Abstract
Clostridium difficile is a nosocomial pathogen that causes a serious toxin-mediated enteric disease in humans. Reducing C. difficile toxin production could significantly minimize its pathogenicity and improve disease outcomes in humans. This study investigated the efficacy of two, food-grade, plant-derived compounds, namely trans-cinnamaldehyde (TC) and carvacrol (CR) in reducing C. difficile toxin production and cytotoxicity in vitro. Three hypervirulent C. difficile isolates were grown with or without the sub-inhibitory concentrations of TC or CR, and the culture supernatant and the bacterial pellet were collected for total toxin quantitation, Vero cell cytotoxicity assay and RT-qPCR analysis of toxin-encoding genes. The effect of CR and TC on a codY mutant and wild type C. difficile was also investigated. Carvacrol and TC substantially reduced C. difficile toxin production and cytotoxicity on Vero cells. The plant compounds also significantly down-regulated toxin production genes. Carvacrol and TC did not inhibit toxin production in the codY mutant of C. difficile, suggesting a potential codY-mediated anti-toxigenic mechanism of the plant compounds. The antitoxigenic concentrations of CR and TC did not inhibit the growth of beneficial gut bacteria. Our results suggest that CR and TC could potentially be used to control C. difficile, and warrant future studies in vivo.
Keywords: plant compounds, Clostridium difficile, toxins, gene expression
1. Introduction
Clostridium difficile is a gram-positive, spore-forming, anaerobic bacterium that causes a toxin-mediated enteric disease in humans [1,2]. More than 300,000 cases of C. difficile-associated disease (CDAD) are reported annually in the United States, resulting in approximately US$3 billion of health care costs [3,4]. The emergence of a hypervirulent strain, NAP1/ribotype 027, that produces increased levels of toxins and a severe form of the disease in humans has been reported [5–7]. C. difficile predominantly affects long-term hospital inpatients and the elderly undergoing prolonged antibiotic therapy [6]. Prolonged antibiotic therapy results in the disruption of the normal enteric microflora, leading to the germination of C. difficile spores and pathogen colonization in the intestine with subsequent production of toxins [1,8]. The toxins, TcdA and TcdB, act as glucosyl transferases that inactivate the Rho family GTPases associated with F-actin regulation, and consequently cause disruption of the cytoskeleton and intestinal epithelial tight junctions [9,10]. This leads to an inflammatory response with the release of cytokines and leukotrienes, causing pseudomembrane formation and watery diarrhea [6,7,11]. The genes tcdA and tcdB, which encode the C. difficile toxins TcdA and TcdB, respectively, along with the genes encoding TcdR, an RNA polymerase sigma factor needed for maximal expression of tcdA and tcdB, TcdC, an antagonist of TcdR, and TcdE, a holin-like protein needed for toxin excretion, constitute the “pathogenicity locus” in C. difficile [11]. Expression of the pathogenicity locus is controlled by a number of environmental factors, including the availability of rapidly metabolizable carbon sources, a regulation mediated by the global regulator CcpA [12,13] and the intracellular pools of the branched-chain amino acids and GTP, mediated by the global regulator CodY [14,15]. Although exposure to broad-spectrum antibiotics predisposes patients to CDAD by disrupting the normal gut flora [16,17], antibiotics are still the primary line of treatment for patients who have contracted the disease. In addition, the emergence of antibiotic resistance in hypervirulent strains of C. difficile is increasingly reported worldwide [18,19]. Since C. difficile toxins are the major virulence factors responsible for the pathogenesis of CDAD, identification of alternative therapeutic agents that inhibit C. difficile toxin production without affecting the normal gastrointestinal flora or exacerbating bacterial antibiotic resistance could be a potentially viable approach for controlling CDAD.
Historically, plants have been used for treating various diseases in traditional medicine [20]. Carvacrol (CR) is a monoterpenoid phenol present in oregano and thyme oils. Diverse pharmacological actions of CR, including antimicrobial and anti-inflammatory properties, have been previously demonstrated [21]. Trans-cinnamaldehyde (TC) is an aromatic aldehyde present as a major component of the bark extract of cinnamon. Previous research conducted in our laboratory revealed that sub-inhibitory concentrations (SICs; the concentrations that do not inhibit bacterial growth) of TC and CR increased the sensitivity of multi-drug resistant Salmonella Typhimurium DT 104 to antibiotics by down-regulating antibiotic resistance genes and the efflux pump, tolC [20]. In addition, we previously observed that TC inhibited biofilm synthesis and virulence in uropathogenic Escherichia coli [22,23]. The objective of this study was to investigate the effect of sub-inhibitory concentrations (SICs) of TC and CR on toxin production and cytotoxicity of C. difficile in vitro, and delineate the potential mechanism(s) behind their effect.
2. Results and Discussion
2.1. Results
2.1.1. Sub-Inhibitory Concentrations of CR and TC
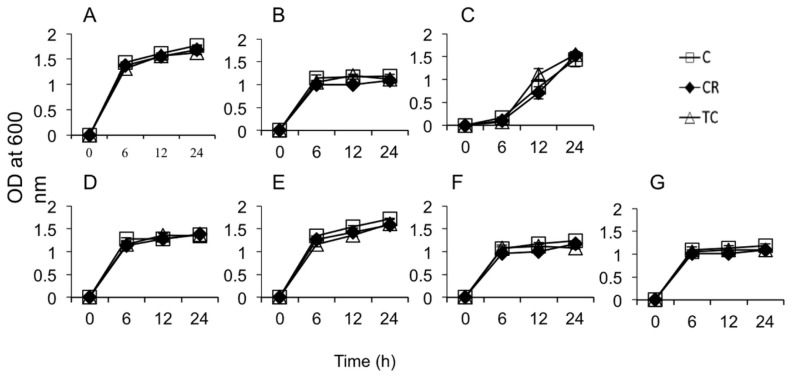
The SICs of CR and TC were found to be 0.60 mM (0.10 mg/mL) and 0.38 mM, (0.05 mg/mL), respectively. These concentrations of plant compounds did not inhibit the growth of all C. difficile isolates including CodY mutant and parental strains after 24 h incubation at 37 °C. In addition, the OD600 values for the seven selected beneficial bacterial isolates cultured in the presence of SICs of CR and TC were not significantly different from their respective controls (bacteria grown without CR or TC, Figure 1). This indicated that the concentrations of CR and TC used in this study were non-inhibitory to the growth of C. difficile (As shown in the Supplementary Figures S1–S3) as well as the seven selected beneficial bacteria.
Figure 1.
Effect of Sub-inhibitory concentrations of carvacrol (CR) and trans-cinnamaldehyde (TC) on beneficial gut bacterial growth. Seven selected beneficial gut bacteria (Lactobacillus brevis (A); L. reuterii (B); L. delbrueckii bulgaricus (C); L fermentum (D); L. plantarum (E); Bifidobacterium bifidum (F); and Lactococcus lactis lactis (G)) were grown in de Man, Rogosa and Sharpe broth in anaerobic condition at 37 °C with and without SICs of CR and TC (Control-Ctrl, open square, Carvacrol-CR, closed diamond, trans-cinnamaldehyde-TC, open triangle) for 24 h. The bacterial growth was monitored by measuring optical density at 600 nm measured at 6, 12 and 24 h. TC- or CR-treated gut bacterial populations did not change significantly from the controls (p > 0.05).
2.1.2. Effect of CR and TC on C. difficile Toxin Production
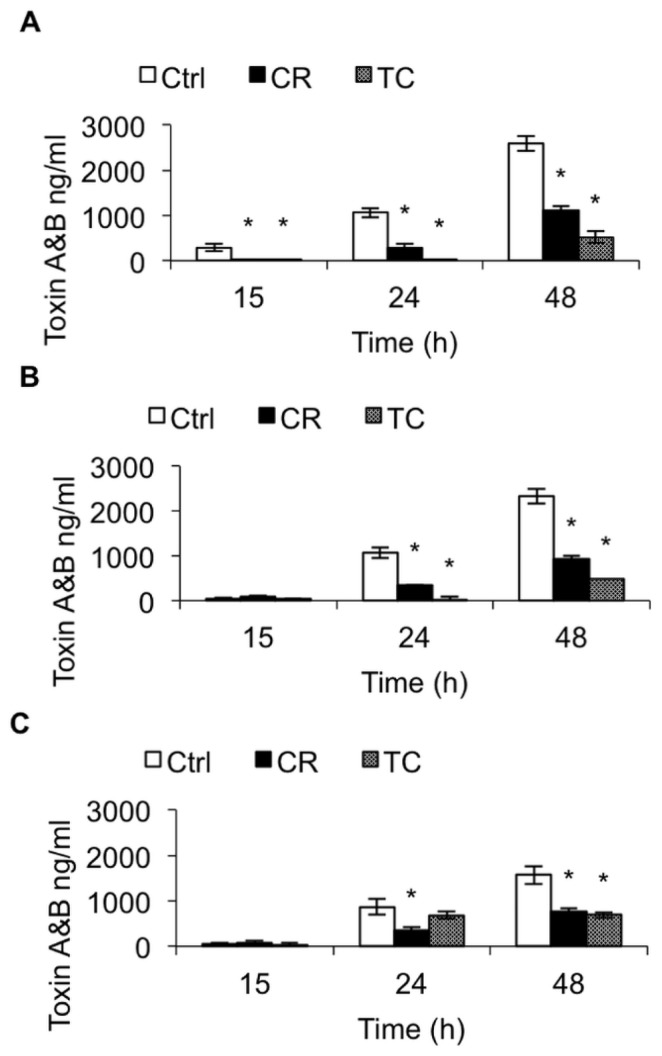
The effect of CR and TC on C. difficile toxin production was determined by ELISA, as described previously [24]. Carvacrol and TC significantly reduced toxin production in all three C. difficile isolates at 24 and 48 h (p < 0.05) compared to their controls. At 48 h, CR and TC inhibited the toxin production by approximately 60% and 80%, respectively in isolates BAA 1870 and 1053 (Figure 2A,B). In isolate BAA 1805, CR and TC reduced toxin production by approximately 55% at 48 h, compared to the control (Figure 2C). In isolates BAA 1870 and 1503, TC was more effective in reducing the toxin production than CR (p < 0.05) (Figure 2A,B), whereas both compounds exerted a similar inhibitory effect on toxin production in BAA 1805 after 48 h of incubation (Figure 2C).
Figure 2.
Effect of carvacrol (CR) and trans-cinnamaldehyde (TC) on C. difficile toxin production. * p < 0.05 vs. CR.
Brain heart infusion with or without the Sub-inhibitory concentration (SIC) of TC or CR, (0.38 and 0.60 mM respectively) was inoculated (104 CFU/mL) separately with three hypervirulent C. difficile isolates, ATCC BAA 1870 (A), ATCC BAA 1053 (B) or ATCC BAA 1805 (C), and incubated anaerobically at 37 °C for 72 h. The culture supernatant from groups TC, CR and Control (Ctrl) was collected at 15, 24, and 48 h of incubation for total toxin A and B quantitation by ELISA. * Treatments significantly differed from the control (p < 0.05).
2.1.3. Effect of CR and TC on C. difficile Toxin-Mediated Cytotoxicity
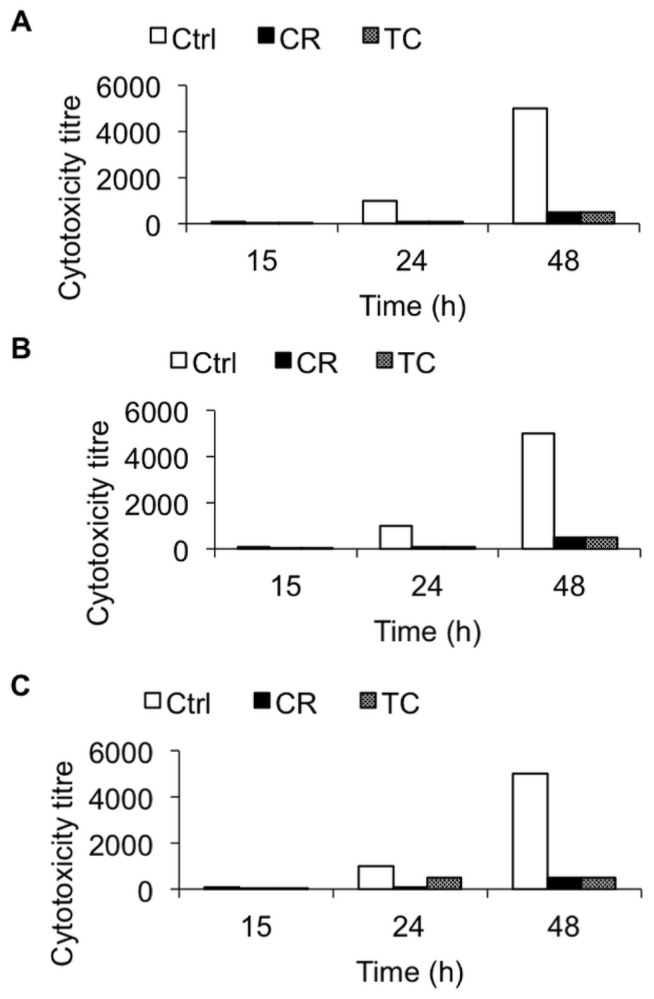
To determine the efficacy of TC and CR in reducing C. difficile-induced cytopathic effects, a cytotoxicity assay was conducted on Vero cells following a previously published protocol [25]. Exposure to TC and CR significantly reduced the ability of C. difficile culture supernatant to produce cytotoxicity on Vero cells compared to culture supernatants of untreated cells (p < 0.05). The cytotoxicity titer in the cells inoculated with CR- and TC-treated C. difficile culture supernatant was reduced by ~90% at 48 h in comparison to that in the control (Figure 3).
Figure 3.
Effect of carvacrol (CR) and trans-cinnamaldehyde (TC) on C. difficile induced cytotoxicity on Vero cells.
Brain heart infusion with or without the Sub-inhibitory concentration (SIC) of TC or CR, (0.38 and 0.60 mM respectively) was inoculated (105 CFU/mL) separately with three hypervirulent C. difficile isolates, ATCC BAA 1870 (A), ATCC BAA 1053 (B) or ATCC BAA 1805 (C), and incubated anaerobically at 37 °C for 72 h. The culture supernatant from groups TC, CR and Control (Ctrl) were collected at 15, 24, and 48 h of and the cytotoxicity titer on Vero cells was determined. Serially diluted C. difficile culture supernatants were added to the monolayers in 96-well plates and incubated at 37 °C under 5% CO2 for 48 h. The cell morphology was examined under an inverted microscope for characteristic rounding as an indication of cytotoxicity.
2.1.4. Effect of CR and TC on C. difficile Toxin Genes
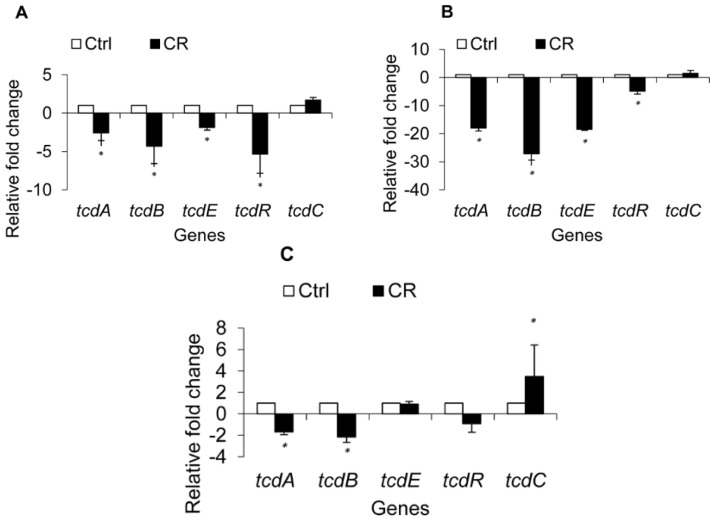
To investigate if the observed reduction in C. difficile toxin production was due to modulation of the expression of toxin-associated genes, transcriptional analysis by RT-qPCR (Real time-quantitative polymerase chain reaction) of the genes known to be involved in toxin production and regulation was performed. The data revealed a modulation of C. difficile toxin regulatory genes by CR and TC at early stationary phase (12 h) (Figures 4 and 5). Carvacrol significantly down-regulated (p < 0.05) the expression of toxin genes tcdA and tcdB in all three isolates of C. difficile (p < 0.05) by ~2.5 fold (Figure 4A,C). However, in isolate BAA 1053, the genes were down-regulated by 15 to 25 fold (Figure 4B). Similar results were obtained in C. difficile isolates exposed to TC (Figure 5A,B). However, in isolate BAA 1805, no significant difference was observed in comparison to the control (Figure 5C). A significant down-regulation was also observed in the expression of the toxin excretion gene tcdE and the positive regulator tcdR in two C. difficile isolates (ATCC # BAA 1870, 1053) treated with CR and TC (Figures 4 and 5). In isolates BAA 1870 and 1805, trans-cinnamaldehyde treatment increased expression of tcdC, which encodes the TcdR antagonist (Figure 5A,B). Up-regulation of tcdC was observed in CR-treated BAA 1870 and 1053 as well (Figure 4A,B).
Figure 4.
Effect of carvacrol (CR) on C. difficile toxin regulatory genes. Brain heart infusion with or without the sub-inhibitory concentration (SIC) of CR, (0.60 mM) was inoculated (105 CFU/mL) separately with three hyper virulent C. difficile isolates, ATCC BAA 1870 (A); ATCC BAA 1053 (B) or ATCC BAA 1805 (C), and incubated anaerobically at 37 °C for 72 h. Bacterial pellets were harvested at 6 and 12 h for RNA isolation and RT-qPCR for toxin regulatory genes. * Treatments significantly differed from the controls (p < 0.05).
Figure 5.
Effect of trans-cinnamaldehyde (TC) on C. difficile toxin regulatory genes. Brain heart infusion with or without the sub-inhibitory concentration (SIC) of TC, (0.38 mM) was inoculated (105 CFU/mL) separately with three hyper virulent C. difficile isolates, ATCC BAA 1870 (A); ATCC BAA 1053 (B) or ATCC BAA 1805 (C), and incubated anaerobically at 37 °C for 72 h. Bacterial pellets were harvested at 6 and 12 h for RNA isolation and RT-qPCR for toxin regulatory genes. * Treatments significantly differed from the controls (p < 0.05).
2.1.5. Effect of CR and TC on a codY Mutant of C. difficile
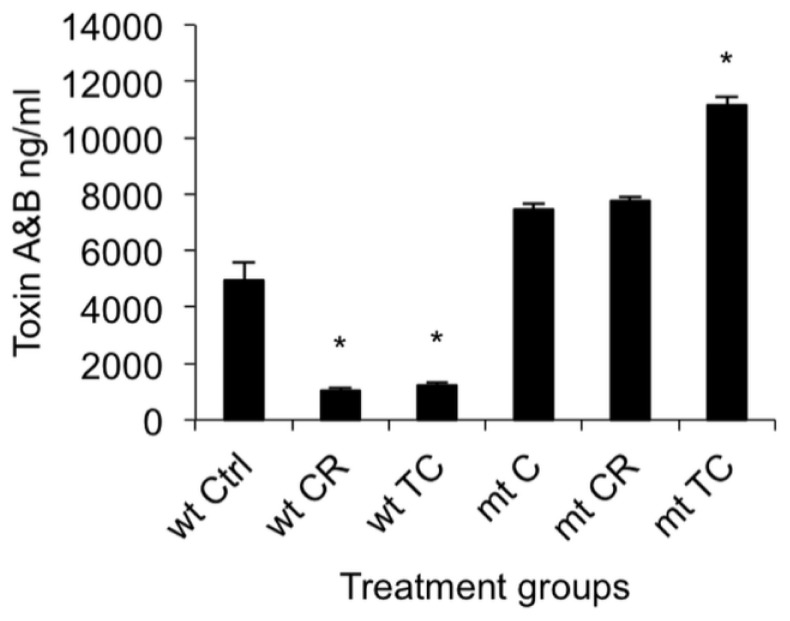
In this experiment, we treated the C. difficile codY mutant (LB-CD16) and its parental strain, UK-1, with the SIC of CR or TC, and determined their effects on toxin production and toxin gene expression. The total toxin quantitation by ELISA (Enzyme Linked Immunosorbent Assay) revealed a significant reduction in toxin production in CR- or TC-treated UK-1 (p < 0.05) compared to the control (Figure 6). However, in a codY mutant, the level of toxin production in the absence of drug was higher than in the parent strain and neither CR nor TC caused any inhibition of toxin production. In fact, TC treatment enhanced the toxin production in the codY mutant strain significantly (p < 0.05) compared to the untreated control. Thus, in the absence of functional CodY protein, neither CR nor TC could inhibit toxin production.
Figure 6.
Effect of carvacrol (CR) and trans-cinnamaldehyde (TC) on codY mutant and wild type C. difficile toxin production. Brain heart infusion with or without the sub-inhibitory concentration (SIC) of TC or CR, (0.38 and 0.60 mM respectively) was inoculated (105 CFU/mL) separately with UK1 and LB-CD16 (codY mutant) C. difficile isolates, and incubated anaerobically at 37 °C for 24 h. The culture supernatants from groups TC, CR and Control (Ctrl) were collected at 24 h of incubation for total toxin A and B quantitation by ELISA. * Treatments significantly differed from respective controls (p < 0.05).
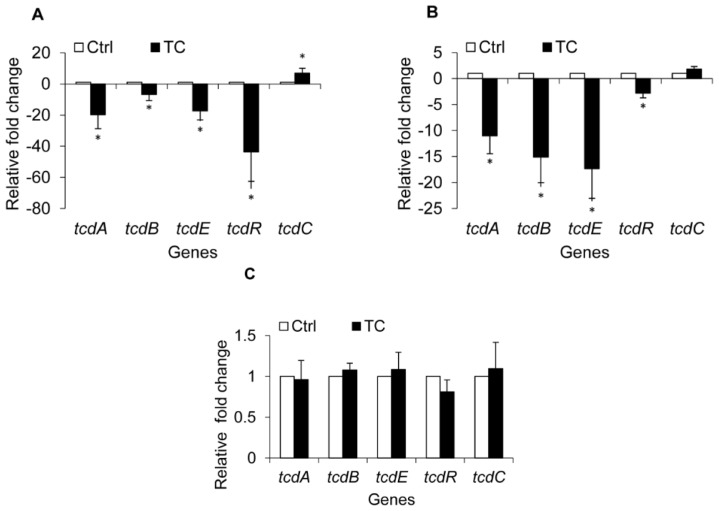
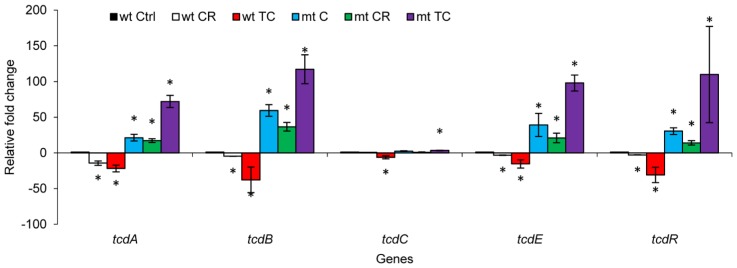
RT-qPCR analysis of the toxin genes in the codY mutant strain and its parent revealed a trend in gene expression similar to that seen for toxin production (Figure 7). A significant down-regulation of tcdA and tcdB was observed in strain UK-1 treated with CR or TC (p < 0.05). However, consistent with the ELISA results, CR and TC did not cause any down-regulation of the tcdA and tcdB genes in the codY mutant (p < 0.05). Specifically, no significant change in the expression of tcdA, tcdB, tcdC and tcdR was observed in the codY mutant treated with CR. However, TC treatment resulted in a significant up-regulation of tcdA and tcdB in the mutant (p < 0.05). These data collectively suggest that the SICs of CR and TC were unable to exert any inhibitory effect on the toxin-encoding genes in the absence of a functional codY.
Figure 7.
Effect of carvacrol (CR) and trans-cinnamaldehyde (TC) on codY mutant and wild type C. difficile toxin genes. Brain heart infusion with or without the sub-inhibitory concentration (SIC) of TC or CR, (0.38 and 0.60 mM respectively) was inoculated (105 CFU/mL) separately with wild type (wt) UK-1 and its codY mutant (mt) C. difficile isolates, and incubated anaerobically at 37 °C for 72 h. Bacterial pellets were harvested at 6 and 12 h for RNA isolation and RT-qPCR for toxin regulatory genes. * Treatments significantly differed from respective controls (p < 0.05).
2.2. Discussion
C. difficile colonizes the large intestine of patients undergoing prolonged antibiotic therapy, and produces toxins TcdA and TcdB, resulting in CDAD [1]. C. difficile toxins lead to inflammation in the intestine, increased epithelial permeability [26–28], enhanced cytokine and chemokine production [26,28], neutrophil infiltration [29] and the release of reactive oxygen intermediates [26], thereby causing direct damage to the intestinal mucosa [1]. Therefore, reducing toxin production by C. difficile is critical in controlling CDAD.
A potential alternate strategy for controlling microbial infections is the use of anti-virulence drugs. This class of drugs aims at reducing bacterial virulence rather than inhibiting bacterial growth [30] and presents a lesser selective pressure for development of bacterial antimicrobial resistance [31–33]. Therefore, in the current study, we investigated the efficacy of SICs of CR and TC as alternative therapeutic agents that can ameliorate CDAD pathology by reducing C. difficile toxin production without affecting the growth of the normal intestinal flora. Since the SICs of antimicrobials, including antibiotics, can modulate bacterial gene expression and physico-chemical functions, they have been used for studying effects on bacterial gene expression and virulence [34].
In the current study, we found that the SICs of CR and TC for C. difficile growth did not inhibit the growth of seven different species of bacteria commonly found in the human gastrointestinal tract. The tested gut bacteria included Lactobacilli and Bifidobacteria, which are the major groups of beneficial gut flora that play a significant role in maintaining normal gut health [35]. Previous studies have reported that CR and TC exerted no adverse effects on endogenous bacterial populations, including Lactobacilli and Bifidobacteria, in pigs and poultry [36,37].
Our ELISA results indicated that both plant compounds decreased the total toxin production in all three C. difficile isolates obtained from the ATCC, compared to their untreated control cultures (Figure 2). In addition, culture supernatants from TC- or CR-treated cells of all three strains had significantly reduced cytotoxic effects on Vero cells (Figure 3), confirming the ELISA results. Previously, Ultee and Smid (2001) reported that sub-MIC concentrations of CR decreased toxin production in Bacillus cereus [38]. Moreover, prior research conducted in our laboratory revealed that plant compounds, including CR and TC, reduce the virulence of Listeria monocytogenes by decreasing its attachment to and invasion of cultured human intestinal epithelial and brain micro-vascular endothelial cells by modulating the expression of several virulence factors, including the global regulator, prfA [39]. Since SICs of CR and TC were used in the experiments, the reduction observed in C. difficile toxin production in the treated samples was not due to growth inhibition of the bacterium by the plant compounds, but could be due to their potential abilities to modulate the expression of virulence genes associated with toxin production. Therefore, we used RT-qPCR to determine the effect of TC and CR on the transcription of major genes reported to play a role in toxin synthesis and secretion. Our RT-qPCR data revealed a 3-to-20-fold reduction in the expression of toxin-encoding genes tcdA and tcdB in strains BAA 1870 and BAA 1053. Furthermore, tcdR, which encodes a factor essential for high-level toxin production, was down-regulated 3-to-40-fold by CR and TC in those strains (Figures 4 and 5). Down-regulation of TcdR is known to lead to reduced toxin production [40]. Down-regulation of tcdE was also observed in these C. difficile isolates treated with CR or TC (Figures 4 and 5). Multiple studies have demonstrated that tcdE is critical for toxin release from C. difficile, helping to explain the reduced amount of toxins in CR- and TC-treated culture supernatants. In strain BAA 1805, however, the correlation between toxin and holin gene expression and toxin production was less clear suggesting that in this strain the modulation of toxin synthesis is mediated by a different mechanism than in other strains. Our results show that the compounds block toxin production in all strains, albeit to different extents, and that this effect can be attributed to decreases in toxin gene transcription in all strains except in the case of strain BAA 1805 in presence of TC.
CodY, a global regulatory protein that senses the intracellular levels of branched-chain amino acids and GTP, monitors the nutritional status and regulates metabolism and stress responses in Gram-positive bacteria [14,41,42]. In addition, CodY acts as a global regulator of virulence; a codY deletion results in enhanced virulence in Staphylococcus aureus and other species [42–45]. In C. difficile, CodY is a potent repressor of tcdR, thereby preventing expression of the Paloc genes when cells are growing rapidly [14,15]. Previous studies have shown increased expression of toxin genes in a codY mutant of C. difficile [14,15]. Our results show that CR and TC reduce toxin production and pathogencity locus gene expression in NAP1/027 strain UK-1, but not in a codY null mutant of UK-1. In fact, TC caused hyperexpression of toxin locus genes in the codY mutant strain. In a codY mutant strain, the compounds lose their effectiveness as inhibitors, suggesting that the compounds work by a mechanism that may affect either CodY synthesis or activity. CodY activity would be reduced if the compounds caused the pools of branched chain amino acids or GTP to drop [14,15]. These data collectively suggest that the antitoxigenic effect of CR and TC is mediated at least in part through CodY.
3. Experimental Section
3.1. Bacterial Strains and Culture Conditions
Four hypervirulent C. difficile isolates (ATCC# BAA 1870, 1053, 1805 and UK1) were grown in brain heart infusion broth (BHI) supplemented with 5% yeast extract (Difco, Sparks, MD, USA) in a Whitley A35 anaerobic work station (Microbiology Inc., Frederick, MD, USA) in the presence of 80% nitrogen, 10% carbon dioxide and 10% hydrogen at 37 °C for 24 h. The bacterial population was titered by plating 0.1 mL portions of appropriate dilutions on BHI agar and Clostridium difficile moxalactum norfloxacin (CDMN) agar (Oxoid, Hampshire, UK) supplemented with 5% horse blood, under strict anaerobic conditions at 37 °C for 24 h. In addition, seven selected beneficial enteric bacteria obtained from the USDA-ARS culture collection, Peoria, IL (Lactobacillus brevis, L. reuterii, L. delbrueckii bulgaricus, L. fermentum, L. plantarum, Bifidobacterium bifidum, and Lactococcus lactis lactis) were separately grown in de Man, Rogosa and Sharpe (MRS) broth (Difco, Sparks, MD, USA) under anaerobic conditions at 37 °C. The titers of the cultures were determined by plating 0.1 mL portions of appropriate dilutions on MRS agar (Difco, Sparks, MD, USA) with incubation at 37 °C for 24 h. The cultures were sedimented by centrifugation (3600× g, 15 min, 4 °C), and the pellets were washed twice, and resuspended in sterile phosphate-buffered saline (pH 7.3), and used as the inoculum.
3.2. Plant Compounds and SIC Determination
The SIC of TC and CR was determined as previously described [20]. Fifty ml of BHI supplemented with 5% yeast extract was inoculated separately with ~5.0 log10 CFU of each C. difficile isolate, followed by the addition of 1 to 10 μL of TC or CR (Sigma-Aldrich, St. Louis, MO, USA) with an increment of 1 μL. The cultures were incubated at 37 °C for 24 h, and bacterial growth was determined by measuring the optical density at 600 nm. The highest concentration of each plant compound that did not inhibit bacterial growth after 24 h of incubation was selected as its respective SIC (Sub Inhibitory Concentration) for this study.
Similarly, the effect of SIC of TC and CR on the growth of the aforementioned beneficial gut bacteria was determined by culturing them separately in 10 mL of MRS broth under anaerobic conditions at 37 °C with or without the plant compounds for 24 h. The growth of each culture was determined by measuring optical density at 600 nm, and plating on MRS agar.
3.3. Effect of Plant Compounds on C. difficile Toxin Production and Cytotoxicity
Brain Heart infusion broth, with or without the SIC of TC or CR was inoculated (105 CFU/mL) separately with each C. difficile isolate, and incubated at 37 °C for 48 h anaerobically as before. The culture supernatant was collected at 15, 24, and 48 h of incubation for total toxin A and B quantitation by ELISA [24] and for determining cytotoxicity on Vero cells. The bacterial pellets were harvested at 6 and 12 h for RNA isolation and RT-qPCR analysis of C. difficile genes associated with toxin synthesis.
3.4. ELISA for Total Toxin A and B
The amount of toxin in the culture supernatant was quantified using the Wampole Tox A/B II kit (TechLabs, Inc., Blacksburg, VA, USA), as described by Merrigan et al. [24]. Purified toxin B (Sigma Aldrich, St. Louis, MO, USA) was used to plot a standard curve. The culture supernatants were diluted and ELISA was performed according to the manufacturer’s instructions. The optical density was measured at 450 nm, compared with the linear range of standard curve, and total toxin concentration was estimated.
3.5. Cytotoxicity Assay
The effect of CR and TC on the cytotoxicity of C. difficile culture supernatant was estimated by Vero cell cytotoxicity assay, as described previously [25]. C. difficile culture supernatant was serially diluted (1:10) and added onto Vero cell monolayers in 96-well microtiter plates. The plates were incubated at 37 °C in a 5% CO2 environment for 48 h, and examined under an inverted microscope. Positive reactions were indicated by the characteristic rounding of Vero cells accompanied by parallel neutralization of cytotoxicity with Clostridium sordellii antitoxin (TechLabs, Inc., Blacksburg, VA, USA). The cytotoxicity titer was expressed as the reciprocal of the highest dilution that caused more than 80% cell rounding.
3.6. Real Time Quantitative PCR (RT-qPCR)
In order to determine the effect of CR and TC on C. difficile genes involved in toxin synthesis, total RNA was isolated from the early stationary phase (12 h) cultures [5]. The culture supernatant was harvested by centrifugation at 3000× g for 10 min at 4 °C. The bacterial pellet was resuspended in RNAwiz solution (Ambion, Austin, TX, USA), flash frozen in liquid nitrogen, and stored at −80 °C. Total RNA extraction was performed using the Ambion RiboPure Bacteria RNA kit (Ambion, Austin, TX, USA) according to the manufacturer’s instructions, followed by DNase I digestion using Turbo DNase I (Ambion). The RNA obtained after each DNase I digestion was purified further using the Qiagen RNeasy RNA column purification kit, according to the manufacturer’s instructions (Qiagen, Germantown, MD, USA). The cDNA was synthesized using the Bio-Rad iScript cDNA synthesis kit (Bio-Rad, Hercules, CA, USA). RT-qPCR analysis of the genes associated with toxin production was performed using published primers for Paloc genes [46] normalized against 16S rRNA gene expression. Twenty-five μL reactions were performed in triplicate using iTaq SYBR (Bio-Rad, Hercules, CA, USA). The relative fold change in gene expression was calculated using the 2−ΔΔCt method [47].
3.7. Construction of a codY Mutant of C. difficile Strain UK-1 and the Effect of Plant Compounds on codY Mutant and Parental Strains
The pJS107 plasmid [48] was used as a TargeTron vector to introduce mutations into the NAP1/027 C. difficile strain UK-1. The group II intron insertion sites for C. difficile codY were identified using an algorithm that can be found at http://dna.med.monash.edu.au/~torsten/intron_site_finder/. The intron fragment was generated by PCR using primers oLB178 (TGAACGCAAGTTTCTA ATTTCGATTGTAGTTCGATAGAGGAAAGTGTCT), oLB179 (AAAAAAGCTTATAATTATCCT TAACTACCGTAGTAGTGCGCCCAGATAGGGTG), oLB180 (CAGATTGTACAAATGTGGTGA TAACAGATAAGTCGTAGTATATAACTTACCTTTCTTTGT) and EBS-Universal (CGAAATTA GAAACTTGCGTTCAGTAAAC), and a 1:1 mixture of pBL64 and pBL65 as template [49] and then cloned at the HindIII and BsrGI sites of pJS107, yielding pBL103. pBL103 was then introduced into B. subtilis strain BS49 by transformation. Strain BS49 carries a conjugative transposon, Tn916, integrated into its chromosome and serves as a donor in conjugation with C. difficile. Conjugation experiments were carried out as described previously [50]. C. difficile transconjugants were selected on thiamphenicol (10 μg/mL) and then screened for the presence of Tn916 by assaying tetracycline resistance (10 μg/mL). Thiamphenicol-resistant, tetracycline-sensitive (plasmid-containing, transposon-negative) transconjugants were selected for further use. Potential TargeTron mutants were identified by plating on lincomycin (20 μg/mL) and screening for the insertion of the intron into C. difficile codY by PCR using primers specific for full-length C. difficile codY. (oLB205, TGAGCATGCTTAATGATGATGATGATGATGTTGATTGTTTTTTAATTTTTTTAATTCATC; and oLB206, TATCCGGAATTCTGAGGAGATGATTAAATGGC), the 5′ intron insertion site and the 3′ intron insertion site and a positive clone, strain LB-CD16, was identified. A Southern blot was performed as previously described [49] to confirm that the mutant contained a single insertion of the intron.
The role of codY on the anti-toxigenic effect mediated by the plant compounds was investigated by quantifying toxin production and expression of toxin-associated genes in CR- and TC-treated codY mutant and wild type strains of C. difficile. Brain Heart infusion broth, with or without the SIC of TC or CR was separately inoculated (105 CFU/mL) with UK1 or LB16, and incubated at 37 °C for 48 h, as described previously. The culture supernatants were collected at 24 and 48 h of incubation for total toxin A and B quantitation by ELISA. In addition, the bacterial pellets were harvested at 12 h for RNA isolation and RT-qPCR analysis.
3.8. Statistical Analysis
All experiments had duplicate samples and the study was repeated three times. The data were analysed using the PROC-MIXED procedure of SAS v. 9.3 (SAS Institute Inc., Cary, NC, USA). Differences between the means were considered significantly different at p < 0.05.
4. Conclusions
To conclude, our study demonstrated that CR and TC were effective in significantly reducing C. difficile toxin production. Although there were variations in the expression of toxin encoding genes, both plant molecules reduced toxin production in all strains (BAA 1870, BAA1053, BAA1805 and UK-1). The major toxin encoding genes, toxA, and toxB were significantly down-regulated in all the tested strains by CR, whereas TC down-regulated these genes in all strains except one (BAA 1805). Moreover, the two plant compounds did not inhibit the growth of major gut microflora in humans. The anti-toxigenic effect of CR and TC appears to be mediated through CodY. The results suggest the potential use of TC and CR to attenuate C. difficile virulence; in vivo studies on the effect of CR and TC on CDAD are thus warranted.
Supplementary Information
Acknowledgments
This research was supported by grants to Kumar Venkitanarayanan from the United States Department of Agriculture-National Institute of Food and Agriculture (USDA-NIFA) Critical and Emerging Food Safety Issues program (Grant # 2010-03567) and to Abraham Sonenshein from the National Institute of General Medical Sciences (Grant # R01 GM042219).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Author Contributions
Kumar Venkitanarayanan conceived the idea. Kumar Venkitanarayanan and Shankumar Mooyottu designed the experiment. Shankumar Mooyottu, Anup Kollanoor-Johny, Genevieve Flock and Abhinav Upadhyay performed the experiment. Laurent Bouillaut and Abraham Sonenshein designed and constructed the mutant. Anup Kollanoor-Johny and Shankumar Mooyottu analysed the data. Shankumar Mooyottu and Kumar Venkitanarayanan wrote the manuscript.
References
- 1.Bartlett J.G. Clostridium difficile infection: Pathophysiology and diagnosis. Semin. Gastrointest. Dis. 1997;8:12–21. [PubMed] [Google Scholar]
- 2.Weese J.S. Clostridium difficile in food—Innocent bystander or serious threat? Clin. Microbiol. Infect. 2010;16:3–10. doi: 10.1111/j.1469-0691.2009.03108.x. [DOI] [PubMed] [Google Scholar]
- 3.Ghose C., Kalsy A., Sheikh A., Rollenhagen J., John M., Young J., Rollins S.M., Qadri F., Calderwood S.B., Kelly C.P., et al. Transcutaneous immunization with Clostridium difficile toxoid A induces systemic and mucosal immune responses and toxin a-neutralizing antibodies in mice. Infect. Immun. 2007;75:2826–2832. doi: 10.1128/IAI.00127-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilkins T.D., Lyerly D.M. Clostridium difficile testing: After 20 years still challenging. J. Clin. Microbiol. 2003;41:531–534. doi: 10.1128/JCM.41.2.531-534.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blossom D.B., McDonald L.C. The challenges posed by reemerging Clostridium difficile infection. Clin. Infect. Dis. 2007;45:222–227. doi: 10.1086/518874. [DOI] [PubMed] [Google Scholar]
- 6.Hookman P., Barkin J.S. Clostridium difficile associated infection diarrhea and colitis. World J. Gastroenterol. 2009;15:1554–1580. doi: 10.3748/wjg.15.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sunenshine R.H., McDonald L.C. Clostridium difficile-associated disease: New challenges from an established pathogen. Clevel. Clin. J. Med. 2006;73:187–197. doi: 10.3949/ccjm.73.2.187. [DOI] [PubMed] [Google Scholar]
- 8.Dial S., Delaney J.A., Barkun A.N., Suissa S. Use of gastric acid-suppressive agents and the risk of community-acquired Clostridium difficile-associated disease. JAMA. 2005;294:2989–2995. doi: 10.1001/jama.294.23.2989. [DOI] [PubMed] [Google Scholar]
- 9.Keel M.K., Songer J.G. The comparative pathology of Clostridium difficile-associated disease. Vet. Pathol. 2006;43:225–240. doi: 10.1354/vp.43-3-225. [DOI] [PubMed] [Google Scholar]
- 10.Von Eichel-Streiber C., Zec-Pirnat I., Grabnar M., Rupnik M. A nonsense mutation abrogates production of a functional enterotoxin A in Clostridium difficile toxinotype VIII strains of serogroups F and X. FEMS Microbiol. Lett. 1999;178:163–168. doi: 10.1111/j.1574-6968.1999.tb13773.x. [DOI] [PubMed] [Google Scholar]
- 11.McDonald L.C., Owings M., Jernigan D.B. Clostridium difficile infection in patients discharged from US short-stay hospitals 1996–2003. Emerg. Infect. Dis. 2006;12:409–415. doi: 10.3201/eid1203.051064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Antunes A., Martin-Verstraete I., Dupuy B. CcpA-mediated repression of Clostridium difficile toxin gene expression. Mol. Microbiol. 2011;79:882–899. doi: 10.1111/j.1365-2958.2010.07495.x. [DOI] [PubMed] [Google Scholar]
- 13.Antunes A., Camiade E., Monot M., Courtois E., Barbut F., Sernova N.V., Rodionov D.A., Martin-Verstraete I., Dupuy B. Global transcriptional control by glucose and carbon regulator CcpA in Clostridium difficile. Nucleic Acids Res. 2012;40:10701–10718. doi: 10.1093/nar/gks864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dineen S.S., McBride S.M., Sonenshein A.L. Integration of metabolism and virulence by Clostridium difficile CodY. J. Bacteriol. 2010;192:5350–5362. doi: 10.1128/JB.00341-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dineen S.S., Villapakkam A.C., Nordman J.T., Sonenshein A.L. Repression of Clostridium difficile toxin gene expression by CodY. Mol. Microbiol. 2007;66:206–219. doi: 10.1111/j.1365-2958.2007.05906.x. [DOI] [PubMed] [Google Scholar]
- 16.Bartlett J.G. Antibiotic-associated diarrhea. Clin. Infect. Dis. 1992;15:573–581. doi: 10.1093/clind/15.4.573. [DOI] [PubMed] [Google Scholar]
- 17.O’Connor K.A., Kingston M., O’Donovan M., Cryan B., Twomey C., O’Mahony D. Antibiotic prescribing policy and Clostridium difficile diarrhoea. QJM. 2004;97:423–429. doi: 10.1093/qjmed/hch076. [DOI] [PubMed] [Google Scholar]
- 18.Prabaker K., Weinstein R.A. Trends in antimicrobial resistance in intensive care units in the United States. Curr. Opin. Crit. Care. 2011;17:472–479. doi: 10.1097/MCC.0b013e32834a4b03. [DOI] [PubMed] [Google Scholar]
- 19.Spigaglia P., Barbanti F., Mastrantonio P. European study group on Clostridium difficile (ESGCD) Multidrug resistance in European Clostridium difficile clinical isolates. J. Antimicrob. Chemother. 2011;66:2227–2234. doi: 10.1093/jac/dkr292. [DOI] [PubMed] [Google Scholar]
- 20.Johny A.K., Hoagland T., Venkitanarayanan K. Effect of Subinhibitory concentrations of plant-derived molecules in increasing the sensitivity of multidrug-resistant salmonella enterica serovar typhimurium DT104 to antibiotics. Foodborne Pathog. Dis. 2010;7:1165–1170. doi: 10.1089/fpd.2009.0527. [DOI] [PubMed] [Google Scholar]
- 21.Baser K.H. Biological and pharmacological activities of carvacrol and carvacrol bearing essential oils. Curr. Pharm. Des. 2008;14:3106–3119. doi: 10.2174/138161208786404227. [DOI] [PubMed] [Google Scholar]
- 22.Amalaradjou M.A., Narayanan A., Venkitanarayanan K. Trans-cinnamaldehyde decreases attachment and invasion of uropathogenic escherichia coli in urinary tract epithelial cells by modulating virulence gene expression. J. Urol. 2011;185:1526–1531. doi: 10.1016/j.juro.2010.11.078. [DOI] [PubMed] [Google Scholar]
- 23.Amalaradjou M.A., Narayanan A., Baskaran S.A., Venkitanarayanan K. Antibiofilm effect of trans-cinnamaldehyde on uropathogenic Escherichia coli. J. Urol. 2010;184:358–363. doi: 10.1016/j.juro.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 24.Merrigan M., Venugopal A., Mallozzi M., Roxas B., Viswanathan V.K., Johnson S., Gerding D.N., Vedantam G. Human hypervirulent Clostridium difficile strains exhibit increased sporulation as well as robust toxin production. J. Bacteriol. 2010;192:4904–4911. doi: 10.1128/JB.00445-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baines S.D., Freeman J., Wilcox M.H. Effects of piperacillin/tazobactam on Clostridium difficile growth and toxin production in a human gut model. J. Antimicrob. Chemother. 2005;55:974–982. doi: 10.1093/jac/dki120. [DOI] [PubMed] [Google Scholar]
- 26.He D., Sougioultzis S., Hagen S., Liu J., Keates S., Keates A.C., Pothoulakis C., Lamont J.T. Clostridium difficile toxin A triggers human colonocyte IL-8 release via mitochondrial oxygen radical generation. Gastroenterology. 2002;122:1048–1057. doi: 10.1053/gast.2002.32386. [DOI] [PubMed] [Google Scholar]
- 27.Feltis B.A., Wiesner S.M., Kim A.S., Erlandsen S.L., Lyerly D.L., Wilkins T.D., Wells C.L. Clostridium difficile toxins A and B can alter epithelial permeability and promote bacterial paracellular migration through HT-29 enterocytes. Shock. 2000;14:629–634. doi: 10.1097/00024382-200014060-00010. [DOI] [PubMed] [Google Scholar]
- 28.Castagliuolo I., Keates A.C., Wang C.C., Pasha A., Valenick L., Kelly C.P., Nikulasson S.T., LaMont J.T., Pothoulakis C. Clostridium difficile toxin A stimulates macrophage-inflammatory protein-2 production in rat intestinal epithelial cells. J. Immunol. 1998;160:6039–6045. [PubMed] [Google Scholar]
- 29.Kelly C.P., Becker S., Linevsky J.K., Joshi M.A., O’Keane J.C., Dickey B.F., LaMont J.T., Pothoulakis C. Neutrophil recruitment in Clostridium difficile toxin A enteritis in the rabbit. J. Clin. Investig. 1994;93:1257–1265. doi: 10.1172/JCI117080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rasko D.A., Sperandio V. Anti-virulence strategies to combat bacteria-mediated disease. Nat. Rev. Drug Discov. 2010;9:117–128. doi: 10.1038/nrd3013. [DOI] [PubMed] [Google Scholar]
- 31.Cegelski L., Marshall G.R., Eldridge G.R., Hultgren S.J. The biology and future prospects of antivirulence therapies. Nat. Rev. Microbiol. 2008;6:17–27. doi: 10.1038/nrmicro1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hung D.T., Shakhnovich E.A., Pierson E., Mekalanos J.J. Small-molecule inhibitor of vibrio cholerae virulence and intestinal colonization. Science. 2005;310:670–674. doi: 10.1126/science.1116739. [DOI] [PubMed] [Google Scholar]
- 33.Hughes D.T., Sperandio V. Inter-Kingdom signalling: Communication between bacteria and their hosts. Nat. Rev. Microbiol. 2008;6:111–120. doi: 10.1038/nrmicro1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsui W.H., Yim G., Wang H.H., McClure J.E., Surette M.G., Davies J. Dual effects of MLS antibiotics: Transcriptional modulation and interactions on the ribosome. Chem. Biol. 2004;11:1307–1316. doi: 10.1016/j.chembiol.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 35.Rupa P., Mine Y. Recent advances in the role of probiotics in human inflammation and gut health. J. Agric. Food Chem. 2012;60:8249–8256. doi: 10.1021/jf301903t. [DOI] [PubMed] [Google Scholar]
- 36.Jamroz D., Wiliczkiewicz A., Wertelecki T., Orda J., Skorupinska J. Use of active substances of plant origin in chicken diets based on maize and locally grown cereals. Br. Poult. Sci. 2005;46:485–493. doi: 10.1080/00071660500191056. [DOI] [PubMed] [Google Scholar]
- 37.Si W., Gong J., Chanas C., Cui S., Yu H., Caballero C., Friendship R.M. In Vitro assessment of antimicrobial activity of carvacrol thymol and cinnamaldehyde towards salmonella serotype typhimurium DT104: Effects of pig diets and emulsification in hydrocolloids. J. Appl. Microbiol. 2006;101:1282–1291. doi: 10.1111/j.1365-2672.2006.03045.x. [DOI] [PubMed] [Google Scholar]
- 38.Ultee A., Smid E.J. Influence of carvacrol on growth and toxin production by bacillus cereus. Int. J. Food Microbiol. 2001;64:373–378. doi: 10.1016/s0168-1605(00)00480-3. [DOI] [PubMed] [Google Scholar]
- 39.Upadhyay A., Johny A.K., Amalaradjou M.A., Ananda Baskaran S., Kim K.S., Venkitanarayanan K. Plant-derived antimicrobials reduce listeria monocytogenes virulence factors in vitro and down-regulate expression of virulence genes. Int. J. Food Microbiol. 2012;157:88–94. doi: 10.1016/j.ijfoodmicro.2012.04.018. [DOI] [PubMed] [Google Scholar]
- 40.Mani N., Lyras D., Barroso L., Howarth P., Wilkins T., Rood J.I., Sonenshein A.L., Dupuy B. Environmental response and autoregulation of Clostridium difficile TxeR a sigma factor for toxin gene expression. J. Bacteriol. 2002;184:5971–5978. doi: 10.1128/JB.184.21.5971-5978.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guedon E., Serror P., Ehrlich S.D., Renault P., Delorme C. Pleiotropic transcriptional repressor CodY senses the intracellular pool of branched-chain amino acids in lactococcus lactis. Mol. Microbiol. 2001;40:1227–1239. doi: 10.1046/j.1365-2958.2001.02470.x. [DOI] [PubMed] [Google Scholar]
- 42.Pohl K., Francois P., Stenz L., Schlink F., Geiger T., Herbert S., Goerke C., Schrenzel J., Wolz C. CodY in staphylococcus aureus: A regulatory link between metabolism and virulence gene expression. J. Bacteriol. 2009;191:2953–2963. doi: 10.1128/JB.01492-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stenz L., Francois P., Whiteson K., Wolz C., Linder P., Schrenzel J. The CodY pleiotropic repressor controls virulence in gram-positive pathogens. FEMS Immunol. Med. Microbiol. 2011;62:123–139. doi: 10.1111/j.1574-695X.2011.00812.x. [DOI] [PubMed] [Google Scholar]
- 44.Rivera F.E., Miller H.K., Kolar S.L., Stevens S.M., Jr, Shaw L.N. The impact of CodY on virulence determinant production in community-associated methicillin-resistant staphylococcus aureus. Proteomics. 2012;12:263–268. doi: 10.1002/pmic.201100298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Montgomery C.P., Boyle-Vavra S., Roux A., Ebine K., Sonenshein A.L., Daum R.S. CodY deletion enhances in vivo virulence of community-associated methicillin-resistant staphylococcus aureus clone USA300. Infect. Immun. 2012;80:2382–2389. doi: 10.1128/IAI.06172-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vohra P., Poxton I.R. Comparison of toxin and spore production in clinically relevant strains of Clostridium difficile. Microbiology. 2011;157:1343–1353. doi: 10.1099/mic.0.046243-0. [DOI] [PubMed] [Google Scholar]
- 47.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 48.Francis M.B., Allen C.A., Shrestha R., Sorg J.A. Bile acid recognition by the Clostridium difficile germinant receptor CspC is important for establishing infection. PLoS Pathog. 2013;9:e1003356. doi: 10.1371/journal.ppat.1003356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bouillaut L., Self W.T., Sonenshein A.L. Proline-dependent regulation of Clostridium difficile stickland metabolism. J. Bacteriol. 2013;195:844–854. doi: 10.1128/JB.01492-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carter G.P., Douce G.R., Govind R., Howarth P.M., Mackin K.E., Spencer J., Buckley A.M., Antunes A., Kotsanas D., Jenkin G.A., et al. The anti-sigma factor TcdC modulates hypervirulence in an epidemic BI/NAP1/027 clinical isolate of Clostridium difficile. PLoS Pathog. 2011;7:e1002317. doi: 10.1371/journal.ppat.1002317. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.