Abstract
Rice is a staple food yet is a significant dietary source of inorganic arsenic, a class 1, nonthreshold carcinogen. Establishing the location and speciation of arsenic within the edible rice grain is essential for understanding the risk and for developing effective strategies to reduce grain arsenic concentrations. Conversely, selenium is an essential micronutrient and up to 1 billion people worldwide are selenium-deficient. Several studies have suggested that selenium supplementation can reduce the risk of some cancers, generating substantial interest in biofortifying rice. Knowledge of selenium location and speciation is important, because the anti-cancer effects of selenium depend on its speciation. Germanic acid is an arsenite/silicic acid analogue, and location of germanium may help elucidate the mechanisms of arsenite transport into grain. This review summarises recent discoveries in the location and speciation of arsenic, germanium, and selenium in rice grain using state-of-the-art mass spectrometry and synchrotron techniques, and illustrates both the importance of high-sensitivity and high-resolution techniques and the advantages of combining techniques in an integrated quantitative and spatial approach.
Keywords: Arsenic, Selenium, Germanium, Rice grain, Speciation, Location
Introduction
Rice, the staple food for over half the world's population [1], is a significant dietary source of inorganic arsenic, a class 1, nonthreshold carcinogen [2]. Understanding the location and speciation of arsenic within the edible rice grain is essential to understanding the risk presented by arsenic in rice [3] and to establishing effective strategies to reduce arsenic concentrations. Germanic acid is an arsenite/silicic acid analogue, and location of germanium may help elucidate mechanisms of arsenite transport into grain. Conversely, selenium is an essential micronutrient in which up to 1 billion people worldwide are deficient, causing a range of health disorders [4]. Several studies have suggested that selenium supplementation can reduce the risk of some cancers [5–9], although a recent review, which re-evaluated the findings of 55 epidemiological studies, concluded that there is currently no convincing evidence that selenium reduces cancer risk [10]. The authors observed that although the incidence of cancers such as bladder or prostate cancer had been found to be lower in people who had higher intakes, or blood levels, of selenium, other factors, for example overall better nutritional status or living conditions, could not be discounted [10]. Nevertheless, selenium is essential for humans and, in a recent global survey of the selenium content of rice, levels were too low in most samples to meet the nutritional requirements of populations depending on rice consumption for their dietary selenium intake [4]. Understanding the mechanisms of selenium accumulation in the filling rice grain would help improve the efficiency of biofortification programmes, and direct the breeding of high-selenium rice cultivars [11, 12]. Furthermore, selenium speciation is particularly important, because human health outcomes are determined by the levels of individual selenium compounds rather than total selenium [6–8]. Organic selenium compounds are better assimilated by the human body and the possible putative anticarcinogenic benefits are conferred by organic species, specifically selenomethionine (SeMet) and, more so, selenomethylcysteine (SeMeSeCys) [6–8].
Research into the location and speciation of arsenic and selenium, and the location of germanium, in rice grain has advanced rapidly in recent years, primarily because of the high sensitivity of inductively coupled plasma-mass spec-trometry (ICP–MS) coupled with high-performance liquid chromatography (HPLC), and the increasing availability of high-resolution bioimaging techniques, particularly laser ablation–ICP–MS (LA–ICP–MS), nanoscale secondary-ion mass spectrometry (nanoSIMS), and synchrotron techniques, including X-ray absorption spectroscopy (XAS), X-ray fluorescence (XRF) microscopy, and XRF microtomography. Synchrotron techniques have recently come to the forefront of these investigations, because the highly-penetrating power of hard X-rays enables investigation of both whole and sectioned grain specimens. The relatively high energy of the XRF emissions from germanium, arsenic, and selenium further improves these techniques, with in-situ measurements possible at high resolution and sensitivity. X-ray techniques can probe oxidation states and detailed atomic environments.
This review summarises the recent discoveries made using these techniques, highlighting the benefits of using the combined approach of high-throughput, high-sensitivity (HPLC–)ICP–MS analysis with high-resolution spatially resolved bioimaging.
Analysis of arsenic and selenium speciation in rice grain
Arsenic and selenium speciation using HPLC–ICP–MS
ICP–MS has revolutionised quantitative analysis of arsenic and selenium in rice grain. Efficient ionisation in the plasma results in high sensitivity enabling analysis even at the trace (μg L–1) levels found in uncontaminated rice grain, and the multi-element capability of the ICP–MS facilitates the identification of relationships with other elements in the grain [13, 14]. A number of studies have combined the good separation capabilities of HPLC with highly sensitive ICP–MS detection to identify and quantify arsenic species in mature rice grain, detecting arsenite, arsenate, and dimethylated arsenic (DMA) with, occasionally, trace amounts of monomethylated arsenic (MMA) [15–20]. The relatively high sample throughput of ICP–MS and HPLC–ICP–MS enables rapid comparative analysis of total arsenic and arsenic speciation across rice cultivars and field sites. For instance, it has been established that the proportion of inorganic-to-organic arsenic varies geographically and genotypically; rice in the USA contains proportionately more DMA and rice in Asia contains proportionately more inorganic arsenic [2, 17, 19–24]. Although a certified reference material (CRM) for arsenic species is still lacking, a range of studies have speciated arsenic in the rice flour CRM NIST 1568a, thereby establishing it as acceptable for comparative quality control. Anion-exchange chromatography is frequently selected for arsenic speciation because it enables good separation of these species. However, Kohlmeyer et al. [25] used ion-pair chromatography to simultaneously separate anionic, neutral, and cationic arsenic species and identified low levels of trimethylarsine oxide (TMAO) in one of their analysed rice samples, in addition to arsenite, arsenate, and DMA [25]. In a recent study, Hansen et al. [26] used cation-exchange chromatography for analysis of rice grain samples that had previously been analysed by anion-exchange chromatography and identified the presence of tetramethylarsonium ions, not previously documented in rice grain, accounting for up to 5.8% of total grain arsenic [26].
With regard to selenium, several studies have reported that species in rice grain are predominantly organic [27–32]. Speciation of mature polished rice grain by HPLC–ICP–MS by Sun et al. [29] revealed that selenium was present primarily as SeMet with lower levels of SeMeSeCys and SeCys. HPLC–ICP–MS analysis of grain selenium by Fang et al. [29] after foliar application of inorganic selenium showed that 87% of grain selenium was present as SeMet. Li et al. [31] determined that soil fertilization with selenate or selenite, and water management treatments, did not alter the percentage of selenium speciated as SeMet.
Although HPLC–ICP–MS has, undoubtedly, significantly advanced our understanding of arsenic and selenium speciation in rice grain, one major disadvantage is that sample preparation requires chemical extraction of the arsenic and/or selenium species into solution. The methods used should, ideally, retain species integrity (both in range and concentration) but the rapid interconversion of arsenite and arsenate mean that often, total “inorganic” arsenic, rather than arsenic and arsenate, is reported. The extraction process also tends to cause decomposition of thiol complexes and this has proven problematic in the study of arsenic speciation, because arsenite, which has high affinity for thiols, may indeed be complexed with thiols in the grain [33, 34]. The extraction process is also often unsuccessful in extracting all of the arsenic and/or selenium from the grain and, additionally, some species may be disproportionately retained on the chromatographic column.
Arsenic and selenium speciation by use of XANES
Recent advances have been made in arsenic and selenium speciation in rice grain by using X-ray absorption near edge spectroscopy (XANES; reviewed by, e.g., Lombi and Susini [35]). Linear combination fitting of the spectra, where XANES spectra of the samples are fitted to those of standards to calculate the relative proportion of each species present, can yield quantitative data although this is subject to limitations because it depends heavily on the choice of standards, which is essentially an a-priori decision, e.g. Lombi and Susini [35]. Furthermore, because XANES spectra are the result of a weighted sum of all the species present in a sample, only species present as at least 5–10% of the total can be ascertained. One of the key benefits of XANES is that there is no need for potentially damaging (and species altering) sample pre-treatment such as that required for HPLC–ICP–MS, even though the possibility of photoreduction during analysis should be considered. Rice grain does not even have to be oven dried before XANES analysis, so if the sample is stored properly speciation integrity can be preserved. “Bulk” XANES has been conducted on powdered bulked grain samples to examine average speciation in whole samples, whereas μXANES analysis of grain slices is a means of detailed in-situ speciation. Lombi et al. [36] advised combining μXANES with bulk XANES analysis of the sample, because although spatially resolved μXANES has been used on sectioned grain to speciate arsenic and selenium in pre-identified hotspots within the grain, speciation of just a few microns may not be truly representative of the region and/or sample of interest.
In the first study of in-situ speciation and location of arsenic in rice grain, Meharg et al. [3] conducted μXANES and HPLC–ICP–MS on whole grain and polished rice from Bangladesh, China, and the USA. For the HPLC–ICP–MS analysis, grain samples were bulked, powdered, and subjected to hot acid extraction which meant arsenite and arsenate could not be reported separately. Approximately 25% of the arsenic in the grain proved unextractable and could therefore not be accounted for in the analysis. The μXANES investigation was conducted on sliced grain, which was not subject to chemical extraction, and focussed on the outer parts of the grain, which XRF arsenic distribution maps had identified as containing the most arsenic. The μXANES technique identified a greater ratio of inorganic arsenic (mainly arsenite) to organic arsenic (DMA) than the HPLC–ICP–MS analysis which, the authors suggested, may be because the μXANES was concentrated on the external part of the grain, which is not representative of the grain as a whole, or because of the unextractable portion that could not be accounted for in the HPLC–ICP–MS analysis.
Lombi et al. [33] used XANES to analyse arsenic speciation in bulked samples of mature rice grain and rice grain fractions that had previously been analysed by Sun et al. [18] using acid extraction and HPLC–ICP–MS analysis. Linear combination fitting of the bulk XANES spectra revealed that arsenic was present primarily in the form of arsenite–thiol complexes in the rice bran and endosperm. Lombi et al [33] also reported that the percentage of DMA in the endosperm, identified to be 36% by HPLC–ICP–MS analysis [18], was calculated to be only 26%. Carey et al. [34] used XANES analysis of bulk grain to investigate arsenic speciation in the developing rice grain from excised panicles that had been supplied with a pulse of arsenate, arsenite, or DMA through the cut stem during grain fill. The spectra for DMA-fed grain initially appeared to have shifted toward arsenite; however, comparison with XANES spectra previously collected for a range of arsenic standards that included DMA thiol complexes, indicated that DMA was in fact partially thiol-complexed within the grain. This would explain the discrepancies identified by Meharg et al. [3] and Lombi et al. [33]. Had the DMA actually shifted to arsenite, no discrepancy would be present because the HPLC–ICP–MS analysis would also identify arsenite. This study also demonstrated that the redox status of inorganic arsenic seemed to be concentration-dependent; when arsenite was supplied to excised rice panicles at 13.3 μmol L–1, arsenite was oxidized to arsenate within the rice grain, yet under exposure to 133 μmol L–1 arsenite remained stable within the grain as free arsenite [34].
To examine selenium speciation in rice grain, Li et al. [31] conducted both XANES and HPLC–ICP–MS analysis on bulked grain samples. Despite good agreement in the identification of SeMet between the two techniques, bulk XANES identified a small percentage of SeMeSeCys (around 10%) that was not identified by HPLC–ICP–MS. The authors noted that this could be attributed to the selenium portion that was not isolated during the extraction process for HPLC–ICP–MS. Sun et al. [30] also identified low levels (6.9%) of SeMeSeCys in rice grain using HPLC–ICP–MS, despite low extraction efficiency of 62.8% (in both studies grain selenium was extracted into solution by enzymatic hydrolysis before HPLC–ICP–MS analysis with anion-exchange chromatography). Williams et al. [28] reported that XANES analysis of selenium speciation in the separate fractions of mature rice grain demonstrated that selenium in the bran layer was present as an approximately 50:50 mixture of inorganic (53%) and organic (47%), specifically SeMeSeCys, species, whereas, in the endo-sperm, conversely, only 5% of the Se was present as inorganic Se, and 95% was a mixture of SeMet and SeMeSeCys [28]. Subsequent re-analysis of this data, with all Se standards analysed concurrently, revealed that all the organic Se was actually present as SeMet [31].
There are clearly a number of potential issues with XANES speciation. The choice and range of standard compounds selected is critical and the absorption edges for some arsenic and selenium compounds may be very close or even identical. In addition, the high energy of the X-ray beam can potentially damage the sample. For example, Smith et al. [37] and Carey et al. [34] both reported photoreduction of arsenate to arsenite, observed by comparison of replicate spectra, during XANES analysis of rice grain. Nonetheless, the ability to avoid the extraction process, to analyse speciation in fresh unadulterated grain, and, furthermore, to do so in situ, has provided remarkable insights into grain arsenic and selenium speciation and this technique must be exploited further. Artefacts such as those caused by photoreduction can potentially be detected and avoided as long as the experimenters are aware of the possibility of damage occurring.
Location of arsenic, germanium, and selenium species in rice grain
Several studies have analysed arsenic distribution in rice grain by quantifying arsenic or selenium in separate grain fractions [18, 30, 38, 39]; all reported that arsenic was most concentrated in the bran, with levels following the pattern bran>wholegrain rice>polished rice. Sun et al. [30] analysed the distribution of selenium in rice plants grown in a selenium-rich environment and reported that levels in rice decreased in the order rice straw>bran>wholegrain>polished rice>husk, and that, within the grain, selenium was concentrated in the bran, with levels almost twice those of the polished grain. Although these studies have proved immensely useful, the manual or mechanical separation of rice grain fractions is relatively crude and subject to variation among cultivars and techniques (particularly in the extent of removal of the bran, which is important in location studies because the aleurone and sub-aleurone are only a couple of cells thick) and these studies were also unable to include the embryo. State-of-the-art in situ techniques avoid these problems, thereby enabling more reliable assessment of the distribution of an element, and have been shown to work well in tandem with ICP–MS and HPLC–ICP–MS.
Location of arsenic, germanium, and selenium in rice grain by use of synchrotron techniques
Synchrotron XRF microscopy uses a focussed beam (1–10 μm) of synchrotron radiation to excite the core electrons of elements within a sample, causing them to emit fluorescence [36]. The energy of the fluorescence emitted depends on the nuclear charge of an atom and is thus specific to an element [35]. Although absolute quantification is difficult (matrix matched standards, for example, will not be homogenous on the micron scale [40]), relative differences generate a distribution pattern, and these can be collected for multiple elements, enabling spatial relationships between elements to be identified. The technique benefits from high lateral resolution (a few microns) and from high sensitivity (in the mg kg–1 range). Furthermore, XRF can be combined with spatially resolved μXANES to generate speciation and structural information at specific points on the resulting 2D map.
Meharg et al. [3] conducted synchrotron XRF microscopy on mature rice grain from Bangladesh and the USA, bisected across their longitudinal axis, using step sizes between 5 and 25 μm depending on the size of the area mapped and the resolution required. In this study μXANES was also conducted to analyse speciation in situ in the areas identified as having high arsenic (see earlier). Arsenic was localized in the region of the ovular vascular trace (OVT) with slight dispersal around the pericarp/aleurone region and into the endosperm. The OVT is the vascular entry point into the rice grain and contains phloem and xylem cells. Minerals are transported into the nucellar tissue and then, via the apoplast, into the aleurone and the endosperm [41]. This was confirmed by LA–ICP–MS analysis for the Bangladeshi grain (see below). The extent of dispersal throughout the grain varied among samples; the Bangladeshi grain, in particular, had relatively little arsenic in the endosperm compared with the outer layer [3]. In the polished white USA grain sample analysed, arsenic seemed to be uniformly distributed throughout the endosperm [3]. The multielement capability of this technique enabled a number of other elements to be analysed and arsenic was shown to be coincident with manganese, iron, copper, and zinc (in agreement with Ren et al. [38] in their ICP–optical emission spectroscopic analysis of grain fractions) and not with nickel or cadmium. A potential problem with XRF microscopy is that the fluorescence detected at each x,y point represents the entire element in the path of the beam and is therefore, essentially, a 3D image compressed into a 2D image. The grain should therefore be sliced thinly and precisely, because uneven thickness could lead to false hotspots, as could areas of higher density. This was not the case in the work by Meharg et al. [3], which used fractured grains. To reduce this problem, a later study by Lombi et al. [32] developed a technique to prepare thin (70 μm), evenly sliced grain sections. The analysis used spot sizes of 5–10 μm and steps sizes of 5–25 μm as in Meharg et al. [3]. Sections of a Chinese grain that was particularly high in arsenic were prepared and μXANES was performed to assess in-situ speciation where levels were high enough. The results showed that arsenic was significantly localized in the OVT region, with limited diffusion throughout the pericarp region, in agreement with Meharg et al. [3], together with significant arsenic accumulation in the embryo, which μXANES identified as arsenite [33]. This study was complemented by bulk XANES analysis and HPLC–ICP–MS analysis of the husk, bran, and endosperm grain sections. Bulk XANES showed that arsenic was predominantly present as arsenite–thiol groups in the bran and endosperm (which would not survive extraction for HPLC–ICP–MS).
In a recent study, Seyfferth et al [42] conducted XRF microscopy on a thin longitudinal slice of a rice grain (embedded in epoxy resin and sliced to 30 μm). Using a beam of 2 μm×2 μm and a pixel size of 5 μm, the authors observed that arsenic was localized in the bran layer, in agreement with previous studies, with little present in the endosperm. The authors also identified an arsenic hotspot in the embryo, concurrent with Fe and Ca, which they were able to speciate by use of in-situ μXANES. This revealed that arsenic within the embryo was primarily speciated as arsenate (80%) with only 20% present as arsenite, in contrast with the findings of Lombi et al., above [32, 42]. The authors also conducted μXANES on two spots within the bran layer and reported that arsenic was not present as arsenite, or arsenite–thiols, as previously reported but was speciated as arsenate (69–88%) with smaller amounts of DMA (12–31%) [42]. Interestingly, this study also used μXANES to locate arsenic species within a cross section of the rice root and reported that arsenic speciation with the root xylem was predominantly arsenate (86%), with lesser amounts of DMA (14%), and that arsenite and arsenite glutathione were located in a vacuole next to the xylem where they represented 71% and 29%, respectively, of the arsenic present [42].
Zheng et al. [43] used synchrotron XRF microscopy to map the distribution of arsenic in rice grain at different stages throughout its development, in an investigation of the spatial distribution of arsenic and temporal variation of its concentration in rice. They used a lower resolution than Meharg et al. [3] and Lombi et al. [33], with a spot size of 120 μm and step sizes from 100 to 120 μm, but clearly showed that arsenic distribution within the grain is time-dependent. Before anthesis, arsenic was present in the ovary but moved to the base of the caryopsis as the ovary developed. Fourteen days after flowering, arsenic was highest in the centre of the grain; when grain filling was complete arsenic seemed to be localized in the OVT region [43]. This study also included XRF imaging of the leaf, internode, and node of the rice plant, demonstrating that arsenic was localized in the veins, primarily the main veins, of the leaf, in the vascular system of the stem, and in the centre of the node below the peduncle. ICP–MS and HPLC–ICP–MS analysis revealed temporal variations in arsenic concentrations in rice grain (and other tissues) at different stages of growth with the concentration of DMA decreasing during grain filling whereas inorganic arsenic concentrations remained constant [43]. The authors suggested that DMA accumulation in rice grain occurred solely via the re-translocation of DMA that had accumulated in the rice plant before flowering, whereas inorganic arsenic that accumulated in the grain was derived from direct uptake and transport during flowering.
To overcome the challenges of producing thin, evenly sliced sections of fresh rice grain for XRF analysis and the ensuing ambiguities present in 2D images that compress 3D information, Carey et al. [34, 44] used synchrotron XRF microtomography to investigate the spatial unloading of arsenic and selenium species in developing rice grains. Synchrotron XRF microtomography is based on the same principles as XRF microscopy, exciting the atoms in a sample with the synchrotron X-ray beam and detecting the fluorescence emitted, which is specific for each element. However, the microtomography set-up utilizes a sample stage that translates and rotates the sample through the path of the beam, with fluorescence recorded at each point to generate a virtual slice. The fresh developing rice grains required no pre-treatment, and were simply dehusked manually before analysis [34]. The dehusked grain was then suspended vertically, positioned in such a way that the embryo was included in the scan, from the rotational/translational sample stage, and passed through the high intensity X-ray beam, scanned, then rotated by a degree, translated across the x-axis and scanned again. This process was repeated through 180° and the elemental fluorescence detected for each angle and position used to produce a sinogram which was then reconstructed into an x,y map, creating a distribution map for the virtual slice of the grain that included both the OVT and the embryo. The low water content of grains makes them ideal for this type of analysis, because hydrated tissues usually suffer from beam damage [36, 45]. The main limitation of this technique for rice grain analysis is that it is very time-consuming, limiting the number of samples that can be analysed and generally prohibiting analysis of replicates or different genotypes. Furthermore, the sensitivity is often not high enough to measure background arsenic and/or selenium and the resolution is insufficient to enable analysis on the cellular scale. Synchrotron XRF microtomography was conducted on immature rice grains pulsed with a known species of arsenic, specifically arsenite or DMA, via the excised panicle stem during grain fill. The tomography images collected showed that arsenite remained localised in the OVT region (along with iron and manganese, as in previous studies of mature grain) whereas DMA rapidly spread throughout the outer layers and into the endosperm [34]. These findings explain why inorganic arsenic is concentrated in rice bran whereas the endosperm contains higher levels of DMA. Carey et al. [34] also reported that phloem interruption, by girdling the stem with a jet of steam, did not affect the spatial unloading of either arsenite or DMA, suggesting that although the relative mobility of arsenic species in the phloem and xylem pathways, and how quickly they are transported into the phloem, may be important in the rate of translocation to the grain, these factors may not be important with regard to mobility within the grain. Quantitative ICP–MS analysis of grain revealed that DMA was translocated from the shoot to the grain with over an order of magnitude greater efficiency than inorganic arsenic species and that phloem transport accounted for 90% of arsenite and 55% of DMA transported into grain [34].
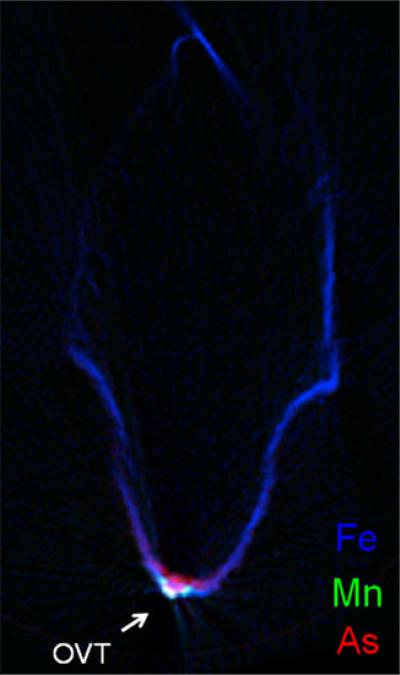
Synchrotron XRF microtomography was later used in a study of the re-translocation of arsenic species from the flag leaf into the filling rice grain, where rice plants were supplied with arsenite, arsenate, MMA, or DMA via the cut flag leaf of intact plants during the period of grain fill [44]. Distribution maps were collected for the dehusked fresh grain for each treatment and compared with those collected for grain exposed to the same arsenic species via the excised panicle stem. The tomography images revealed that MMA, arsenate, and arsenite were all localized in the region of the OVT whereas DMA was extremely mobile within the grain, whether arriving via direct stem-to-grain transport or via re-translocation from flag leaves. ICP–MS analysis revealed that re-translocation from the flag leaf into the filling grain was extremely efficient for DMA and MMA but was relatively poor when flag leaves were exposed to arsenate, and not detectable at all when leaves were fed arsenite. HPLC–ICP–MS analysis of the fresh flag leaves fed either arsenate or arsenite (ground in liquid nitrogen, extracted with chilled buffer solution, and analysed immediately) found that arsenate was rapidly reduced to arsenite within the flag leaf. This study also used 3D XRF microtomography of arsenite-exposed grain, fed via the excised stem (because there was no measurable retranslocation of arsenite from flag leaves) to confirm the location of arsenite within the OVT. This was achieved by collecting 20 XRF tomography sections along the length of the grain from the awn to the rachilla, at intervals of 240 μm, and clearly shows arsenite localized within the OVT, but starting to spread beyond the confines of the OVT, and a 2D scan clearly illustrated that it had, to some extent, spread into the nucellar epidermis [44]. The 2D scans for arsenic, iron, and manganese are overlain in Fig. 1. Iron and manganese, which were clearly localized with arsenic in the OVT, as reported in previous studies [3, 33, 34, 37], appeared to be located to either side of the main concentration of arsenic and had different mobility (iron had also spread throughout the nucellar epidermis, much more so than arsenic, whereas manganese was restricted to the OVT) [44]. Greater spatial resolution is needed to investigate these observations further.
Fig. 1.
Synchrotron X-ray fluorescence microtomography images illustrating the different distributions of arsenic (As; red), manganese (Mn; green), and iron (Fe; blue) for a virtual cross-section of a dehusked immature rice grain pulsed with 133 μmol L–1 arsenite through the excised panicle stem. OVT refers to the ovular vascular trace, the point of grain entry
Zheng et al. [43] suggested that DMA in the rice grain derived solely from the re-translocation of DMA that had accumulated in the rice plant before flowering, whereas inorganic arsenic was primarily transported to the grain during flowering. Carey et al. [44] demonstrated that DMA is actively retranslocated from flag leaves into the filling rice grain, in contrast with arsenite for which measurable re-translocation was not observed. The authors suggested that this rapid translocation of DMA from flag leaves will cause any DMA accumulated within the plant before panicle development to swiftly partition into the panicle and ovary, leading to a high proportion of DMA in the initial stages of grain fill which will then be diluted by the carbohydrate filling the grain, and, because the initial plant uptake via rice roots is so slow for DMA [46], the amount of DMA taken into the plant during grain fill will be comparatively low [44]. Both studies agreed that inorganic arsenic in the rice grain is derived largely from direct shoot-to-grain trans-location occurring during grain fill and not from vegetative stores, and this has significant implications for arsenic mitigation. However, it should be noted that a recent study did identify some arsenite retranslocation from the flag leaves to the filling rice grain by use of a radioactive tracer, and this should clearly be investigated further [47].
The techniques used by Carey et al. [34, 44] were also applied to selenium (Carey et al. 2011 submitted) to investigate the translocation and spatial unloading into the filling rice grain of the main selenium species that had been identified in the mature rice grain in previous speciation studies. Selenomethionine (SeMet), selenomethylcysteine (SeMeS-eCys), selenite, and selenate were supplied via cut flag leaves of intact plants, and via excised panicle stems subjected to a±stem-girdling treatment, during the grain-fill period. Quantitative ICP–MS analysis of the grain and flag leaves revealed that SeMet and SeMeSeCys were rapidly transported into the phloem whereas inorganic species were transported to the grain in both phloem and xylem, and were translocated to the filling grain far more efficiently than inorganic species whether from flag leaf stores or direct stem-to-grain transport (Carey et al. 2011 submitted). Synchrotron XRF microtomography images illustrated that spatial unloading was significantly different for selenite, SeMet, and SeMeSeCys-fed grain (time constraints prevented imaging of selenate-treated grain). In SeMet and SeMeSeCys-fed grain, selenium rapidly diffused throughout the external layers and into the endosperm and, for SeMeS-eCys, had also spread into the embryo, whereas for selenite fed grain selenium was retained in the OVT (Carey et al. 2011 submitted). As observed for arsenic species, these species-specific patterns were sustained whether arriving from fed leaves or from direct translocation through the stem.
Williams et al. [28] conducted a synchrotron XRF microscopy analysis of the selenium distribution in a mature rice grain slice and demonstrated that selenium was present throughout the endosperm but was particularly concentrated in the OVT region, the pericarp/aleurone layer, and the embryo.
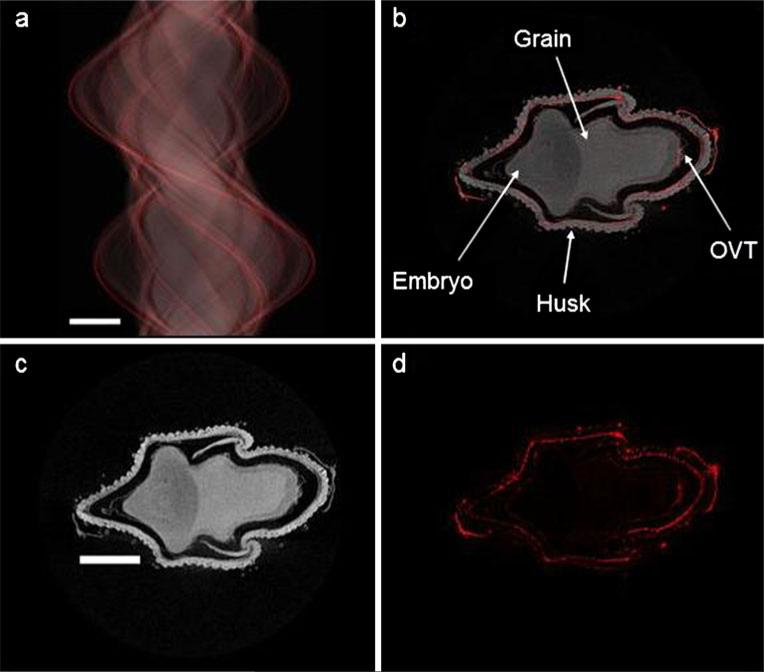
Recent advances in XRF detector technology have significantly improved the speed (in pixels per second) at which such detectors can operate [48–51]. The Maia detector system developed by BNL and CSIRO, and in use at the Australian synchrotron, uses a novel annular geometry to increase the solid angle of the detector, and thus its detection efficiency. The combination of high speed and sensitivity enables the collection of XRF images at extremely high definition, or, in XRF microtomography, quicker measurements [52] or higher resolution through finer sampling. Figure 2 shows the distribution of germanium in a fresh developing husked rice grain, exposed to germanic acid via the excised panicle stem during grain fill (as in Carey et al. [44]), recently imaged at the XFM beamline of the Australian synchrotron [53] using the Maia detector system [49–51]. X-ray fluorescence was acquired with the sample stage continually moving laterally (“on the fly”) with an effective dwell time of 1.95 ms per 2-micron pixel. As with previous synchrotron XRF microtomography, no sample preparation was required; however, fluorescence data were collected for 2000 rotations over 360 degrees (rather than the conventional 100–200 rotations over 180 degrees), generating a much higher-resolution image. The reconstruction resolution is conservatively estimated at better than approximately five microns. Data were acquired over the full 360 degrees, because this renders the reconstruction less susceptible to self absorption artefacts and enables their identification [54]; however, in this case a small motion of the specimen prevented us from using the entire measured sinogram in the reconstruction. The scan took just under 3 h to complete.
Fig. 2.
Synchrotron X-ray fluorescence microtomography images for a virtual cross section of a husked immature rice grain pulsed with 133 μmol L–1 germanic acid through the excised panicle stem. The sinogram (a) was reconstructed to generate maps of the grain density (Compton signal, c) and germanium distribution (d). Grain density and germanium distribution maps are overlain in (b). Bars are 600 μm. OVT refers to the ovular vascular trace, the point of grain entry
In addition to the germanium map, this figure also shows a reconstruction of the Compton scatter image. Compton scattering is strongly biased toward the lighter elements in a system, and so this image quite clearly maps the grain's ultrastructure. The ~30 μm diameter hairs on the outside of the husk can be identified and are clearly hollow. The fluorescence data for germanium (Fig. 2) shows that germanium is present throughout the husk, and in the husk hairs, with high concentrations in the rachilla, and is localized in the OVT of the edible grain. As an analogue for arsenite and silicic acid [55, 56] this distribution in the husk and husk hairs is to be expected, as is the lack of mobility within the edible grain. Localization in the OVT matches the distribution of arsenite. Arsenite enters the rice plant through the silicic acid pathway [57, 58], but how silicic acid transporters are involved in arsenite transport into the grain remains unknown. Norton et al. [59] found no correlation between shoot silicon and grain arsenic levels and a recent competition experiment found that germanic acid had no significant effect on shoot-to-grain translocation of arsenite [44]. The fact that germanium and arsenite have similar location patterns within the grain could suggest that silicic acid transporters are involved in the transport of arsenite into the rice grain. This higher resolution XRF microtomography can now be used to generate clearer images of the distribution of arsenic and/or selenium species.
Location of arsenic and selenium in grain by use of mass spectrometry
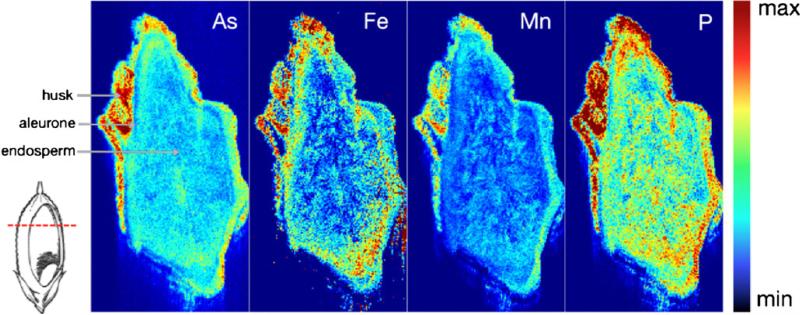
Despite its lower spatial resolution than the synchrotron XRF microscopy analysis conducted in the same study, Meharg et al. [3] conducted LA–ICP–MS on a cross section of a mature wholegrain of rice to confirm their interpretation of the 2D XRF images of arsenic distribution. In LA–ICP– MS, areas on the surface of the grain are vaporized by a focussed laser beam and the volatilised material is transported via a carrier gas into the connected ICP–MS [36, 60]. Sample pre-treatment is minimal; in Meharg et al. [3] the rice grain was embedded in resin, sectioned and polished, before placement on the x,y,z translation stage for analysis. The sensitivity of this technique is very high; with the ICP–MS as the detector, a wide range of elements, including arsenic and selenium, can be measured at trace levels. The relative concentrations between ablated areas can be determined and raster scanning enables the distribution of an element to be mapped [3]. Spot resolution ranges from 5 to 400 μm with the overall resolution varying according to how many spots are analysed [36, 60]. Meharg et al. [3] used a spot size of 200 μm whereas in Fig. 3 the resolution was 12 μm, generating a significantly more detailed distribution map. Here, immature rice grain was leaf-fed 333 μmol L–1 DMA, following procedures described by Carey et al. [44], and hand-sectioned to approximately 250 μm immediately before analysis. With the nanosecond laser; specifically a New Wave UP213 laser (Fremont, CA, USA), set to 70% power with a repeat rate of 10 Hz, one section of rice grain took 4.5 h to image. The accuracy of LA–ICP–MS depends on the specific spots ablated representing the region/sample of interest, as in the microprobe synchrotron techniques of μXANES and XRF microscopy, and it is often regarded as a semi-quantitative technique because of the problem of element fractionation (with enhanced signal intensities exhibited by some elements compared with less volatile elements), although reference to matrix-matched standards can overcome this problem [60, 61]. For example, Jackson et al. [61] found good correlation between arsenic and selenium in environmental samples (tail clips of the banded water snake, Nerodia fasciata) measured with LA–ICP–MS compared with levels measured in their acid digested counterparts via ICP–MS. In a recent review, Husted et al. [14] commented that as resolution increases, the lower cost and greater accessibility of LA–ICP–MS compared with synchrotron techniques mean that it is likely to be increasingly used in spatial studies of rice (and other) grain [14].
Fig. 3.
Laser ablation–ICP–MS images illustrating the distributions of arsenic (As), iron (Fe), manganese (Mn), and phosphorus (P) in transverse slices (250 μm) of a husked immature rice grain leaf-fed 333 μmol L–1 DMA during grain fill
Nanoscale secondary-ion mass spectrometry (Nano-SIMS) is another mass spectrometry technique that has been used effectively in the spatial analysis of arsenic and selenium in grain. It uses a focussed primary ion beam followed by detection of secondary ions and, although it is not as sensitive as LA–ICP–MS, its resolution is significantly higher [14, 36]. Examples of Nanosims images are given by Moore et al. in this issue. In a NanoSIMS study of arsenic in rice grain and selenium in wheat grain the lateral resolution was <100 nm [62]. Whole grains of rice and wheat were experimentally contaminated with arsenic and selenium, respectively, to ensure detectable levels for analysis [62]. As with synchrotron XRF, damaging chemical pretreatment was unnecessary for the mature whole grain, although NanoSIMS does require that the sample be completely moisture-free and moisture removal could destroy location patterns in highly hydrated specimens (details of sample preparation are given by Moore et al. in this issue). In Moore et al. [62] the low moisture content of the mature grains meant that storing grains under vacuum for one week before analysis to remove water absorbed during storage was sufficient. Each grain was then mounted using resin on to steel rings and sliced transversely with a razor blade to yield a flat surface for analysis. Analysis of a rice grain prepared using high-pressure freezing, an ultra-rapid cryo-fixation technique, gave similar results, confirming that the sample preparation had not introduced any artefacts. With such high resolution, NanoSIMS was able to reveal arsenic localization in hotspots within the OVT region of the rice grain that could not be identified by the larger step sizes of LA–ICP–MS and XRF microtomography, and in the protein matrix of the subaleurone layer that surrounds the endo-sperm (for a rice grain containing ca 30% inorganic As and 70% organic As). NanoSIMS is not effective for scanning large areas, because it is very time consuming and Moore et al. [62] therefore used synchrotron XRF micro-tomography to identify areas with high concentrations (using a beam focussed to 2 μm and step size of 15 μm) and then targeted those regions for high resolution NanoSIMS analysis, thus combining both techniques to capitalize on their respective strengths (and overcome their weaknesses).
Effective bioimaging presents a number of challenges. For example, despite the increasing availability of synchrotron techniques, time constraints have prohibited analysis of the large sample numbers necessary for comparative genomic studies and, with detection limits of the order of mg kg–1, clear synchrotron XRF maps of arsenic and selenium are often only achievable for contaminated grain. Furthermore, resolution of a few microns, while effective in identifying arsenic and/or selenium distribution in grain tissues, does not allow location on the cellular scale [40, 62] and this is necessary if we are to identify spatial differences in arsenic and/or selenium unloading among genotypes etc. The dual approach of Moore et al. [62] of targeting Nano-SIMS at areas of interest identified with the lower resolution, but faster, synchrotron XRF, is an advantageous one that should be adopted to investigate the embryo.
The future
The combination of ICP–MS and HPLC–ICP–MS with state-of-the-art in-situ techniques has significantly advanced our knowledge of the location and speciation of arsenic and selenium in rice grain. In situ bioimaging has revealed significant differences in localization patterns of arsenic and selenium species, suggesting markedly different mobility within the grain. Lombi et al. [33] suggested that transport of material from the maternal to the filial tissues of the rice grain could be a physiological barrier which arsenic species cross with different efficiency, and the findings of Carey et al. [34, 44] support this suggestion. Furthermore, the same may be true for selenium (Carey et al., 2011 submitted) and this could be exploited to control entry of arsenic and selenium into the commonly consumed polished grain (to further facilitate entry of SeMet and SeMeSeCys into the endosperm and to restrict inorganic arsenic to the OVT). The ultimate objective must be to identify the genes responsible for the mechanisms that enable arsenic/selenium species to accumulate, and to disperse, within the edible rice grain, so that these can be manipulated by targeted breeding programmes. However, we must first identify those mechanisms because, currently, substantial gaps in our understanding remain. Spatial studies must be extended to the whole plant, to monitor, in real time, from the point of initial plant uptake through to grain release, at different stages during the plants’ development, and to identify genotypic differences. High-resolution time-course synchrotron XRF tomography studies, coupled with quantitative ICP–MS analysis, are also required to definitively establish the translocation pathways of arsenic species during vegetative and reproductive growth, and to identify the critical time points/periods during plant and/or grain development must be a focus of future work. Achieving this will require bioimaging techniques with faster detection, higher sensitivity, and greater lateral resolution. Nevertheless, recent, ongoing advances in synchrotron fluorescence detector technology (e.g. the Maia detector at the Australian synchrotron) make this a very real possibility [63] as illustrated in the tomography image of germanium distribution in rice grain in Fig. 2 and, for example, in Lombi et al. [52] where fast detection has enabled successful tomography of metal distributions in hydrated plant roots. Spatial resolution of the order of nanometres will increase accuracy and enable analysis on the cellular scale [40]. Although the lower cost and greater accessibility of LA–ICP–MS compared with synchrotron techniques mean that it is likely to become increasing used in spatial studies of rice (and other) grain [14], the promising advances in synchrotron detectors mean that fast 3D XRF microtomography of fresh rice tissues on the cellular scale is likely to be possible in the very near future.
Acknowledgements
This work was supported by a Biotechnology and Biological Sciences Research Council Doctoral Training Grant. Part of this research was undertaken on the X-ray fluorescence microscopy beamline at the Australian synchrotron, Victoria, Australia. The authors thank Drs David Paterson and Daryl Howard for their help in collecting the germanium microtomography images.
Glossary of terms
- Aleurone
The aleurone cells house hormones and enzymes instrumental in the germination and development of the grain and are rich in phytic acid. Phytic acid, or phytate, a salt of phytic acid, is the main grain store of phosphorus and is known to chelate cations including those of many micronutrients, for example iron, zinc, and calcium. The aleurone is understood to specifically accumulate phytate and metals for germination and growth of the rice seed.
- Anthesis
Anthesis refers to the period of flowering. The point of anthesis is the point at which the anthers have emerged, demonstrating that the flower has reached the reproductive stage.
- Bran
The bran comprises the pericarp, testa, and aleurone cells and is rich in micro and macronutrients, B vitamins, protein, and fibre.
- Caryopsis
The caryopsis is the whole grain (not including the husk/hull).
- Embryo
The embryo, or germ, is the point at which the new plant develops and contains abundant proteins, lipids, and vitamins. In the processing of rice grain for consumption, the bran and embryo are removed together, leaving the sub-aleurone and endosperm to make up the polished (white) rice that is commonly consumed.
- Endosperm
The inner starchy part of a grain which serves as food reserves for the seed's development. In contrast with the embryo and bran layer, the endosperm contains relatively less protein, lipids, and nutrients and is mainly composed of starch and non-starch polysaccharides.
- Husk
The husk, or hull, is the outermost layer of a rice grain, a silica-rich envelope that protects against predation and disease.
- Internode
The node is the section of stem between two nodes.
- Node
Nodes are joints along the plant stem at which leaves and buds develop. They are also the points at which vascular contents are transferred to leaf and/or panicle.
- Nucellar
The nucellar tissue is the tissue that initially tissue surrounds the embryo. As the starchy endosperm develops and expands, the nucellar tissue becomes stretched, and is eventually crushed in the mature grain.
- Panicle
The panicle is the flower head of the rice plant. Technically, a panicle is an inflorescence in which each flower is on its own stalk.
- Peduncle
The peduncle is the panicle stem, i.e. the stem supporting the inflorescence (group of flowers).
- Pericarp
Pericarp is the ripened ovary wall. In the rice grain this thin, fibrous layer offers protection against moulds and oxidation.
Footnotes
Published in the special paper collection Elemental Imaging and Speciation in Plant Science with guest editors J. Feldmann and E. Krupp.
Contributor Information
Anne-Marie Carey, Institute of Biological and Environmental Sciences, University of Aberdeen, Cruickshank Building, St Machar Drive, Aberdeen AB24 3UU, UK.
Enzo Lombi, Centre for Environmental Risk Assessment and Remediation, University of South Australia, Building X, Mawson Lakes Campus, Mawson Lakes, South Australia 5095, Australia.
Erica Donner, Centre for Environmental Risk Assessment and Remediation, University of South Australia, Building X, Mawson Lakes Campus, Mawson Lakes, South Australia 5095, Australia.
Martin D. de Jonge, Australian Synchrotron, X-ray Fluorescence Microscopy, 800 Blackburn Road, Clayton, Victoria 3168, Australia
Tracy Punshon, Department of Biological Sciences, Dartmouth College, Hanover, NH 03755, USA.
Brian P. Jackson, Department of Biological Sciences, Dartmouth College, Hanover, NH 03755, USA
Mary Lou Guerinot, Department of Biological Sciences, Dartmouth College, Hanover, NH 03755, USA.
Adam H. Price, Institute of Biological and Environmental Sciences, University of Aberdeen, Cruickshank Building, St Machar Drive, Aberdeen AB24 3UU, UK
Andrew A. Meharg, Institute of Biological and Environmental Sciences, University of Aberdeen, Cruickshank Building, St Machar Drive, Aberdeen AB24 3UU, UK
References
- 1.Fageria NK. J Plant Nutr. 2007;30:843. [Google Scholar]
- 2.Meharg AA, Williams PN, Adomako E, Lawgali YY, Deacon C, Villada A, Cambell RCJ, Sun G, Zhu YG, Feldmann J, Raab A, Zhao F, Islam R, Hossain S, Yanai J. Environ Sci Technol. 2009;43:1612–1617. doi: 10.1021/es802612a. [DOI] [PubMed] [Google Scholar]
- 3.Meharg AA, Sun G, Williams PN, Adomako E, Deacon C, Zhu YG, Feldmann J, Raab A. Environ Pollut. 2008;152:746–749. doi: 10.1016/j.envpol.2008.01.043. [DOI] [PubMed] [Google Scholar]
- 4.Combs GF., Jr Br J Nutr. 2001;85:517. doi: 10.1079/bjn2000280. [DOI] [PubMed] [Google Scholar]
- 5.Clark LC, Combs GF, Jr, Turnbull BW, Slate EH, Chalker DK, Chow J, Davis LS, Glover RA, Graham GF, Gross EG, Krongrad A, Lesher JL, Jr, Park HK, Sanders BB, Jr, Smith CL, Taylor JR. J Am Med Assoc. 1996;276:1957–1963. [PubMed] [Google Scholar]
- 6.Ellis DR, Salt DE. Curr Opin Plant Biol. 2003;6:273–279. doi: 10.1016/s1369-5266(03)00030-x. [DOI] [PubMed] [Google Scholar]
- 7.Whanger PD. Br J Nutr. 2004;91:11. doi: 10.1079/bjn20031015. [DOI] [PubMed] [Google Scholar]
- 8.Rayman MP, Infante HG, Sargent M. Br J Nutr. 2008;100:238. doi: 10.1017/S0007114508922522. [DOI] [PubMed] [Google Scholar]
- 9.Tsubura A, Lai Y, Kuwata M, Uehara N, Yoshizawa K. Anti-Cancer Agent ME. 2011;11:249–253. doi: 10.2174/187152011795347441. [DOI] [PubMed] [Google Scholar]
- 10.Dennert G, Zwahlen M, Brinkman M, Vinceti M, Zeegers MP A, Horneber M. Cochrane Database of Systematic Reviews. 2011;(5) doi: 10.1002/14651858.CD005195.pub2. Art. No.: CD005195. doi: 10.1002/14651858.CD005195.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu YG, Pilon-Smits EAH, Zhao FJ, Williams PN, Meharg AA. Trends Plant Sci. 2009;14:436–442. doi: 10.1016/j.tplants.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 12.Zhao FJ, McGrath SP. Curr Opin Plant Biol. 2009;12:373–380. doi: 10.1016/j.pbi.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 13.Feldmann J, Salaün P, Lombi E. Environ Chem. 2009;6:275–289. [Google Scholar]
- 14.Husted S, Persson DP, Laursen KH, Hansen TH, Pedas P, Schiller M, Hegelund JN, Schjoerring JK. J Anal At Spectrom. 2011;26:52–79. [Google Scholar]
- 15.Schoof RA, Yost LJ, Eickhoff J, Crecelius EA, Cragin DW, Meacher DM, Menzel DB. Food Chem Toxicol. 1999;37:839–846. doi: 10.1016/s0278-6915(99)00073-3. [DOI] [PubMed] [Google Scholar]
- 16.Heitkemper DT, Vela NP, Stewart KR, Westphal CS. J Anal At Spectrom. 2001;16:299–306. [Google Scholar]
- 17.Williams PN, Price AH, Raab A, Hossain SA, Feldmann J, Meharg AA. Environ Sci Technol. 2005;39:5531–5540. doi: 10.1021/es0502324. [DOI] [PubMed] [Google Scholar]
- 18.Sun GX, Williams PN, Carey AM, Zhu YG, Deacon C, Raab A, Feldmann J, Islam RM, Meharg AA. Environ Sci Technol. 2008;42:7542–7546. doi: 10.1021/es801238p. [DOI] [PubMed] [Google Scholar]
- 19.Norton GJ, Islam MR, Deacon CM, Zhao F, Stroud JL, Mcgrath SP, Islam S, Jahiruddin M, Feldmann J, Price AH, Meharg AA. Environ Sci Technol. 2009;43:6070–6075. doi: 10.1021/es901121j. [DOI] [PubMed] [Google Scholar]
- 20.Norton GJ, Duan G, Dasgupta T, Islam MR, Lei M, Zhu Y, Deacon CM, Moran AC, Islam S, Zhao F, Stroud JL, Mcgrath SP, Feldmann J, Price AH, Meharg AA. Environ Sci Technol. 2009;43:8381–8386. doi: 10.1021/es901844q. [DOI] [PubMed] [Google Scholar]
- 21.Williams PN, Islam MR, Adomako EE, Raab A, Hossain SA, Zhu YG, Feldmann J, Meharg AA. Environ Sci Technol. 2006;40:4903–4908. doi: 10.1021/es060222i. [DOI] [PubMed] [Google Scholar]
- 22.Zavala YJ, Gerads R, Gürleyük H, Duxbury JM. Environ Sci Technol. 2008;42:3861–3866. doi: 10.1021/es702748q. [DOI] [PubMed] [Google Scholar]
- 23.Zhu YG, Sun GX, Lei M, Teng M, Liu YX, Chen NC, Wang LH, Carey AM, Deacon C, Raab A, Meharg AA, Williams PN. Environ Sci Technol. 2008;42:5008–5013. doi: 10.1021/es8001103. [DOI] [PubMed] [Google Scholar]
- 24.Adomako EE, Solaiman ARM, Williams PN, Deacon C, Rahman GKMM, Meharg AA. Environ Int. 2009;35:476–479. doi: 10.1016/j.envint.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 25.Kohlmeyer U, Jantzen E, Kuballa J, Jakubik S. Anal Bioanal Chem. 2003;377:6–13. doi: 10.1007/s00216-003-2064-1. [DOI] [PubMed] [Google Scholar]
- 26.Hansen HR, Raab A, Price AH, Duan G, Zhu Y, Norton GJ, Feldmann J, Meharg AA. J Environ Monit. 2011;13:32–34. doi: 10.1039/c0em00460j. [DOI] [PubMed] [Google Scholar]
- 27.Beilstein MA, Whanger PD, Yang GQ. Biomed Environ Sci. 1991;4:392–398. [PubMed] [Google Scholar]
- 28.Williams PN, Lombi E, Sun GX, Scheckel K, Zhu YG, Feng X, Zhu J, Carey AM, Adomako E, Lawgali Y, Deacon C, Meharg AA. Environ Sci Technol. 2009;43:6024–6030. doi: 10.1021/es900671m. [DOI] [PubMed] [Google Scholar]
- 29.Fang Y, Zhang Y, Catron B, Chan Q, Hu Q, Caruso JA. J Anal At Spectrom. 2009;24:1657–1664. [Google Scholar]
- 30.Sun GX, Liu X, Williams PN, Zhu YG. Environ Sci Technol. 2010;44:6706–6711. doi: 10.1021/es101843x. [DOI] [PubMed] [Google Scholar]
- 31.Li HF, Lombi E, Stroud JL, McGrath SP, Zhao FJ. J Agric Food Chem. 2010;58:11837–11843. doi: 10.1021/jf1026185. [DOI] [PubMed] [Google Scholar]
- 32.Zhao Y, Zheng J, Yang M, Yang G, Wu Y, Fu F. Talanta. 2011;84:983–988. doi: 10.1016/j.talanta.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 33.Lombi E, Scheckel KG, Pallon J, Carey AM, Zhu YG, Meharg AA. New Phytol. 2009;184:193–201. doi: 10.1111/j.1469-8137.2009.02912.x. [DOI] [PubMed] [Google Scholar]
- 34.Carey A, Scheckel KG, Lombi E, Newville M, Choi Y, Norton GJ, Charnock JM, Feldmann J, Price AH, Meharg AA. Plant Physiol. 2010;152:309–319. doi: 10.1104/pp.109.146126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lombi E, Susini J. Plant Soil. 2009;320:1–35. [Google Scholar]
- 36.Lombi E, Scheckel KG, Kempson IM. Environ Exp Bot. 2011;72:3–17. [Google Scholar]
- 37.Smith E, Kempson I, Juhasz AL, Weber J, Skinner WM, Gräfe M. Chemosphere. 2009;76:529–535. doi: 10.1016/j.chemosphere.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 38.Ren XL, Liu QL, Wu DX, Shu Q. Rice Sci. 2006;13:170–178. [Google Scholar]
- 39.Rahman MA, Hasegawa H, Rahman MM, Rahman MA, Miah MAM. Chemosphere. 2007;69:942–948. doi: 10.1016/j.chemosphere.2007.05.044. [DOI] [PubMed] [Google Scholar]
- 40.Punshon T, Guerinot ML, Lanzirotti A. Ann Bot- London. 2009;103:665–672. doi: 10.1093/aob/mcn264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krishnan S, Dayanandan P. J Biosci. 2003;28:455–469. doi: 10.1007/BF02705120. [DOI] [PubMed] [Google Scholar]
- 42.Seyfferth AL, Webb SM, Andrews JC, Fendorf S. Geochim Cosmochim Acta. 2011;75:6655–6671. [Google Scholar]
- 43.Zheng MZ, Cai C, Hu Y, Sun GX, Williams PN, Cui HJ, Li G, Zhao FJ, Zhu YG. New Phytol. 2011;189:200–209. doi: 10.1111/j.1469-8137.2010.03456.x. [DOI] [PubMed] [Google Scholar]
- 44.Carey A, Norton GJ, Deacon C, Scheckel KG, Lombi E, Punshon T, Guerinot ML, Lanzirotti A, Newville M, Choi Y, Price AH, Meharg AA. New Phytol. 2011;192:87–98. doi: 10.1111/j.1469-8137.2011.03789.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim SA, Punshon T, Lanzirotti A, Li A, Alonso JM, Ecker JR, Kaplan J, Guerinot ML. Science. 2006;314:1295–1298. doi: 10.1126/science.1132563. [DOI] [PubMed] [Google Scholar]
- 46.Abedin MJ, Feldmann J, Meharg AA. Plant Physiol. 2002;128:1120–1128. doi: 10.1104/pp.010733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao FJ, Stroud JL, Khan MA, McGrath SP. Plant Soil. 2011 In press doi: 10.1007/s11104-011-0926-4. [Google Scholar]
- 48.Rivers M. 4,000 Spectra or 4,000,000 ROIs per Second: EPICS Support for High-Speed Digital X-ray Spectroscopy with the XIA xMap. [3-Dec-2010];XRM 2010, 10th International Conference on X-ray Microscopy. 2010 2010 See also http://cars9.uchicago.edu/software/epics/dxp.html for DXP 3.0 release.
- 49.Ryan CG, Siddons DP, Moorhead G, Kirkham R, De Geronimo G, Etschmann B E, Dragone A, Dunn P A, Kuczewski A, Davey P, Jensen M, Ablett JM, Kuczewski J, Hough R, Paterson D. J Physics: Conference Series. 2009;186:012013. [Google Scholar]
- 50.Ryan CG, Kirkham R, Hough RM, Moorhead G, Siddons DP, de Jonge MD, Paterson DJ, De Geronimo G, Howard DL, Cleverley JS. Nucl Instr Meth A. 2010;619:37–43. [Google Scholar]
- 51.Kirkham R, Dunn PA, Kucziewski A, Siddons DP, Dodanwela R, Moorhead G, Ryan CG, De Geronimo G, Beuttenmuller R, Pinelli D, Pfeffer M, Davey P, Jensen M, Paterson D, de Jonge MD, Kusel M, McKinlay J. AIP Conf Ser. 2010;1234:240. [Google Scholar]
- 52.Lombi E, de Jonge MD, Donner E, Kopittke PM, Howard DL, Kirkham R, Ryan CG, Paterson D. PLoS One. 2011;6:e20626. doi: 10.1371/journal.pone.0020626. doi:10.1371/journal.pone.0020626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Paterson DJ, Boldeman JW, Cohen DD, Ryan CG. AIP Conf Proc. 2007;879:864–867. [Google Scholar]
- 54.de Jonge MD, Vogt S. Curr Op Str Biol. 2010;20:606–614. doi: 10.1016/j.sbi.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 55.Ma JF, Tamai K, Ichii M, Wu GF. Plant Physiol. 2002;130:2111–2117. doi: 10.1104/pp.010348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nikolic M, Nikolic N, Liang Y, Kirkby EA, Romheld V. Plant Physiol. 2007;143:495–503. doi: 10.1104/pp.106.090845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ma JF, Yamaji N. Cell Mol Life Sci. 2008;65:3049–3057. doi: 10.1007/s00018-008-7580-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ma F, Yamaji N, Mitani N, Xu XY, Su YH, McGrath SP, Zhao FJ. Proc Nat Acad Sci USA. 2008;105:9931–9935. doi: 10.1073/pnas.0802361105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Norton GJ, Dasgupta T, Islam MR, Islam S, Deacon CM, Zhao F, Stroud JL, McGrath SP, Feldmann J, Price AH, Meharg AA. Environ Sci Technol. 2010;44:8284–8288. doi: 10.1021/es101487x. [DOI] [PubMed] [Google Scholar]
- 60.Punshon T, Jackson BP, Bertsch PM, Burger J. J Environ Monitor. 2004;6:153–159. doi: 10.1039/b310878c. [DOI] [PubMed] [Google Scholar]
- 61.Jackson BP, Hopkins WA, Baionno J. Environ Sci Technol. 2003;37:2511–2515. doi: 10.1021/es026416p. [DOI] [PubMed] [Google Scholar]
- 62.Moore KL, Schröder M, Lombi E, Zhao FJ, McGrath SP, Hawkesford MJ, Shewry PR, Grovenor CRM. New Phytol. 2010;185:434–445. doi: 10.1111/j.1469-8137.2009.03071.x. [DOI] [PubMed] [Google Scholar]
- 63.Lombi E, de Jonge MD, Donner E, Ryan CG, Paterson D. Anal Bioanal Chem. 2011;400:1637–1644. doi: 10.1007/s00216-011-4829-2. [DOI] [PubMed] [Google Scholar]