 HCC cell line. We found that 296 genes were differentially expressed in HOTAIR knockdown cells, including 167 up-regulated and 129 down-regulated. GO and pathway analysis indicated the differentially expression genes were significantly enriched in processes, such as cell metabolism, cell proliferation, growth, cell adhesion, and so on, providing important clues for understanding molecular pathogenesis of HCC. Furthermore, several RNA binding proteins involved in this biological process caught our attention, such as QKI and RBM38. Microarray analysis revealed that QKI and RBM38 expression increased in siHOTAIR groups compared with control groups, which were selected and subsequently validated by qRT-PCR and Western blot. These findings suggest RNA binding proteins were downstream molecules repressed by HOTAIR. Interestingly, we found that TGF-β signaling pathway might play an important role in the network according to the integrated KEGG pathway analysis. Our findings are in agreement with Alves’s reports [25], in which HOTAIR acted as a regulator in TGF-β inducing epithelial-to-mesenchymal transition process.
Additionally, in vivo experiments have shown that the expression levels of RBM38, not QKI, were decreased in human HCC tissues compared with paired noncancerous tissues. Moreover, the expression of RBM38 negatively correlated with serum AFP level. These findings suggest that RBM38 may serve as a potential tumor suppressor in HCC. Consistently, Leveille et al. found the reduced expression of RBM38 in cohorts of human breast cancer [26]. RBM38 has also been described as a target of p53, and induces cell cycle arrest in G1 by stabilizing the CDK inhibitor p21 [27].
To confirm the functional relevance between HOTAIR and RBM38, we compared the cell motility among HCC cells with siRBM38, siHOTAIR, and co-transfected with siHOTAIR and siRBM38. Silencing of RBM38 could restore cell motility, while knockdown of HOTAIR significantly reduced cell motility. Furthermore, HCC cells co-transfected with siHOTAIR and siRBM38 could rescue cell motility inhibition by HOTAIR knockdown. These findings suggest that HOTAIR knockdown could reduce cell motility probably by increasing RBM38, which indicate RBM38 as a repressed target of HOTAIR.
Many lncRNAs have been documented to promote tumor progression and metastasis via several different pathways: transcriptional silencing through recruitment of epigenetic complex to specific loci [28]; posttranscriptional gene regulation [29,30]; and decoys for miRNAs and splicing factors [31,32]. Gupta et al. reported that HOTAIR could recruit PRC2 to HOXD locus, trimethylated H3K27me3, increasing breast cancer invasiveness and metastasis [14]. Similarly, Ge et al. reported that HOTAIR directly promoted H3K27me3 in the promoter region of WIF-1, a key regulator in Wnt/β-catenin signaling pathway, and thereby decreased its expression to promote the ESCC cell invasion [22]. Our studies focused on the regulatory roles of RBM38 in HCC cell migration and invasion, and its functional relevance with HOTAIR. However, further studies are needed to confirm whether HOTAIR promoted H3K27me3 in the promoter region of RBM38 to suppress its expression.
In summary, we profiled the gene expression pattern by microarray analysis of HOTAIR loss in Bel-7402 HCC cell line. The expression levels of QKI, CD82, and RBM38 were increased after HOTAIR knockdown in microarray data, which were further validated by qRT-PCR and Western blot. Moreover, the expression levels of RBM38, not QKI or CD82, were significantly lower in HCC tissues than paired adjacent noncancerous tissues. In addition, knockdown of HOTAIR resulted in a decrease of cell migration and invasion, which could be specifically rescued by down-regulation of RBM38. Taken together, HOTAIR could promote migration and invasion of HCC cells by inhibiting RBM38, which indicated critical roles of HOTAIR and RBM38 in HCC progression.
3. Experimental Section
3.1. Patient Samples
Fifty-three cases of patients who underwent surgery between August 2007 and May 2011, in the First Affiliated Hospital, Zhejiang University School of Medicine were enrolled in this study. This study was approved by the Ethics Committee of the hospital and was performed after obtaining written informed consent of patients. Specimens were obtained immediately after surgical resection and stored at −80 °C for further analysis. There were 44 men and 9 women, ranging in age from 29 to 78 years, with an average age of 51.6 years.
3.2. Materials
Liver cancer cell lines HepG2 and Bel-7402 were purchased from Shanghai Institute of Cell Biology (Shanghai, China). All of the cell lines were maintained in the recommended culture condition and incubated at 37 °C in a humidified environment containing 5% CO2.
3.3. Microarray Experiments and Data Analysis
Briefly, Bel-7402 cells were transfected with HOTAIR siRNA or siGFP in triplicate. Forty-eight hours after transfection, total RNA was extracted using the Trizol reagent (Invitrogen, Carlsbad, CA, USA) and integrity assessed with an Agilent Bioanalyzer 2100 (Agilent Technologies, Palo Alto, CA, USA). The RNA was then labeled and hybridized to U133 Plus 2.0 arrays (Affymetrix, Santa Clara, CA, USA) according to the manufacturer’s instructions. Raw microarray data were normalized using Affymetrix Expression Console Software Version 4.0 (Affymetrix, Santa Clara, CA, USA) and Log 2-transformed. The transformed data of two groups were compared using Welch’s t-test. The threshold set for up- and down-regulated genes was a fold change >2.0 and a p value <0.05. GO analysis and KEGG analysis were applied to determine the roles of these differentially expressed mRNAs. Finally, Hierarchical clustering was performed to display the distinguishable genes’ expression pattern among samples.
3.4. RNA Isolation and Real-Time Quantitative Reverse-Transcription Polymerase Chain Reaction (qRT-PCR)
Total RNA from cells was extracted using Trizol reagent (Invitrogen) and cDNA was synthesized with M-MLV Reverse Transcriptase (Promega, Madison, WI, USA). The primers for qRT-PCR were synthesized from Genepharma Inc. (Shanghai, China). QRT-PCR reactions were performed by ABI7500 Fast (Applied Biosystems, Carlsbad, CA, USA) with the SYBR Premix Dimmer Eraser kits (TaKaRa, Dalian, China). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an internal control to normalize target mRNA levels. The relative expression was calculated by the 2−ΔΔCt method. The nucleotide sequences of the primers were as follows:
GAPDH-F: ATGGGGAAGGTGAAGGTCG
GAPDH-R: GGGGTCATTGATGGCAACAATA
CD82-F: CTGCAGGATGCCTGGGACTA
CD82-R: CTCAGCGTTGTCTGTCCAGTTGTA
QKI-F: GATGCAGCTGATGAACGACAAG
QKI-R: CAGCATCAGGCAATTCTGCAC
RBM38-F: GATACTGCCGCCACCAAGA
RBM38-R: CGAGCCGGCAGACACTTTATTATAC
3.5. Small Interfering RNA Transfection
Untreated cells were plated at 3 × 105/well in 2 mL of medium for 24 h before transfection. HepG2 and Bel-7402 cells were transfected with 50 nM siRNAs targeting genes or siGFP (GenePharma, Shanghai, China) using Lipofectamine2000 transfection reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions, The mRNA of HOTAIR, QKI, RBM38 and CD82 was harvest after siRNA transfection for 48 h. The siRNA sequences were listed as follows:
siQKI: CCUUGAGUAUCCUAUUGAACCUAGU
siRBM38: GACACCACGUUCACCAAGA
siCD82: UAUUUGGUGACUUUGAUACAGGCUG
siGFP: CUACAACAGCCACAACGUCTT
siHOTAIR: GAACGGGAGUACAGAGAGAUU
3.6. Cell Proliferation Assay
Cell proliferation was detected using a cell counting kit-8 (Dojindo Laboratories, Kumamoto, Japan) according to the manufacturer’s instructions after transfecting with siRNA in 6-well plates, and were transferred to 96-well plates with 3000 cells per well. After 24, 48, and 72 h incubation in 96-well plates, the relative numbers of viable cells were represented by the absorbance optical density at 450 nm using a microplate reader (Elx800; BioTek, Winooski, VT, USA).
3.7. Cell Migration and Invasion Assay
After being washed with PBS, HepG2 or Bel-7402 cells were detached with trypsin, and resuspended in serum free medium. Then, 200 μL of cell suspensions (2.5 × 105 cells/mL) was added to the upper chamber with non-coated membrane (24-well insert; 8-mm pore size; Millipore, Billerica, MA, USA) or coated with Matrigel (BD Bioscience, Bedford, MA, USA) for the transwell migration or invasion assays, respectively. Culture medium containing 10% FBS was added to the bottom wells of the chambers. The cells were incubated for 24 h (migration assay) or 48 h (invasion assay). After 24 or 48 h, the cells that had still stayed in the upper face of the filters were removed using cotton swabs, and the cells that had migrated to the lower face of the filters were fixed with 100% methanol and stained with 0.2% crystal violet and counted. The mean of triplicate assays for each experimental condition was used.
3.8. Protein Extraction and Western Blot Analysis
After incubation, cells were collected and lysed in lysis buffer (50 mL Tris (pH 7.4), 150 mL sodium chloride, 1% Triton X-100, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate, sodium orthovanadate, sodium fluoride, EDTA, and leupeptin supplemented with 1 mm phenylmethanesulfonyl fluoride) at 4 °C. The lysate was vibrated three times for 10 min each and sonicated, followed by centrifugation at 13,000 rpm for 20 min at 4 °C. Soluble proteins were collected and stored at −80 °C. The protein concentrations were determined using a BCA Protein Assay Kit (Pierce, Rockford, IL, USA) by Bradford assay (Bio-Rad, Hercules, CA, USA). 25~30 μg of total protein was applied for detection of QKI, CD82, and RBM38 protein. After denaturation of proteins by heating at 70 °C for 10 min, the proteins were separated by gel electrophoresis using 12% SDS-PAGE, and blotted onto polyvinylidene fluoride membrane (Millipore, Billerica, MA, USA). Then, the membrane was blocked with 5% nonfat milk in Tris-buffered saline with 0.05% Tween-20 (TBST) and incubated overnight with the following primary antibodies: rabbit anti-QKI (Epitomics, Inc., Burlingame, CA, USA), rabbit anti-CD82 (Epitomics, Inc., Burlingame, CA, USA); rabbit anti-RBM38 (Abcam, Cambridge, UK); and mouse anti-β-actin (Sigma-Aldrich, St. Louis, MO, USA). After incubation with horseradish peroxidase (HRP)-conjugated secondary antibody (Jackson Immuno Research, West Grove, PA, USA), antibody binding was visualized with SuperSignal West Pico Chemiluminescent Substrate (Pierce, Billerica, MA, USA). β-actin was tested as a control.
3.9. Statistical Analysis
The association between RBM38 expression and clinicopathological variables was assessed by two-sided Pearson chi-square test. All statistical analysis was performed using SPSS 16.0 (Chicago, IL, USA) and GraphPad Prism 5.0 (GraphPad Software Inc., San Diego, CA, USA). The experimental data were expressed as median with range or mean ± SD where applicable. Differences between groups were determined by Student’s t test, and p < 0.05 was set to be statistically significant.
4. Conclusions
Our findings suggest that HOTAIR could promote migration and invasion of HCC cells by inhibiting RBM38, which indicated critical roles of HOTAIR and RBM38 in HCC progression.
Supplementary Information
Acknowledgments
This study supported by grants from National High Technology Research and Development Program 863 of China (No.2012AA021002); Special Fund for Health Research in the Public Welfare (No.201302009); Zhejiang Provincial Natural Science Foundation for Young Distinguished Scholars (No.R2110125); National S&T Major Project (No.2012ZX10002017); and Foundation for Innovative Research Groups of the National Natural Science Foundation of China (Grant No.81121002).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Author Contributions
Zheng S.S. and Wu J. conceived and designed the experiments; Ding C.F., Cheng S.B., Yang Z., Lv Z., Xiao H. performed the experiments; Ding C.F., Cheng S.B., Du C.L, Peng C.H., Xie H.Y., Zhou L. analyzed the data; Ding C.F., Cheng S.B. wrote the manuscript.
References
- 1.Jemal A., Bray F., Center M.M., Ferlay J., Ward E., Forman D. Global cancer statistics. CA Cancer J. Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.El-Serag H.B., Rudolph K.L. Hepatocellular carcinoma: Epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 3.Tang Z.Y., Ye S.L., Liu Y.K., Qin L.X., Sun H.C., Ye Q.H., Wang L., Zhou J., Qiu S.J., Li Y., et al. A decade’s studies on metastasis of hepatocellular carcinoma. J. Cancer Res. Clin. Oncol. 2004;130:187–196. doi: 10.1007/s00432-003-0511-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang Y., Nagano H., Ota H., Morimoto O., Nakamura M., Wada H., Noda T., Damdinsuren B., Marubashi S., Miyamoto A., et al. Patterns and clinicopathologic features of extrahepatic recurrence of hepatocellular carcinoma after curative resection. Surgery. 2007;141:196–202. doi: 10.1016/j.surg.2006.06.033. [DOI] [PubMed] [Google Scholar]
- 5.Boyault S., Rickman D.S., de Reynies A., Balabaud C., Rebouissou S., Jeannot E., Herault A., Saric J., Belghiti J., Franco D., et al. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology. 2007;45:42–52. doi: 10.1002/hep.21467. [DOI] [PubMed] [Google Scholar]
- 6.Pogribny I.P., Rusyn I. Role of epigenetic aberrations in the development and progression of human hepatocellular carcinoma. Cancer Lett. 2014;342:223–230. doi: 10.1016/j.canlet.2012.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang J.L., Zheng L., Hu Y.W., Wang Q. Characteristics of long noncoding RNA and its relation to hepatocellular carcinoma. Carcinogenesis. 2013 doi: 10.1093/carcin/bgt405. [DOI] [PubMed] [Google Scholar]
- 8.Deng G., Sui G. Noncoding RNA in oncogenesis: A new era of identifying key players. Int. J. Mol. Sci. 2013;14:18319–18349. doi: 10.3390/ijms140918319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iorio M.V., Croce C.M. MicroRNA dysregulation in cancer: Diagnostics monitoring and therapeutics: A comprehensive review. EMBO Mol. Med. 2012;4:143–159. doi: 10.1002/emmm.201100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mercer T.R., Dinger M.E., Mattick J.S. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 11.Lee J.T., Bartolomei M.S. X-inactivation imprinting and long noncoding RNAs in health and disease. Cell. 2013;152:1308–1323. doi: 10.1016/j.cell.2013.02.016. [DOI] [PubMed] [Google Scholar]
- 12.Gibb E.A., Brown C.J., Lam W.L. The functional role of long non-coding RNA in human carcinomas. Mol. Cancer. 2011;10:38:1–38:17. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsai M.C., Manor O., Wan Y., Mosammaparast N., Wang J.K., Lan F., Shi Y., Segal E., Chang H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta R.A., Shah N., Wang K.C., Kim J., Horlings H.M., Wong D.J., Tsai M.C., Hung T., Argani P., Rinn J.L., et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rinn J.L., Kertesz M., Wang J.K., Squazzo S.L., Xu X., Brugmann S.A., Goodnough L.H., Helms J.A., Farnham P.J., Segal E., et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu L., Zhu G., Zhang C., Deng Q., Katsaros D., Mayne S.T., Risch H.A., Mu L., Canuto E.M., Gregori G., et al. Association of large noncoding RNA HOTAIR expression and its downstream intergenic CpG island methylation with survival in breast cancer. Breast Cancer Res. Treat. 2012;136:875–883. doi: 10.1007/s10549-012-2314-z. [DOI] [PubMed] [Google Scholar]
- 17.Sorensen K.P., Thomassen M., Tan Q., Bak M., Cold S., Burton M., Larsen M.J., Kruse T.A. Long non-coding RNA HOTAIR is an independent prognostic marker of metastasis in estrogen receptor-positive primary breast cancer. Breast Cancer Res. Treat. 2013;142:529–536. doi: 10.1007/s10549-013-2776-7. [DOI] [PubMed] [Google Scholar]
- 18.Yang Z., Zhou L., Wu L.M., Lai M.C., Xie H.Y., Zhang F., Zheng S.S. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann. Surg. Oncol. 2011;18:1243–1250. doi: 10.1245/s10434-011-1581-y. [DOI] [PubMed] [Google Scholar]
- 19.Kogo R., Shimamura T., Mimori K., Kawahara K., Imoto S., Sudo T., Tanaka F., Shibata K., Suzuki A., Komune S., et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71:6320–6326. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 20.Kim K., Jutooru I., Chadalapaka G., Johnson G., Frank J., Burghardt R., Kim S., Safe S. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. 2013;32:1616–1625. doi: 10.1038/onc.2012.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu X.H., Liu Z.L., Sun M., Liu J., Wang Z.X., De W. The long non-coding RNA HOTAIR indicates a poor prognosis and promotes metastasis in non-small cell lung cancer. BMC Cancer. 2013;13:464:1–464:10. doi: 10.1186/1471-2407-13-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ge X.S., Ma H.J., Zheng X.H., Ruan H.L., Liao X.Y., Xue W.Q., Chen Y.B., Zhang Y., Jia W.H. HOTAIR a prognostic factor in esophageal squamous cell carcinoma inhibits WIF-1 expression and activates Wnt pathway. Cancer Sci. 2013;104:1675–1682. doi: 10.1111/cas.12296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li X., Wu Z., Mei Q., Guo M., Fu X., Han W. Long non-coding RNA HOTAIR a driver of malignancy predicts negative prognosis and exhibits oncogenic activity in oesophageal squamous cell carcinoma. Br. J. Cancer. 2013;109:2266–2278. doi: 10.1038/bjc.2013.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gutschner T., Diederichs S. The hallmarks of cancer: A long non-coding RNA point of view. RNA Biol. 2012;9:703–719. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alves C.P., Fonseca A.S., Muys B.R., de Barros E.L.B.R., Burger M.C., de Souza J.E., Valente V., Zago M.A., Silva W.A., Jr The lincRNA Hotair is required for epithelial-to-mesenchymal transition and stemness maintenance of cancer cells lines. Stem Cells. 2013;31:2827–2832. doi: 10.1002/stem.1547. [DOI] [PubMed] [Google Scholar]
- 26.Leveille N., Elkon R., Davalos V., Manoharan V., Hollingworth D., Oude Vrielink J., le Sage C., Melo C.A., Horlings H.M., Wesseling J., et al. Selective inhibition of microRNA accessibility by RBM38 is required for p53 activity. Nat. Commun. 2011;2:513:1–513:11. doi: 10.1038/ncomms1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shu L., Yan W., Chen X. RNPC1 an RNA-binding protein and a target of the p53 family is required for maintaining the stability of the basal and stress-induced p21 transcript. Genes Dev. 2006;20:2961–2972. doi: 10.1101/gad.1463306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mercer T.R., Mattick J.S. Structure and function of long noncoding RNAs in epigenetic regulation. Nat. Struct. Mol. Biol. 2013;20:300–307. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- 29.Gumireddy K., Li A., Yan J., Setoyama T., Johannes G.J., Orom U.A., Tchou J., Liu Q., Zhang L., Speicher D.W., et al. Identification of a long non-coding RNA-associated RNP complex regulating metastasis at the translational step. EMBO J. 2013;32:2672–2684. doi: 10.1038/emboj.2013.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoon J.H., Abdelmohsen K., Srikantan S., Yang X., Martindale J.L., De S., Huarte M., Zhan M., Becker K.G., Gorospe M. LincRNA-p21 suppresses target mRNA translation. Mol. Cell. 2012;47:648–655. doi: 10.1016/j.molcel.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poliseno L., Salmena L., Zhang J., Carver B., Haveman W.J., Pandolfi P.P. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tripathi V., Ellis J.D., Shen Z., Song D.Y., Pan Q., Watt A.T., Freier S.M., Bennett C.F., Sharma A., Bubulya P.A., et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.