Abstract
Marfan syndrome (MFS) is a dominantly inherited connective tissue disorder caused by mutations in the gene encoding fibrillin-1 (FBN1) and is characterized by aortic dilatation and dissection, which is the primary cause of death in untreated MFS patients. However, disease progression-associated changes in gene expression in the aortic lesions of MFS patients remained unknown. Using a mouse model of MFS, FBN1 hypomorphic mouse (mgR/mgR), we characterized the aortic gene expression profiles during the progression of the MFS. Homozygous mgR mice exhibited MFS-like phenotypic features, such as fragmentation of elastic fibers throughout the vessel wall and were graded into mgR1–4 based on the pathological severity in aortic walls. Comparative gene expression profiling of WT and four mgR mice using microarrays revealed that the changes in the transcriptome were a direct reflection of the severity of aortic pathological features. Gene ontology analysis showed that genes related to oxidation/reduction, myofibril assembly, cytoskeleton organization, and cell adhesion were differentially expressed in the mgR mice. Further analysis of differentially expressed genes identified several candidate genes whose known roles were suggestive of their involvement in the progressive destruction of aorta during MFS. This study is the first genome-wide analysis of the aortic gene expression profiles associated with the progression of MFS. Our findings provide valuable information regarding the molecular pathogenesis during MFS progression and contribute to the development of new biomarkers as well as improved therapeutic strategies.
Keywords: Microarray, mgR mice, Marfan syndrome
INTRODUCTION
Marfan syndrome (MFS) is a dominantly inherited connective tissue disorder with prominent ocular, skeletal, and cardiovascular abnormalities. Approximately, 1 in 5,000 individuals are affected by this syndrome. The primary cause of morbidity and mortality in MFS is the dilatation and dissection of ascending aortic roots, which is often asymptomatic until a life-threatening aortic rupture occurs (Dietz et al., 1991).
MFS is caused by mutations in the gene encoding fibrillin-1 (FBN1), the major structural constituent of extracellular microfibrils. Microfibrils are abundant in the elastic fibers of the aorta. Therefore, FBN1 mutations have been thought to contribute to aberrant aortic microfibril assembly. Such an aberrant assembly of microfibrils could contribute to the fragmentation of elastic fibers and increase the mechanical strain on the aortic walls, leading to aortic dilation and dissection. However, this “weak connective tissue” theory was challenged by the recent findings that microfibrils modulate the activity of transforming growth factor beta (TGF-β) superfamily and that FBN1 deficiency promotes the release of extracellular matrix-bound TGF-β, thereby promoting the downstream TGF-β signaling pathways (Neptune et al., 2003). Indeed, several studies have found significantly higher activation of Smad2 in the aortic tissues of human MFS patients than in those of controls, implying that dysregulation of TGF-β signaling pathway is also involved in the development and/or progression of MFS (Matt et al., 2009; Kim et al., 2013). Although previous studies have proven that MFS is a monogenetic disorder caused by mutations in FBN1, the precise molecular mechanism leading to aortic dilation and dissection in MFS remain unknown. Additionally, there is considerable inter- and intra-familial variability in the severity of MFS (Pyeritz, 2000). The variable and unpredictable progression of MFS impedes its clinical management and prediction of the risk for aortic rupture. Therefore, it is important to understand how MFS progresses in the aortic tissues.
The FBN1 hypomorphic mouse (mgR/mgR) has been used as a murine model of MFS (Pereira et al., 1999). While FBN1 knockout mice die soon after birth and show morphologically normal elastic fibers in the vasculature, hypomorphic mgR mice live long enough to display histopathological changes reminiscent of that in patients with MFS. Further, homozygous mgR mice gradually develop MFS-like manifestation during their lifespan, and their phenotypic severity depends on the residual expression level of FBN1 protein. In this regard, hypomorphic mgR mice are suitable for investigating the progressive changes in the gene expression profile of the aortic walls as MFS advances. Although previous studies have used murine models or human samples to investigate the differences in the gene expression between normal and MFS subjects, little is known about the dynamic changes in gene expression profiles associated with the disease progression. Therefore, we sought to characterize the transcriptomes of aortic tissues at successive pathological stages of MFS by using hypomorphic mgR mice.
MATERIALS AND METHODS
Animals
Homozygous mgR mice and their wild type (WT) siblings (Jackson Laboratory, Bar Harbor, ME) were used in all the experiments. All protocols were approved by the Institutional Animal Care and Use Committee of Ajou University. The animals were cared in accordance with the Guide for the Care and Use of Laboratory Animals published by the United States National Institutes of Health. For the surgical procedures, the mice were anesthetized by intraperitoneal injection of ketamine and xylazine (79.5 mg/kg and 9.1 mg/kg, respectively). The adequacy of the anesthesia was assessed by monitoring the pedal withdrawal reflex response.
Quantitative real-time polymerase chain reaction analysis
Total RNA was extracted from cells or tissues using Trizol reagent (Invitrogen, Carlsbad, CA), and cDNA was synthesized using Superscript first-strand synthesis kit (Invitrogen). Real time PCR was performed using SYBR-Green PCR master mix (Applied Biosystems, Carlsbad, CA) on a StepOnePlusTM Real-Time PCR System (Applied Biosystems). Data analysis was performed using the ΔΔ Ct method, and values for GAPDH were used for normalization. All the reactions were performed in triplicate. The following primers were used for the reactions: FBN, forward 5′-GGAATGACATCAGCAGGCAC-3′, reverse 5′-TACACAAATCCTTTGGGGCA-3′; GAPDH, forward 5′-CGTGGAAGGACTCATGAC-3′, reverse 5′-CAAATTCGTTGTCATACCAG-3′.
Aortic wall histopathology
After systemic perfusion fixation using 4% paraformaldehyde, the thoracic aorta was excised and embedded in paraffin. Cross-sections of the aortic tissue (5 μm in thickness) were dewaxed, rehydrated in ethanol, and stained with hematoxylin-eosin and Movat’s pentachrome to visualize fragmented elastic fibers.
cDNA microarray
For gene expression analysis, the mice were sacrificed at the age of 12–20 weeks, and their thoracic aorta were snap frozen and pestle ground under liquid nitrogen. Trizol reagent (Invitrogen) was used for the extraction of RNA from the powdered tissue. The gene expression profiling of the tissues from homozygous mgR and WT mice was performed using Affymetrix Genechip Mouse Gene 1.0 ST microarray (Affymetrix, Santa Clara, CA). Briefly, biotinylated cDNAs were synthesized from equal amounts of total RNA. After cDNA probes were hybridized as described in the Gene Chip whole transcript sense target labeling assay manual (Affymetrix), the chips were scanned using a Genechip Array scanner 3000 7G (Affymetrix) and the scanned images were analyzed using the Affymetrix Command Console software (version1.1). Probes with p value of <0.05 were used for analysis and were normalized using the Robust Multi-array Average normalization method.
Gene ontology (GO) analysis
We first mapped all the probe sets to NCBI official gene symbols and used an average expression value of a probe set for a corresponding gene. To identify differentially expressed genes (DEGs) across WT and four homozygous mgR mice, we used a 2-fold change threshold for all the comparisons. We first identified gene differentially expressed in WT and each of the four homozygous mgR mice, and combined all the DEGs as an input for hierarchical clustering. Heat maps were drawn using the MeV4 software. We used DAVID 6.7 for GO term enrichment analysis of identified DEGs.
Statistical analysis
All the data are expressed as mean ± standard error of the mean (SEM). Comparisons of parameters between groups were made using the post-hoc Student’s t-tests for unpaired observations. p<0.05 was regarded as statically significant.
RESULTS
Histopathological evaluation of aortic media of WT and homozygous mgR mice
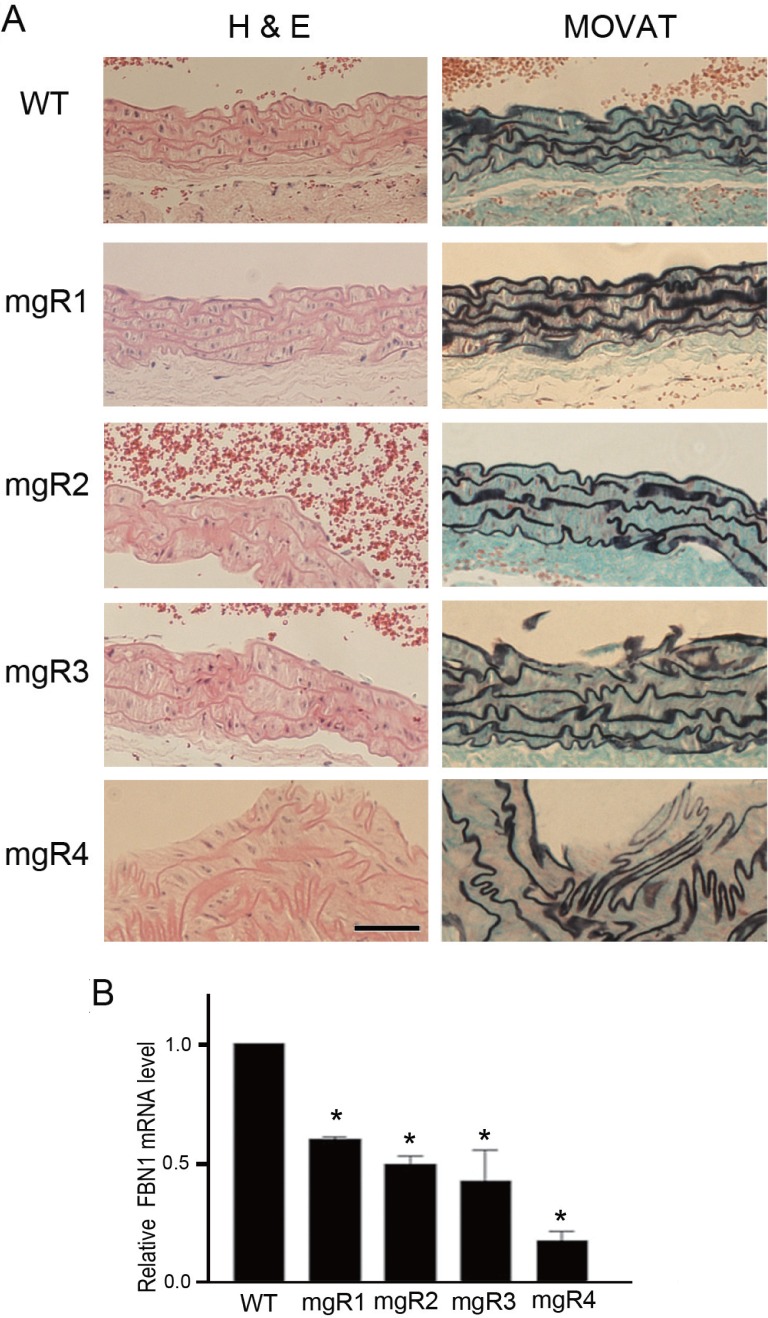
Hypomorphic FBN1 mutant mgR/mgR mice exhibited several skeletal deformities similar to those seen in MFS patients. The mice displayed overgrowth of the ribs, severe kyphosis, and diaphragm hernia, which was associated with their death approximately 3 to 5 months after birth (data not shown). As expected, vascular tissue of the homozygous mgR mice showed MFS-like characteristics, including the loss of elastin content and fragmentation as well as disorganization of elastic fibers throughout the vessel wall (Fig. 1A). Based on the increasing histopathological severity in the aortic walls, homozygous mgR mice were graded into mgR1, mgR2, mgR3, and mgR4. The mgR1 mouse showed intact elastic laminae. However, mgR2 and mgR3 mice showed several breaks and smoothened curvature of elastic fibers in the aortic walls. Further, the mgR4 mouse showed the most severe kinking and breaking of the elastic fibers and suffered from intimal hyperplasia that is often associated with deposition of collagen and amorphous matrix. Thus, the four mgR mice used in this study represented various stages of MFS with progressive alterations in aortic histopathology. Additionally, quantitative real-time PCR analysis revealed that the expression levels of FBN1 was substantially lower in the aortic tissues of mgR mice than in those of the WT animals (Fig. 1B). Of all the mgR mice examined, FBN1 mRNA level was lowest in the aortic tissue of the mgR4 mouse.
Fig. 1.
Aortic histopathology and FBN1 expression level of normal wild type and homozygous mgR mice. (A) Representative images of H & E and Movat-stained sections of thoracic aortas from wild type (WT) and homozygous mgR (mgR) mice. In the images of Movat-stained sections, elastic fibers are stained black. Aortic media of WT mouse show long parallel and closely packed arrays of intact elastic fibers. However, aortic media from mgR mice display various degrees of elastic fiber fragmentation and disorganization. Based on the histopathological severity, mgR mice were graded into 1 (least severe) to 4 (most severe). Vascular lumen is at the top in all the panels. Scale bar is 50 μm. (B) Quantitative real-time RT-PCR analysis of FBN1 expression in the aortic tissues of WT and homozygous mgR mice. FBN1 mRNA levels in mgR mice relative to its levels in WT (set as 1) was estimated (mean ± SEM, *p<0.05; triplicate experiments).
cDNA microarray of WT and homozygous mgR mice
To characterize the temporal changes in gene expression associated with the disease progression, cDNA microarray analysis was performed with aortic tissue samples harvested from WT, mgR1, mgR2, mgR3, and mgR4 mice. Comparative gene expression profiling revealed that the expression of 1,137 genes was altered during the course of disease progression. The heat map shown in Fig. 2 indicated that gene expression profiles of the four mgR mice were markedly different from that of the WT animals. The gene expression profile of the mgR1 mouse that showed only minor changes in the aortic lesion was similar to that of the WT mice. Notably, the gene expression profile of mgR4 mouse with the most severe pathological condition showed the greatest difference from that of the WT mice. The extent of the changes in the gene expression of mgR2 and mgR3 mice with modest phenotypic alteration was more than that of mgR1, but less than that of mgR4. Thus, the severity of aortic histopathology in the four mgR mice was well correlated with the extent of the changes in their transcriptomes from that of the WT mice. GO enrichment analysis of 1,137 DEGs reveals that genes involved in processes such as oxidation/reduction, myofibril assembly, actin cytoskeleton organization, and cell adhesion were highly enriched in the mgR mice (Table 1). These GO terms are highly relevant to the contractile function and survival of vascular smooth muscle cells. Thus, mgR mice seemed to have recapitulated the pathological feature of MFS, i.e., aortic dilatation and dissection.
Fig. 2.
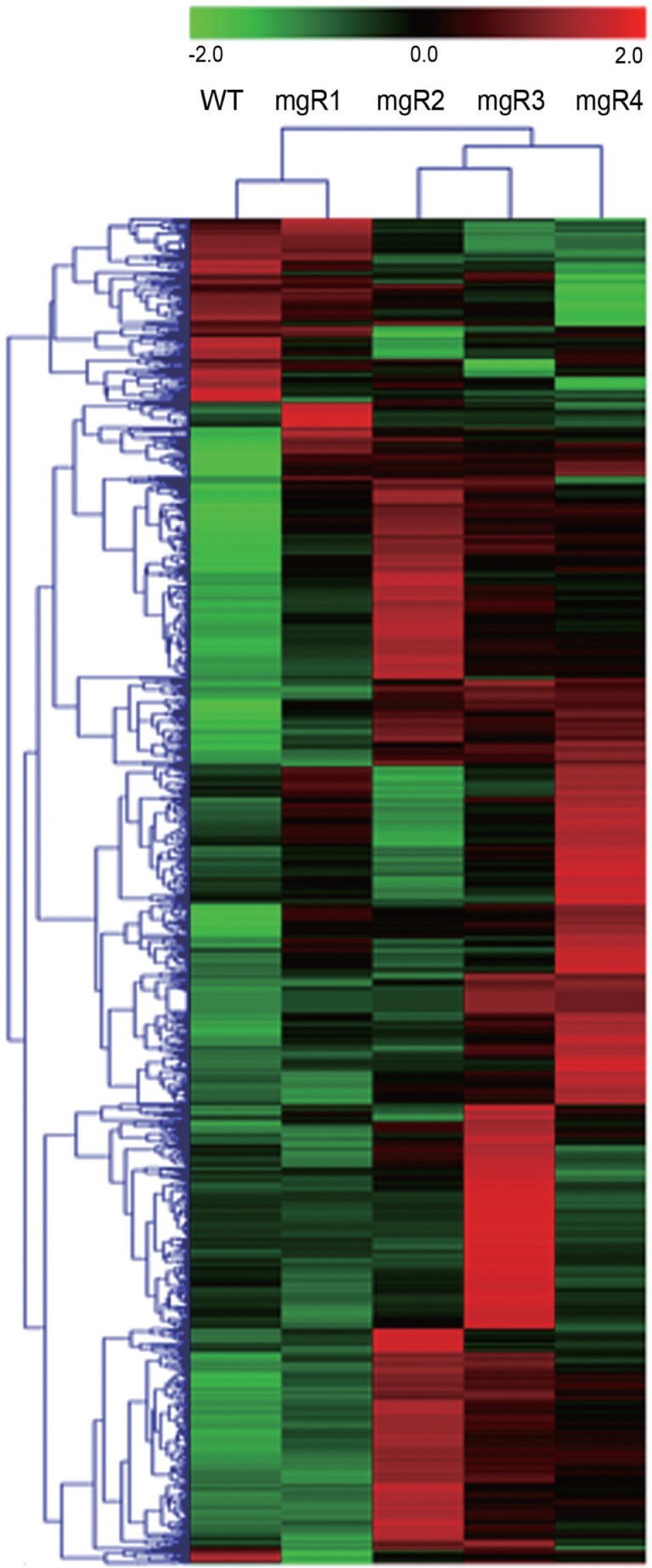
Dynamic changes in the transcriptome of homozygous mgR mice. The hierarchical clustering of 1,137 DEGs (>2-fold change compared to that in WT) is shown as a heat map. Gene expression level is shown in a color scale ranging from red (overexpression) to green (underexpression) according to the scale bar at the top of the heat map.
Table 1.
Major biological processes representing genes that were differentially expressed in the aorta of homozygous mgR mice compared to that of the wild type aorta
| Gene ontology ID | Biological process | p value | Count |
|---|---|---|---|
| GO:0055114 | Oxidation reduction | 1.28E-13 | 82 |
| GO:0030239 | Myofibril assembly | 6.35E-06 | 11 |
| GO:0030036 | Actin cytoskeleton organization | 3.07E-06 | 25 |
| GO:0007155 | Cell adhesion | 0.034 | 35 |
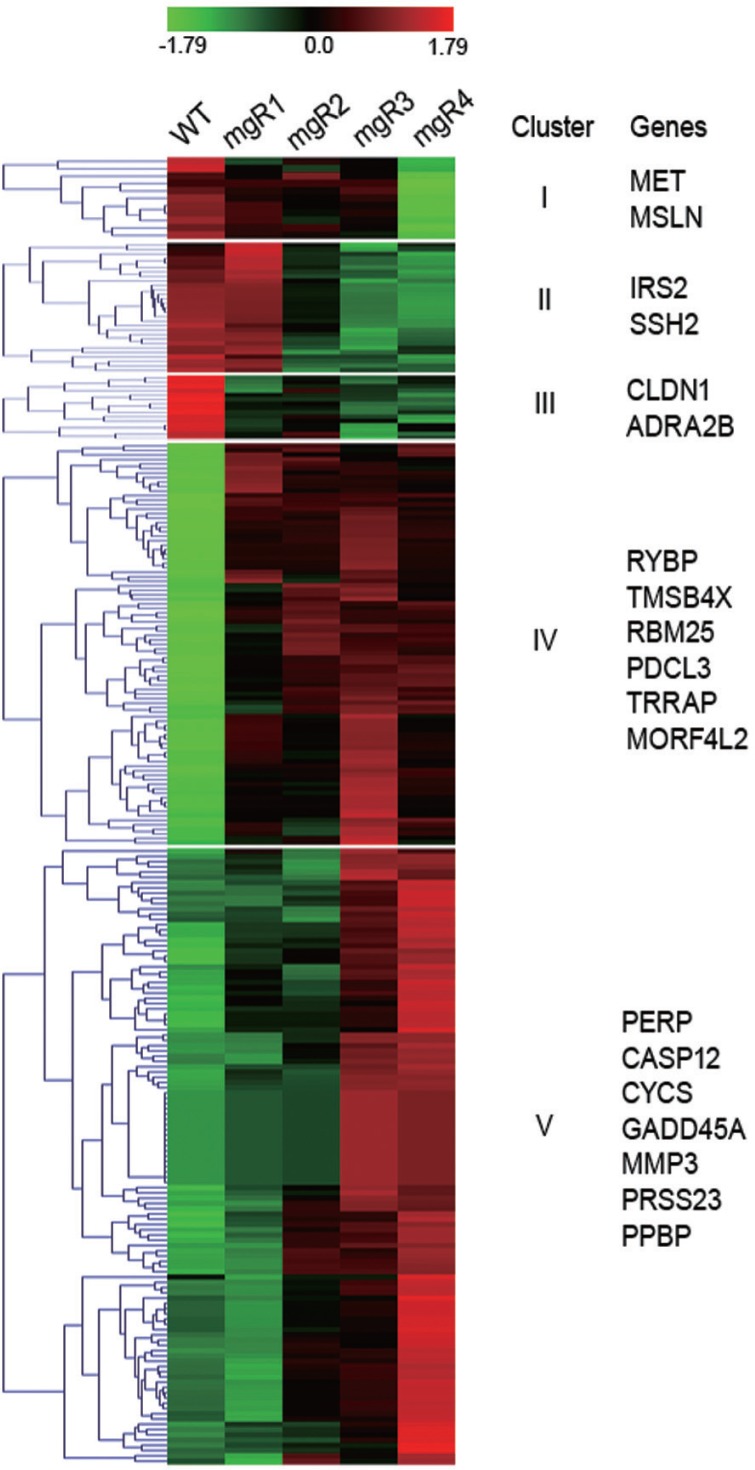
Five Clusters with different gene expression profiles during MFS progression
Since the expression of the genes related to MFS pathogenesis was likely to be persistently elevated or repressed during the disease progression, we grouped 1,137 DEGs into 16 clusters according to their expression patterns by means of K-mean clustering analysis. Out of the 16 clusters, we chose five clusters that represented progressive changes in gene expression as seen in the four mgR mice and listed several genes that were likely to be associated with MFS pathogenesis (Fig. 3). Genes in clusters I, II, and III were expressed in high levels in WT and/or mgR1 mice that showed only minor aortic pathogenesis, but were expressed only at low levels in mgR mice that showed severe pathological conditions in the aortic walls. As MFS progressed, genes involved in cell proliferation (MET, MSLN, IRS2), migration (SSH2), adhesion (CLDN1), and contraction (ADRA2B) were substantially downregulated in the aortic lesions. In contrast, cluster IV contained groups of genes that were expressed at low levels in WT, but were expressed at elevated levels in all mgR mice (1–4). Cluster V genes showed the lowest expression in WT mice, and their expression steadily increased as the disease progressed. Clusters IV and V contained genes involved in apoptosis (RYBP, RBM25, PDCL3, PERP, CASP12, GADD45A, CYCS), DNA damage response (TRRAP, MORF4L2), matrix degradation (PRSS23, MMP3), inflammation (PPBP), and actin polymerization (TMSB4X). Their upregulation likely reflect the aortic phenotype in MFS.
Fig. 3.
Five Clusters with different gene expression profiles during MFS progression. The 1,137 DEGs were clustered on the basis of the gene expression patterns in WT and four mgR mice. Five clusters with different gene expression profiles and the related genes selected from each cluster are shown. Gene expression level is shown in a color scale ranging from red (overexpression) to green (underexpression) according to the scale bar at the top of the heat map. The color scale is shown at the top of the heat map.
DISCUSSION
MFS is a monogenetic connective tissue disorder characterized by skeletal deformities, ocular lens dislocation, and cardiovascular complications such as aortic dilatation and dissection. The underlying molecular basis for MFS pathogenesis is poorly understood. To advance the understanding of molecular pathogenesis, three previous studied analyzed the genes differentially expressed in human MFS samples or in samples from murine MFS models. Yao et al. performed cDNA microarray analysis of gene expression in cultured skin fibroblasts derived from MFS patients and unaffected controls (Yao et al., 2007). However, large sample-to-sample variations in gene expression levels in the unaffected controls made it difficult to identify the genes differentially expressed in MFS. Radonic et al. performed cDNA microarray analysis of gene expression in skin biopsies of 55 MFS patients (Radonic et al., 2012). However, because the disease severity did not differ among the patients whose samples were analyzed, this study failed to identify a distinct gene expression pattern that marked the progression of MFS. Additionally, gene expression profiles of skin tissues may not be relevant to the phenotypic changes of aorta. Schwill et al. founded that genes related to immune system functions were differentially expressed in the aortic tissues of homozygous mgR mice. However, this study did not address the changes in gene expression patterns associated with MFS progression (Schwill et al., 2013). Therefore, using a murine model of MFS, we sought to characterize the genes differentially expressed at various stages during the course of MFS progression.
To this end, we employed four homozygous mgR mice that exhibited progressive severity in the fragmentation and organization of elastin fibers of the aortic walls. Comparison of the gene expression profiles of mgR and WT mice showed that transcripts of genes involved in myofibril assembly, actin cytoskeleton organization, and cell adhesion were highly enriched in the aortic tissues of mgR mice, suggesting that mgR mice recapitulated the aortic phenotype of MFS. Further analysis of DEG whose expression gradually changed with the severity of aortic pathology identified several genes whose known roles were suggestive of their involvement in MFS pathogenesis. As the disorganization of elastic fibers advanced, the expression of many genes was found substantially downregulated in the aortic walls. Among these genes, Met is a hepatocyte growth factor (HGF) receptor, and HGF/Met signaling has been shown to negatively regulate Angiotensin II that induces the premature senescence of vascular smooth muscle cells (VSMCs) (Kunieda et al., 2006; Sanada et al., 2009). Given the therapeutic effect of Angiotensin II blockers such as losartan on the aortic dilatation in MFS, reduced expression of Met in aortic walls is likely to be associated with aortic pathogenesis in MFS (Habashi et al., 2006). MSLN and IRS2 promote cell proliferation and survival by activating phosphatidylinositol 3-kinase/Akt signaling pathway (Bharadwaj et al., 2011; Gui et al., 2013). Downregulation of MSLN and IRS2 may render VSMCs more vulnerable to a variety of MFS-related vascular stresses including oxidative stress and shear stress. Reduced expression levels of genes such as ADRA2B that regulate vascular contraction was seen in the aortic tissues of all mgR mice. ADRA2B is one of the adrenergic receptor group members and plays a key role in the VSMC contraction. In contrast, genes involved in extracellular matrix degradation were substantially upregulated in aortic walls as MFS progressed in mgR mice. It has been reported that MMP3 is upregulated in the aortic lesion of MFS and promotes aortic dilatation and dissection (Palombo et al., 1999). PRSS23 is a novel serine protease that is expressed during the development of cardiovascular system and has been shown to be involved in vascular TGF-β signaling (Chen et al., 2013). Therefore, the upregulation of PRSS23 may be consistent with MFS pathogenesis in the aortic walls. Genes involved in apoptosis were also found to be upregulated in the aortic walls of mgR mice. Among these genes, PDCL3 and CASP12 are known to activate caspase pathway during apoptosis (Yoneda et al., 2001; Wilkinson et al., 2004). Rbm25 was shown to be involved in apoptosis and regulating the expression of proapoptotic factor BCL2L1 (Zhou et al., 2008). PERP and RYBP play key roles in TP53-dependent apoptotic pathway. PERP is the apoptosis-associated transcriptional target of TP53 and RYBP inhibits ubiquitination and subsequent degradation of TP53 (Attardi et al., 2000; Chen et al., 2009). The expression of GADD4A is known to increase under conditions of DNA damage or stressful growth arrest (Kettenhofen et al., 2001). These data suggest that VSMCs in the aortic lesions of mgR mice might be undergoing apoptosis under the conditions of DNA damage and other extracellular stresses. Indeed, genes such as TRRAP and MORF4L2 that are related with DNA damage responses were substantially upregulated in mgR mice. TRRAP is a component of histone acetyltransferase complex and regulates the repair of DNA double strand breaks (Murr et al., 2006). MORF4L2 is also a component of NuA4 histone acetyltransferase complex that participates in DNA damage responses and DNA repair (Squatrito et al., 2006).
Although further studies are required to define the precise roles of several identified candidate genes in the aortic pathogenesis, our data provide valuable information regarding the genes that could contribute to the development and progression of the aortic wall destruction seen in MFS patients. A complete understanding of the precise molecular mechanisms responsible for MFS pathogenesis and progression will contribute to the development of effective therapeutics and prevent patient morbidity and mortality.
Acknowledgments
This work was supported by Basic Science Research Program (2012-0003729) and the Bio & Medical Technology Development Program (2010-0020275) through NRF grant funded by the Korean government.
REFERENCES
- Attardi LD, Reczek EE, Cosmas C, Demicco EG, McCurrach ME, Lowe SW, Jacks T. PERP, an apoptosis-associated target of p53, is a novel member of the PMP-22/gas3 family. Genes Dev. 2000;14:704–718. [PMC free article] [PubMed] [Google Scholar]
- Bharadwaj U, Marin-Muller C, Li M, Chen C, Yao Q. Mesothelin confers pancreatic cancer cell resistance to TNF-alpha-induced apoptosis through Akt/PI3K/NF-kappaB activation and IL-6/Mcl-1 overexpression. Mol Cancer. 2011;10:106. doi: 10.1186/1476-4598-10-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Zhang J, Li M, Rayburn ER, Wang H, Zhang R. RYBP stabilizes p53 by modulating MDM2. EMBO Rep. 2009;10:166–172. doi: 10.1038/embor.2008.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen IH, Wang HH, Hsieh YS, Huang WC, Yeh HI, Chuang YJ. PRSS23 is essential for the Snail-dependent endothelial-to-mesenchymal transition during valvulogenesis in zebrafish. Cardiovasc Res. 2013;97:443–453. doi: 10.1093/cvr/cvs355. [DOI] [PubMed] [Google Scholar]
- Dietz HC, Cutting GR, Pyeritz RE, Maslen CL, Sakai LY, Corson GM, Puffenberger EG, Hamosh A, Nanthakumar EJ, Curristin SM, et al. Marfan syndrome caused by a recurrent de novo missense mutation in the fibrillin gene. Nature. 1991;352:337–339. doi: 10.1038/352337a0. [DOI] [PubMed] [Google Scholar]
- Gui S, Yuan G, Wang L, Zhou L, Xue Y, Yu Y, Zhang J, Zhang M, Yang Y, Wang DW. Wnt3a regulates proliferation, apoptosis and function of pancreatic NIT-1 beta cells via activation of IRS2/PI3K signaling. J Cell Biochem. 2013;114:1488–1497. doi: 10.1002/jcb.24490. [DOI] [PubMed] [Google Scholar]
- Habashi JP, Judge DP, Holm TM, Cohn RD, Loeys BL, Cooper TK, Myers L, Klein EC, Liu G, Calvi C, et al. Losartan, an AT1 antagonist, prevents aortic aneurysm in a mouse model of Marfan syndrome. Science. 2006;312:117–121. doi: 10.1126/science.1124287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettenhofen R, Hoppe J, Eberhard G, Seul C, Ko Y, Sachinidis A. Regulation of Gadd45a mRNA expression in vascular smooth muscle under growth and stress conditions. Cell Signal. 2001;13:787–799. doi: 10.1016/s0898-6568(01)00198-x. [DOI] [PubMed] [Google Scholar]
- Kim KL, Yang JH, Song SH, Kim JY, Jang SY, Kim JM, Kim JA, Sung KI, Kim YW, Suh YL, et al. Positive correlation between the dysregulation of transforming growth factor-beta1 and aneurysmal pathological changes in patients with Marfan syndrome. Circ J. 2013;77:952–958. doi: 10.1253/circj.cj-12-0874. [DOI] [PubMed] [Google Scholar]
- Kunieda T, Minamino T, Nishi J, Tateno K, Oyama T, Katsuno T, Miyauchi H, Orimo M, Okada S, Takamura M, et al. Angiotensin II induces premature senescence of vascular smooth muscle cells and accelerates the development of atherosclerosis via a p21-dependent pathway. Circulation. 2006;114:953–960. doi: 10.1161/CIRCULATIONAHA.106.626606. [DOI] [PubMed] [Google Scholar]
- Matt P, Schoenhoff F, Habashi J, Holm T, Van Erp C, Loch D, Carlson OD, Griswold BF, Fu Q, De Backer J, et al. Circulating transforming growth factor-beta in Marfan syndrome. Circulation. 2009;120:526–532. doi: 10.1161/CIRCULATIONAHA.108.841981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murr R, Loizou JI, Yang YG, Cuenin C, Li H, Wang ZQ, Herceg Z. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat Cell Biol. 2006;8:91–99. doi: 10.1038/ncb1343. [DOI] [PubMed] [Google Scholar]
- Neptune ER, Frischmeyer PA, Arking DE, Myers L, Bunton TE, Gayraud B, Ramirez F, Sakai LY, Dietz HC. Dysregulation of TGF-beta activation contributes to pathogenesis in Marfan syndrome. Nat Genet. 2003;33:407–411. doi: 10.1038/ng1116. [DOI] [PubMed] [Google Scholar]
- Palombo D, Maione M, Cifiello BI, Udini M, Maggio D, Lupo M. Matrix metalloproteinases. Their role in degenerative chronic diseases of abdominal aorta. J Cardiovasc Surg (Torino) 1999;40:257–260. [PubMed] [Google Scholar]
- Pereira L, Lee SY, Gayraud B, Andrikopoulos K, Shapiro SD, Bunton T, Biery NJ, Dietz HC, Sakai LY, Ramirez F. Pathogenetic sequence for aneurysm revealed in mice underexpressing fibrillin-1. Proc Natl Acad Sci USA. 1999;96:3819–3823. doi: 10.1073/pnas.96.7.3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyeritz RE. The Marfan syndrome. Annu Rev Med. 2000;51:481–510. doi: 10.1146/annurev.med.51.1.481. [DOI] [PubMed] [Google Scholar]
- Radonic T, de Witte P, Groenink M, de Waard V, Lutter R, van Eijk M, Jansen M, Timmermans J, Kempers M, Scholte AJ, et al. Inflammation aggravates disease severity in Marfan syndrome patients. PLoS One. 2012;7:e32963. doi: 10.1371/journal.pone.0032963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanada F, Taniyama Y, Iekushi K, Azuma J, Okayama K, Kusunoki H, Koibuchi N, Doi T, Aizawa Y, Morishita R. Negative action of hepatocyte growth factor/c-Met system on angiotensin II signaling via ligand-dependent epithelial growth factor receptor degradation mechanism in vascular smooth muscle cells. Circ Res. 2009;105:667–675. doi: 10.1161/CIRCRESAHA.109.202713. [DOI] [PubMed] [Google Scholar]
- Schwill S, Seppelt P, Grunhagen J, Ott CE, Jugold M, Ruhparwar A, Robinson PN, Karck M, Kallenbach K. The fibrillin-1 hypomorphic mgR/mgR murine model of Marfan syndrome shows severe elastolysis in all segments of the aorta. J Vasc Surg. 2013;57:1628–1636. doi: 10.1016/j.jvs.2012.10.007. [DOI] [PubMed] [Google Scholar]
- Squatrito M, Gorrini C, Amati B. Tip60 in DNA damage response and growth control: many tricks in one HAT. Trends Cell Biol. 2006;16:433–442. doi: 10.1016/j.tcb.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Wilkinson JC, Richter BW, Wilkinson AS, Burstein E, Rumble JM, Balliu B, Duckett CS. VIAF, a conserved inhibitor of apoptosis (IAP)-interacting factor that modulates caspase activation. J Biol Chem. 2004;279:51091–51099. doi: 10.1074/jbc.M409623200. [DOI] [PubMed] [Google Scholar]
- Yao Z, Jaeger JC, Ruzzo WL, Morale CZ, Emond M, Francke U, Milewicz DM, Schwartz SM, Mulvihill ER. A Marfan syndrome gene expression phenotype in cultured skin fibroblasts. BMC Genomics. 2007;8:319. doi: 10.1186/1471-2164-8-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda T, Imaizumi K, Oono K, Yui D, Gomi F, Katayama T, Tohyama M. Activation of caspase-12, an endoplastic reticulum (ER) resident caspase, through tumor necrosis factor receptor-associated factor 2-dependent mechanism in response to the ER stress. J Biol Chem. 2001;276:13935–13940. doi: 10.1074/jbc.M010677200. [DOI] [PubMed] [Google Scholar]
- Zhou A, Ou AC, Cho A, Benz EJ, Jr, Huang SC. Novel splicing factor RBM25 modulates Bcl-x pre-mRNA 5′ splice site selection. Mol Cell Biol. 2008;28:5924–5936. doi: 10.1128/MCB.00560-08. [DOI] [PMC free article] [PubMed] [Google Scholar]