Abstract
H2A.Z is a highly conserved histone variant in all species. The chromatin deposition of H2A.Z is specifically catalyzed by the yeast chromatin remodeling complex SWR1 and its mammalian counterpart SRCAP. However, the mechanism by which H2A.Z is preferentially recognized by non-histone proteins remains elusive. Here we identified Anp32e, a novel higher eukaryote-specific histone chaperone for H2A.Z. Anp32e preferentially associates with H2A.Z-H2B dimers rather than H2A-H2B dimers in vitro and in vivo and dissociates non-nucleosomal aggregates formed by DNA and H2A-H2B. We determined the crystal structure of the Anp32e chaperone domain (186-232) in complex with the H2A.Z-H2B dimer. In this structure, the region containing Anp32e residues 214-224, which is absent in other Anp32 family proteins, specifically interacts with the extended H2A.Z αC helix, which exhibits an unexpected conformational change. Genome-wide profiling of Anp32e revealed a remarkable co-occupancy between Anp32e and H2A.Z. Cells overexpressing Anp32e displayed a strong global H2A.Z loss at the +1 nucleosomes, whereas cells depleted of Anp32e displayed a moderate global H2A.Z increase at the +1 nucleosomes. This suggests that Anp32e may help to resolve the non-nucleosomal H2A.Z aggregates and also facilitate the removal of H2A.Z at the +1 nucleosomes, and the latter may help RNA polymerase II to pass the first nucleosomal barrier.
Keywords: H2A.Z, Anp32e, nucleosome, crystal structure, histone chaperone
Introduction
As the most important protein components of chromatin, histones have been extensively studied. In the past decade, variant histones that differ from canonical histones have been recognized to occupy specific genomic regions and to possess distinct regulatory functions1,2,3,4,5,6. Histone variants are often regulated by their distinct expression timing, unique post-translational modifications and use of specific chaperones1,2,3,4.
H2A.Z is highly conserved among eukaryotes from the yeast S. cerevisiae to human, but shares only 60% sequence identity with the canonical H2A7, suggesting that it may possess a distinct function. Indeed, H2A.Z displays a unique genomic distribution and demarcates the transcription start site (TSS) in S. cerevisiae8,9,10,11,12, Drosophila13, Arabidopsis14 and mammals15,16,17,18,19,20. Recently, H2A.Z was shown to be enriched in enhancers18,19. Although H2A.Z has a role in both gene activation and silencing (see the review paper7), it has been generally agreed that H2A.Z occupancy positively correlates with transcriptional activity in higher eukaryotes13,14,15,16,17,18,19,20.
Like many other histone variants, H2A.Z has dedicated chaperone proteins. The main H2A.Z-specific histone deposition complex SWR1 was first discovered in S. cerevisiae21,22,23; later, the mammalian SRCAP complex, a protein complex closely related to the yeast SWR1 complex, was reported to possess similar activity24,25. Apart from the SWR1 complex and its higher eukaryotic counterparts, to our knowledge, only one additional H2A.Z-specific chaperone protein has been discovered, the yeast Chz1 protein26. Other histone chaperones, such as NAP1 and FACT, can also associate with H2A.Z-H2B dimers; however, unlike Chz1, these chaperones do not display a preference toward H2A.Z-H2B dimers over the canonical H2A-H2B dimers26. Chz1 homologs exist in multiple fungi, but other than the CHZ1 motif, no clear Chz1 homolog has been reported in higher eukaryotes26.
Here, we report the identification of a mammalian H2A.Z-specific chaperone protein, Anp32e, which preferentially associates with H2A.Z-H2B dimers in vitro and in vivo. The co-crystal structure of H2A.Z-H2B in association with Anp32e explains the following biochemical observations: (1) Anp32e, but not other Anp32 family proteins, specifically associates with H2A.Z-H2B dimers; (2) H2A.Z-H2B dimers, but not H2A-H2B dimers, are preferentially recognized by Anp32e; (3) H2A.Z-H2B dimers, but not H2A.Z-containing nucleosomes, are recognized by Anp32e. Finally, the chromatin association pattern of Anp32e recapitulates H2A.Z occupancy at the genome-wide level, suggesting that Anp32e plays a role in regulating H2A.Z deposition.
Results
Purification and identification of an H2A.Z-H2B dimer-specific chaperone
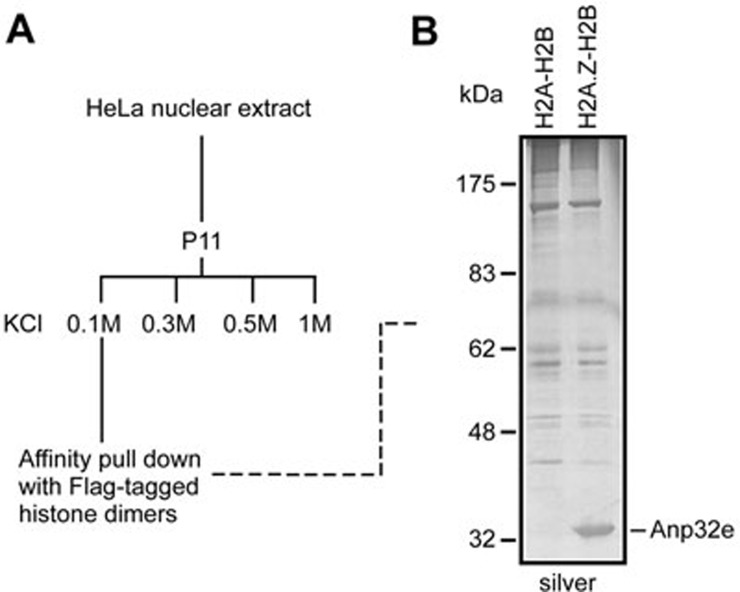
To purify an H2A.Z-H2B-specific chaperone, we first assembled recombinant Flag-tagged H2A-H2B and Flag-tagged H2A.Z-H2B dimers in vitro and then immobilized them with anti-Flag agarose in parallel. We next applied various fractions from HeLa nuclear extracts onto the immobilized beads and then eluted the associated proteins after extensive washing (Figure 1A). The eluates were subjected to SDS-PAGE for comparison. Although most proteins in the eluates appeared to be non-specific, one protein band was remarkably enriched in eluates associated with H2A.Z-H2B dimers but not in eluates associated with H2A-H2B dimers (Figure 1B). This protein was identified as Anp32e by tandem mass spectrometry analysis.
Figure 1.
Identification of Anp32e as an H2A.Z-specific chaperone. (A) Experimental scheme. (B) A silver-stained SDS-PAGE gel displaying proteins associated with H2A-H2B or H2A.Z-H2B dimers.
Anp32e, also known as CPD1 (cerebellum postnatal development gene 1), was first cloned due to its high expression in the postnatal mouse cerebellum27. In some reports, Anp32e has been referred to as LANPL (leucine-rich acidic nuclear protein-like) because of its sequence homology with Anp32a (also known as LANP)28. Anp32a was reported to associate with SET to form the INHAT complex, which inhibits histone acetylation29.
Anp32e preferentially associates with H2A.Z in vitro
The association between Anp32e and H2A.Z has been observed previously25. However, it is not clear whether Anp32e directly interacts with H2A.Z and whether Anp32e can discriminate H2A.Z from canonical H2A.
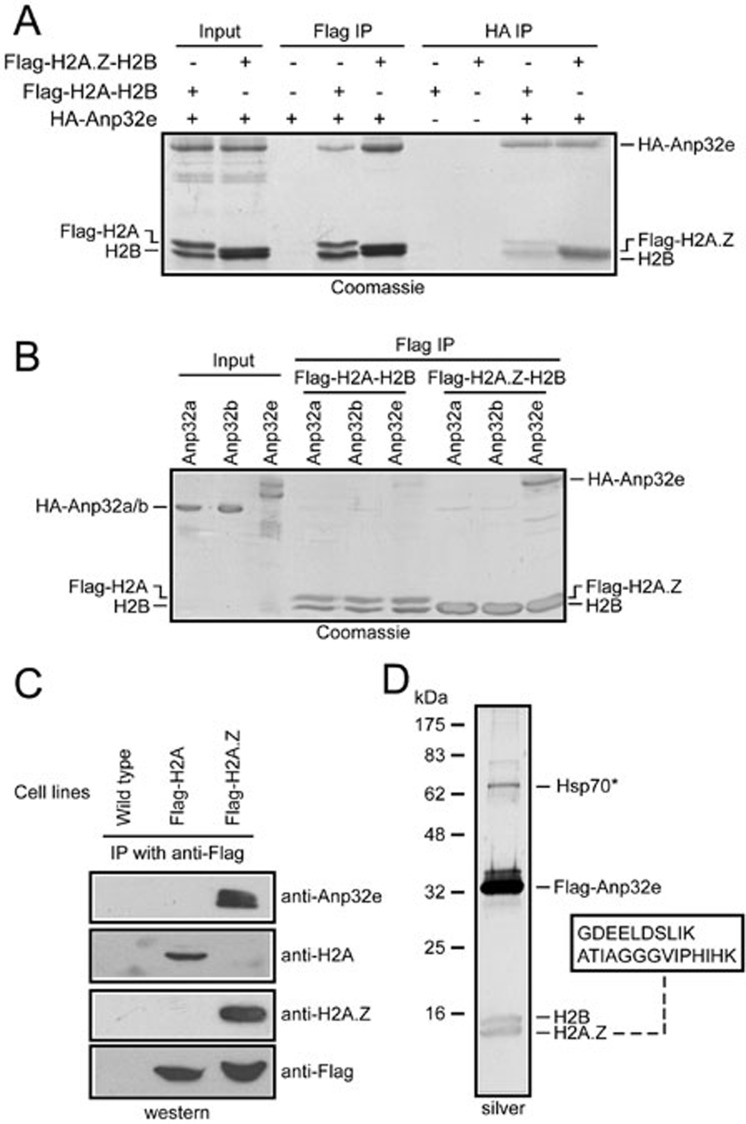
To answer these questions, recombinant HA-tagged Anp32e was expressed and purified from E. coli and then mixed with Flag-tagged H2A.Z-H2B dimers or Flag-tagged H2A-H2B dimers (Figure 2A). The above mixtures were then subjected to immunoprecipitation (IP) with antibodies against Flag or HA. In the Flag IP eluates, similar amounts of Flag-H2A-H2B dimers and Flag-H2A.Z-H2B dimers were obtained. However, much less HA-Anp32e was found in association with Flag-H2A-H2B dimers (Figure 2A). Similarly, in a reciprocal experiment, antibodies against HA precipitated equal amounts of HA-Anp32e, but far fewer Flag-H2A-H2B dimers were found to associate with Anp32e, in comparison to the Flag-H2A.Z-H2B dimers (Figure 2A).
Figure 2.
Anp32e preferentially associates with H2A.Z in vitro and in vivo. (A) Recombinant HA-Anp32e preferentially associates with Flag-H2A.Z-H2B dimers in immunoprecipitation experiments in vitro. (B) Anp32e, but not Anp32a or Anp32b, associates with Flag-H2A.Z-H2B dimers. (C) Immunoprecipitation with antibodies against Flag specifically enriches Anp32e in cells expressing Flag-H2A.Z but not in cells expressing Flag-H2A. (D) Affinity purification from Flag-Anp32e-expressing cells revealed that H2B and H2A.Z associate with Flag-Anp32e. The boxed area indicates peptides unique to H2A.Z histones that were identified using tandem mass spectrometry analysis. *Common contamination proteins.
Considering that Anp32e belongs to a family including several highly similar proteins (Supplementary information, Figure S1) and that Anp32a has been reported to associate with histones in the context of the INHAT complex29, we examined the ability of several Anp32e homologs to associate with H2A.Z-H2B or H2A-H2B dimers. Under the conditions we used, Anp32e, but not Anp32a or Anp32b, exhibited a robust interaction with H2A.Z-H2B dimers (Figure 2B).
These results indicate that Anp32e directly and preferentially interacts with H2A.Z-H2B dimers in vitro.
Anp32e specifically associates with H2A.Z in vivo
To further validate the specificity in vivo, we generated HeLa cells stably expressing Flag-H2A or Flag-H2A.Z and then performed anti-Flag IP experiments with non-chromatic nuclear extracts from these cells. H2A and H2A.Z histones were correctly precipitated from the corresponding cells (Figure 2C). Moreover, Anp32e was robustly detected in IP eluates from the Flag-H2A.Z cells; however, no detectable amount of Anp32e was found in IP eluates from the Flag-H2A cells (Figure 2C). These results suggest that Anp32e exclusively associates with H2A.Z-H2B dimers but not with canonical H2A-H2B dimers in vivo.
Next, we generated HeLa cells stably expressing Flag-Anp32e and performed affinity purification using nuclear extracts from these cells. Flag-Anp32e clearly associates with two small proteins with molecular weights similar to those of histones (Figure 2D). Tandem mass spectrometry analysis identified these two proteins as H2B and H2A.Z. Two peptides unique to H2A.Z (Figure 2D and Supplementary information, Figure S2), but not any other H2A proteins, were revealed by the tandem mass spectrometry analysis. Importantly, no peptide unique to canonical H2A or any other H2A variants was detected.
Taken together, the above results clearly demonstrate that Anp32e is an H2A.Z-specific chaperone protein in vivo.
Anp32e associates with H2A.Z-H2B dimers but not with H2A.Z-containing nucleosomes
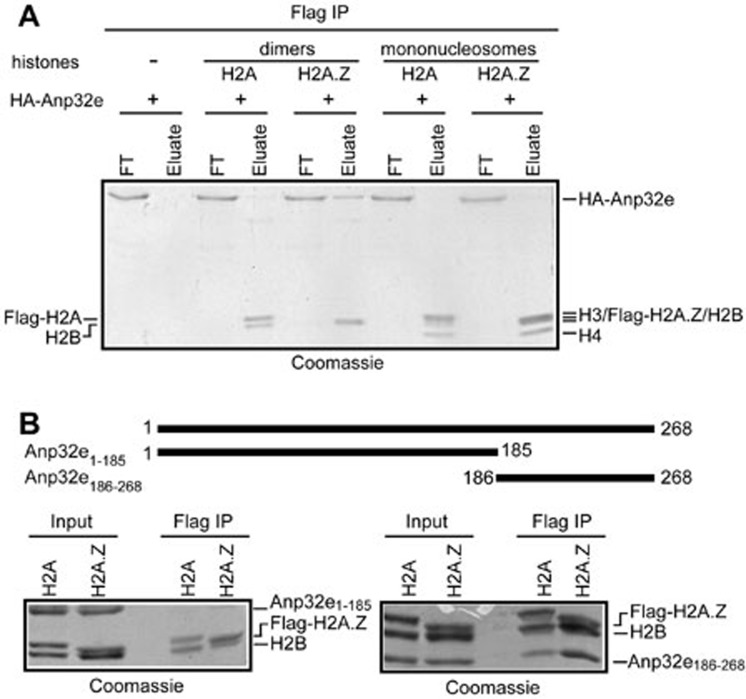
Affinity-purified Flag-Anp32e from HeLa extracts associates with H2A.Z and H2B but not with H3 and H4 histones (Figure 2D). This suggests that Anp32e may not be able to recognize H2A.Z within the nucleosomes. To validate this conclusion, we assembled Flag-H2A- or Flag-H2A.Z-containing mononucleosomes and tested whether they can associate with Anp32e. Antibodies against Flag successfully precipitated Flag-H2A-H2B dimers, Flag-H2A.Z-H2B dimers, Flag-H2A-containing mononucleosomes and Flag-H2A.Z-containing mononucleosomes (Figure 3A). However, only the Flag-H2A.Z-H2B dimers, but not the Flag-H2A.Z-containing nucleosomes, retrieved a large amount of Anp32e (Figure 3A). This result is a clear indication that Anp32e cannot recognize nucleosomal H2A.Z.
Figure 3.
Characterization of Anp32e. (A) Recombinant Anp32e does not interact with nucleosomal H2A.Z in vitro. (B) The C-terminal region, but not the N-terminal region, of Anp32e associates with H2A.Z-H2B dimers.
The C-terminal region of Anp32e associates with H2A.Z-H2B dimers
Anp32e is a small protein without obvious domains, but its C-terminal region is highly acidic (Supplementary information, Figure S1). To characterize the region of Anp32e that associates with H2A.Z-H2B dimers, we expressed and purified recombinant proteins corresponding to the N-terminal (amino acids 1-185, termed Anp32e1-185) and C-terminal (amino acids 186-268, termed Anp32e186-268) regions of Anp32e and tested their ability to associate with H2A.Z-H2B dimers. IP experiments showed that the C-terminal acidic region of Anp32e, but not the N-terminal region of Anp32e, is able to associate with H2A.Z-H2B dimers (Figure 3B).
Structure of the Anp32e fragment in complex with single-chain H2A.Z-H2B
To understand the detailed mechanism by which Anp32e recognizes H2A.Z, Anp32e186-232 and Anp32e186-268 were used to reconstitute Anp32e-lnkH2B-H2A.Z complex for structural study. Here, lnkH2B-H2A.Z represents linked H2B-H2A.Z, in which linked human H2B33-125 and human H2A.Z16-114 were expressed as a single polypeptide chain. The validity of using single-chain H2A.Z-H2B in structural studies has been demonstrated previously30.
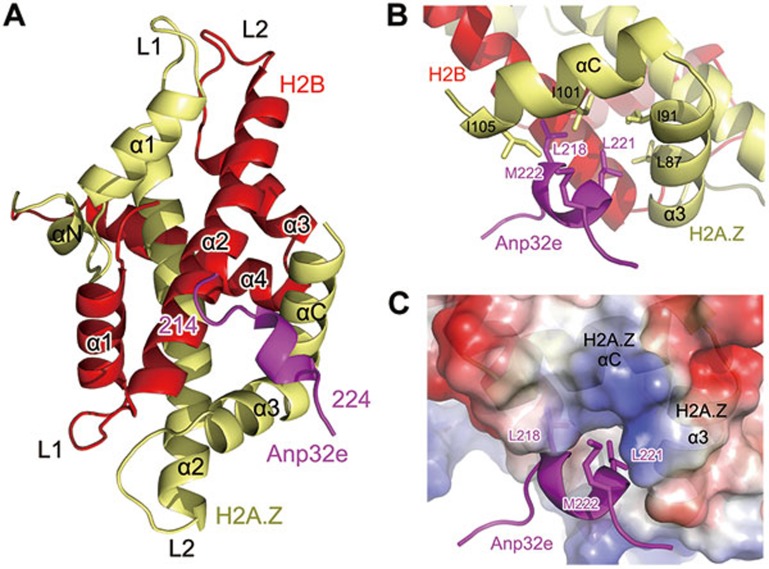
We successfully crystallized the Anp32e186-232-lnkH2B-H2A.Z complex and solved the structure at 2.6 Å resolution (Figure 4 and Supplementary information, Table S1). The structure of lnkH2B-H2A.Z within the Anp32e complex is similar to that of the nucleosomal H2B-H2A.Z dimer (Supplementary information, Figure S3). The mean square deviation (RMSD) of the two structures is 0.828. In the structure, Anp32e residue 214-224 formed a short helix that is tightly packed against the H2A.Z α3 and αC helices, largely through hydrophobic interactions. The hydrophobic residues L218, L221 and M222 of Anp32e interacted with residues L87 and I91 in the α3 helix and residues I101 and I105 in the αC helix of H2A.Z. However, Anp32e residues 186-213 and 225-232 were not observable in the structure because of lack of electron density (Figure 4B). These residues might interact with lnkH2B-H2A.Z in a less rigid conformation that causes poor crystal packing. It is worth noting that the binding site of Anp32e on lnkH2B-H2A.Z is distant from the connection point between H2B and H2A.Z, suggesting that the presence of the link region in lnkH2B-H2A.Z does not perturb the observed binding site (Figure 4A).
Figure 4.
The structure of Anp32e in complex with H2B-H2A.Z. (A) The overall structure of the Anp32e186-232-lnkH2B-H2A.Z complex. The sequence of the observable part of Anp32e in the structure is 214EEVGLSYLMKE224. (B) Major interactions between Anp32e214-224 and H2A.Z α3 and αC helices. (C) Complementary surfaces between Anp32e214-224 and H2A.Z α3 and αC helices
The structure of the complex reveals an unexpected conformational change in the H2A.Z αC helix. In contrast to the three-turn-helix structure observed in structures of H2A.Z nucleosome and H2A.Z-H2B-Chz1 complexes, H2A.Z in the Anp32e complex forms an extended αC helix, which is stabilized by interacting with Anp32e214-224 (Figure 4B, 4C and Supplementary information, Figure S4)31. Interestingly, the asymmetric unit of the crystal structure comprises two free lnkH2B-H2A.Z molecules without binding Anp32e. Extension of the αC helix of H2A.Z in both molecules suggested that the αC helix, which exhibits structural plasticity, might serve as a universal docking site for other proteins to recognize H2A.Z (Supplementary information, Figure S5). We performed sedimentation experiments to validate the complex stoichiometry. The result indicated that Anp32e186-232 and lnkH2B-H2A.Z form a 1:1 complex similar to that of Chz1 (Supplementary information, Figure S5)26.
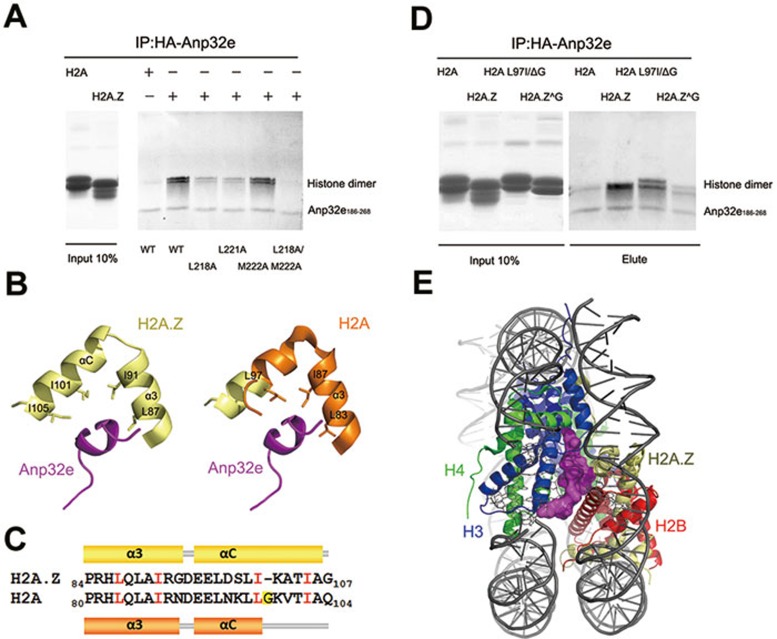
We next mutated the critical residues in Anp32e186-268 to alanine and investigated whether they can associate with H2A.Z-H2B dimer. The IP results show that the Anp32e mutants L218A and L221A displayed significantly reduced binding to H2A.Z-H2B dimer (Figure 5A). Moreover, the double mutation L218A/M222A completely abolished the association between Anp32e and H2A.Z-H2B dimer (Figure 5A). These results, which underscore the importance of residues L218, L221 and M222 in Anp32e, are consistent with the crystal structure.
Figure 5.
The structural basis of specific H2A.Z recognition and the histone chaperone function of Anp32e. (A) Immunoprecipitation analysis of interaction between histone dimer and HA-tagged Anp32e186-286 mutants. Histone dimer represents H2B-H2A.Z or H2B-H2A. (B) Comparison of the C-terminal regions of H2A.Z and H2A. Left: local view of H2A.Z in complex with Anp32e. Right: an overlay of the nucleosomal H2A structure with the H2A.Z structure in complex with Anp32e. Conserved residues that are critical for Anp32e discrimination are highlighted. (C) Secondary structure differences between the C-terminal region of H2A.Z in the Anp32e complex and that of H2A in the nucleosome. Conserved residues in H2A.Z and H2A that are involved in Anp32e discrimination are depicted in red. H2A G98, which is absent in H2A.Z, is highlighted in yellow. (D) Immunoprecipitation analysis of interaction between histone dimer mutants and HA-tagged Anp32e186-286. Histone-mutant dimer represents H2A.Z^G102-H2B and H2A L97IΔG98-H2B. (E) Anp32e (magenta) precludes H2A.Z-H2B from binding nucleosomal H3-H4.
A specific residue of H2A.Z in the αC helix is critical for the preferential recognition of H2A.Z by Anp32e
Anp32e preferentially binds the H2A.Z-H2B dimer despite the apparent sequence similarity between H2A and H2A.Z. We reasoned that a particular sequence of H2A.Z might confer the specificity to recognize H2A.Z by Anp32e. The extended αC helix of H2A.Z in the Anp32e-bound complex was analyzed further (Figure 5B). Surprisingly, H2A.Z residues that appear important for Anp32 interaction, including L87, I91, I101 and I105, are highly conserved in H2A (Figure 5C). Notwithstanding their obvious similarity, G98, which is present in the C-terminal region of H2A, does not exist in H2A.Z. Omission of the glycine appears to be highly conserved in H2A.Z in all species. We speculated that the presence of G98 in H2A perturbs the side-chain packing beyond G98 and prevents the αC helix from extending further, as in H2A.Z. We generated two histone-dimer mutants and tested their association with Anp32e186-268. Notably, H2A.Z G102, a H2A.Z mutant with a glycine insertion at position 102, displayed greatly decreased Anp32e186-268 association. In contrast, when H2A residues L97G98 were mutated to I97ΔG98 to mimic H2A.Z, it gained substantial ability to associate with Anp32e186-268 (Figure 5D). Therefore, we conclude that the omission of glycine confers the αC helix of H2A.Z the ability to strongly interact with Anp32e, and plays a critical role in the specific recognition of H2A.Z by Anp32e.
Anp32e prevents non-specific aggregation between H2A.Z-H2B dimers and DNA
We superimposed H2A.Z in the Anp32e complex structure onto nucleosomal H2A.Z. Interestingly, binding of Anp32e occluded the H2A.Z C-terminal region from interacting with nucleosomal H3-H4 (Figure 5E), which explains our biochemical observations (Figure 3A).
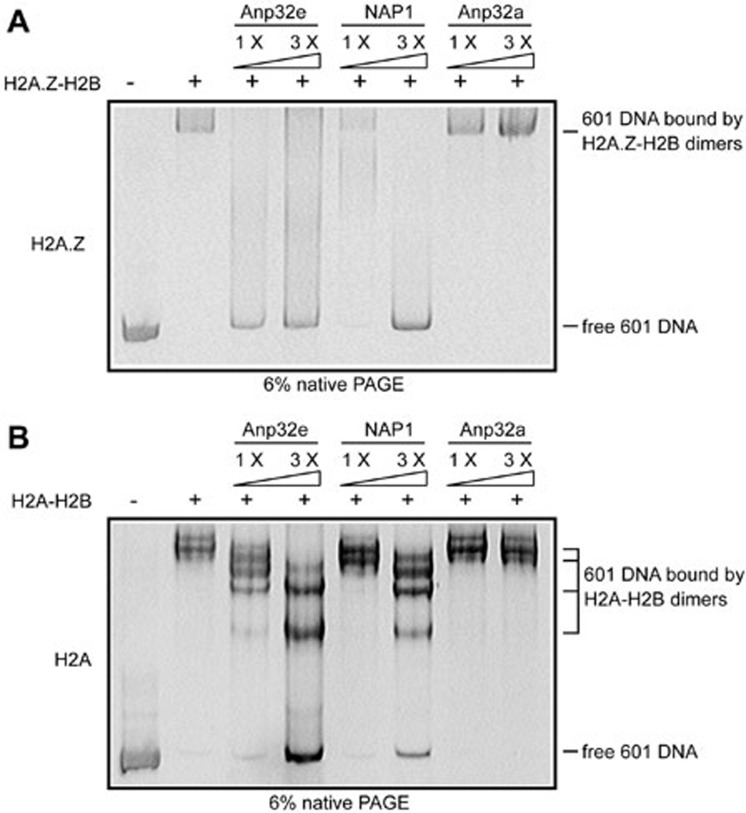
Moreover, because of their highly positive charge, histones display a high affinity toward negatively charged DNA. The interaction between histones and DNA is typically a high-affinity, low-specificity type of binding, which leads to the formation of non-specific aggregates when histones and DNA are mixed at physiological salt concentrations32. One critical criterion for a histone chaperone is that it should possess the ability to prevent the formation of non-specific aggregates by DNA and histones and to dissolve preformed DNA-histone aggregates32. To test whether Anp32e possesses such ability, we mixed 601-sequence DNA33 and H2A-H2B dimers or H2A.Z-H2B dimers to form non-specific aggregates and then titrated them with recombinants Anp32e, NAP1 or Anp32a. Anp32e efficiently dissolved the H2A.Z-H2B-DNA aggregates (Figure 6A). As a positive control, NAP1 was also capable of dissolving these aggregates but required a higher concentration (Figure 6A). By contrast, Anp32a failed to achieve this result (Figure 6A). This is consistent with its inability to interact with H2A.Z-H2B dimers (Figure 2B). In addition, Anp32e partially dissolved the aggregates formed by H2A-H2B and DNA at a higher concentration (Figure 6B).
Figure 6.
Anp32e disrupts non-nucleosomal aggregates formed by DNA and H2A.Z-H2B dimers. H2A.Z-H2B or H2A-H2B dimers were mixed with 601 sequence DNA to form aggregates. One- to three-fold molar ratios of recombinant Anp32e, NAP1 or Anp32a were then titrated into the aggregates to examine their chaperone activity in dissolving non-nucleosomal DNA-histone aggregates. (A) H2A.Z. (B) H2A.
In conclusion, similar to NAP1, which exhibits chaperone activity for H2A-H2B dimers and H2A.Z-H2B dimers32, Anp32e can prevent the formation of non-nucleosomal H2A.Z-H2B-DNA aggregates, supporting its role as an H2A.Z-specific chaperone.
Anp32e co-localizes with H2A.Z and fine-tunes H2A.Z distribution near TTSs
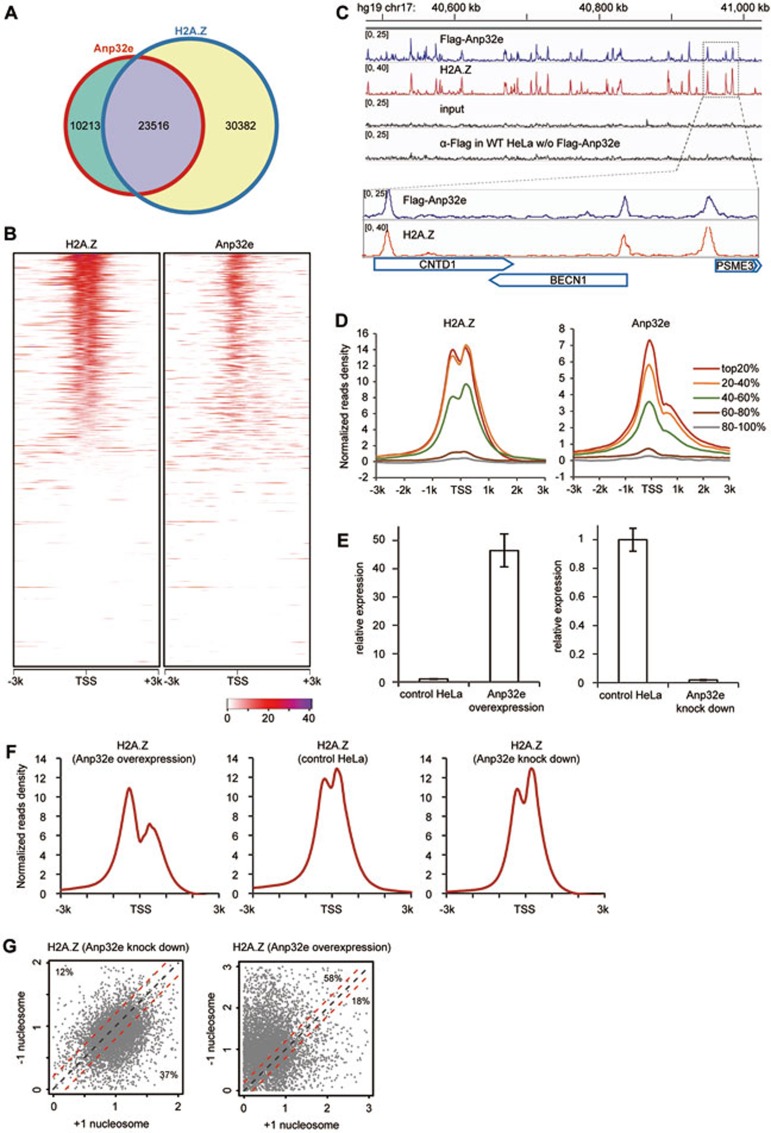
To determine Anp32e's chromatin distribution pattern and to analyze its relationship with H2A.Z at the genome-wide level, we performed ChIP-Seq (chromatin IP-sequencing) experiments using antibodies against Flag with HeLa cells stably expressing Flag-Anp32e. We also performed ChIP-Seq experiments using antibodies against H2A.Z with HeLa cells, HeLa cells with Anp32e knockdown and HeLa cells with Anp32e overexpression.
In total, we identified 33 729 Anp32e peaks and 53 898 H2A.Z peaks (HeLa) at the genome-wide level. Remarkably, close to 70% of the Anp32e peaks overlapped with the H2A.Z peaks (Figure 7A). Next, we sorted all the genes according to their H2A.Z-ChIP-Seq reads at the TSS regions (−300 bp to +300 bp) (Figure 7B, left panel) and then analyzed their Anp32e ChIP-seq reads (Figure 7B, right panel). Again, Anp32e displayed a highly similar profile in comparison with H2A.Z in the TSS regions. The difference is, however, Anp32e tended to be enriched at the TSSs (Figures 7B-7D), whereas H2A.Z often flanked the TSSs and were enriched in +1 and −1 nucleosomes (Figures 7B-7D), in agreement with previous reports9,15,18,19.
Figure 7.
Anp32e and H2A.Z display similar genome-wide profiles. (A) Close to 70% of Anp32e peaks in the genome overlap with H2A.Z peaks. (B) The Anp32e and H2A.Z distribution around the TSSs of all genes. Genes were sorted according to their H2A.Z ChIP-Seq reads in HeLa cells. (C) An example region showing the co-occupancy of Anp32e and H2A.Z. (D) Differential distribution of Anp32e and H2A.Z at genes with different transcriptional activity. Genes were categorized into five groups according to their transcriptional levels. (E) RT-PCR showing relative expression levels of Anp32e in HeLa cells, HeLa cell with Anp32 knockdown and HeLa cell overexpressing Anp32e. (F) Distribution of H2A.Z around TSS regions in HeLa cells overexpressing Anp32e (left), control HeLa cells (middle) and HeLa cells with Anp32e knockdown (right). Top 40% of genes in B that display clear H2A.Z occupancy were selected for the plotting. (G) Scatter plots showing the relative change of H2A.Z densities at +1 nucleosomes and −1 nucleosomes in Anp32e-overexpressing and Anp32e knockdown HeLa cells. The density of each peak was normalized by the corresponding peak in control HeLa cells. The red lines in parallel with the diagonal line represent a difference of +0.2 or −0.2, respectively.
Next, we categorized all genes into five groups according to their RNA abundance using RNA-Seq data of HeLa cells. Similar to H2A.Z, Anp32e was found to be enriched in the active genes (Figure 7D).
Such striking similarity between Anp32e and H2A.Z at the genome-wide level strongly suggests a functional connection between Anp32e and H2A.Z. Therefore, we performed H2A.Z ChIP-Seq experiments in HeLa cells, HeLa cells with Anp32e knockdown and HeLa cells with Anp32e overexpression (Figure 7E), and compared their profiles at the TSS regions of top 40% genes with clear H2A.Z occupancy (Figure 7B). Interestingly, we noticed that in HeLa cells, H2A.Z occupancy at the +1 nucleosomes and the −1 nucleosomes were quite comparable (Figure 7F, middle panel). However, in Anp32e-overexpressing HeLa cells, H2A.Z occupancy at the +1 nucleosomes was significantly lower (Figure 7F, left panel). Consistently, when Anp32e was knocked down, H2A.Z displayed higher occupancy at the +1 nucleosomes (Figure 7F, right panel).
To get a more quantitative comparison, we plotted the H2A.Z densities at the −1 and +1 nucleosomes in Anp32e-knockdown HeLa cells and Anp32e-overexpressing HeLa cells (Figure 7G) for top 40% genes with clear H2A.Z occupancy. In each case, H2A.Z densities at the ± 1 nucleosomes were normalized against the control HeLa cells. In both plots, TSSs with a comparable H2A.Z occupancy at both nucleosomes were distributed within the area enclosed by two red lines (the difference of H2A.Z occupancy is within a factor of 0.2). Notably, in Anp32e-knockdown cells, 37% of TSSs were distributed below the lower red line, whereas only 12% of TSSs were distributed above the higher red line (Figure 7G, left panel), indicating a global H2A.Z increase of occupancy at the +1 nucleosomes. Anp32e-overexpressing cells displayed the opposite change, only 18% of TSSs were distributed below the lower red line, whereas 58% of TSSs were distributed above the higher red line, indicating a global loss of H2A.Z occupancy at the +1 nucleosomes (Figure 7G, right panel). Together, these results suggest that Anp32e may facilitate the removal of H2A.Z at +1 nucleosomes and fine-tune H2A.Z profile at the TSS regions.
Discussion
Our studies of Anp32e introduced the first higher eukaryote-specific histone chaperone for H2A.Z. The Anp32e complex structure revealed an unexpected structural change in the αC helix of H2A.Z, which is specifically recognized by Anp32e residue 214–224. Remarkably, the absence of a glycine in the αC helix of H2A.Z, which is critical for Anp32e preferential recognition, is likely the major contributor to the conformational change. These observations further broaden our knowledge regarding histone variant selection34,35,36,37,38. It has previously been reported that the C-terminal region of H2A.Z is functionally essential in Drosophila and important for preferential binding of H2A.Z to the SWR1 complex and the Chz1 protein26,31. Our studies provide evidence at atomic resolution to support the conclusion that the C-terminal region of H2A.Z confers the specificity for H2A variants.
Anp32e contains long acidic stretches that are rich in Glu/Asp residues. These residues are likely involved in histone interaction, as reported for other histone chaperones26. In our study, Anp32e dissociates non-nucleosomal aggregates formed by DNA and H2A.Z. The structure shows that Anp32e binding prevents H2A.Z-H2B from interacting with H3-H4. In addition, genome-wide ChIP-Seq profiles indicate that the distributions of Anp32e and H2A.Z are correlated. These results are in good agreement with the conclusion that Anp32e has histone chaperone functions. Notably, the acidic stretches of Anp32e are not observable in the complex structure, indicating that they are weakly associated with H2B-H2A.Z. It will be of great interest to understand the roles that these regions play in histone recognition.
In fungi, Chz1 is the only histone chaperone with a demonstrated preference for H2A.Z. Interestingly, the regular H2A histone chaperones Nap1 and FACT, which can recognize most major H2A histones, are able to compensate the chaperone function of Chz126. These results suggest that H2A.Z-specific chaperones may play exclusive roles, although they have functional redundancy with the regular H2A histone chaperone. In agreement with this speculation, it has been reported that chz1 deletion reduces the ubiquitination of subtelomere-associated H2B and decreases gene expression in subtelomeric regions39. Our studies suggest that Anp32e functions in regulating the chromatin deposition of H2A.Z.
The Anp32 protein belongs to a protein family, all members of which contain a highly conserved leucine-rich repeat (LRR) domain at the N-terminal region. It has been proposed that Anp32 family proteins functionally overlap and that the LRR domain is of particular importance27. Instead, Anp32 C-terminal regions, which are enriched with acidic residues and are more diverse in sequence, lack obvious function. In our study, H2A.Z was recognized by the C-terminal residues (amino acids 214-224) of Anp32e; these residues exclusively exist in Anp32e and are not present in other Anp32 family proteins. This finding suggests that the more divergent C-terminal regions of Anp32 family proteins may confer specific functions for each family member.
In addition to Anp32e's role in resolving non-nucleosomal structure formed by H2A.Z-H2B dimers and DNA, we observed an interesting role of Anp32 in fine-tuning the relative occupancy of H2A.Z between the +1 nucleosomes and −1 nucleosomes at TSS regions. Given that the +1 nucleosome is the first barrier that elongating RNA pol II encounters, Anp32e's role in regulating the stability of +1 nucleosome might facilitate the kinetics of RNA pol II in passing the first nucleosome barrier. Mechanistically, it remains a puzzle about how Anp32e achieves this role in the absence of an ability to associate with the nucleosomes. One potential explanation is that Anp32e may work in co-operation with chromatin remodelers that can alter the nucleosome structure and remove H2A.Z from the deformed H2A.Z-containing nucleosomes. Further studies delineating this molecular mechanism will be of great interest.
Materials and Methods
Antibodies
Antibodies against H2A.Z (Abcam, ab4174), H2A (Cell Signaling Technology, L88A6), Flag (Sigma, F3165) and Anp32e (Abcam, ab5993) were purchased commercially.
Reconstitution of histone dimers and mononucleosomes
Flag-H2A or Flag-H2A.Z histones were mixed with H2B in equal molar ratio, dialyzed against refolding buffer (20 mM Tris-Cl pH 8.0, 2 M NaCl, 1 mM DTT) and then separated using a 24-ml size-exclusion column (Superdex-200). Fractions containing the dimers were pooled for further analysis. Mononucleosomes were reconstituted as previously reported40 with purified recombinant histones and 601-sequence DNA33. Briefly, core histones were individually expressed and purified from E. coli and then assembled by dialysis against the refolding buffer. Histone octamers were subsequently purified using size-exclusion chromatography in a 24-ml Superdex-200 column. Histone octamers were then mixed with an equal molar ratio of 601-sequence DNA in 2 M NaCl and then sequentially dialyzed against the following buffers (in order): (1) TE (10 mM Tris, pH 8.0, 1 mM EDTA) supplemented with 1.2 M NaCl, (2) TE supplemented with 1.0 M NaCl, (3) TE supplemented with 0.8 M NaCl, (4) TE supplemented with 0.6 M NaCl, and (5) TE.
Affinity pull-down assays
HeLa cell nuclear extract P11 phosphocellulose fractions or recombinant HA-Anp32 family proteins and recombinant Flag-H2A-H2B or Flag-H2A.Z-H2B dimers were mixed in binding buffer (10 mM Tris-HCl, pH 8.0, 500 mM NaCl, 0.1% NP-40, 1 mM EDTA and 1 mM DTT). Then, anti-Flag M2-agarose (Sigma) was added, and the mixture was incubated for 2 h at 4 °C. After extensive washing with binding buffer, the bound proteins were eluted with elution buffer containing 10 mM Tris-HCl, pH 8.0, 1 mM EDTA and 0.5 mg/ml Flag peptide.
Immunoprecipitation
HeLa cells stably expressing Flag-H2A, Flag-H2A.Z or Flag-Anp32e were established individually. For IP, nuclear extracts from these cells were mixed with anti-Flag M2-agarose (Sigma) and the mixture was rotated for 4 h at 4 °C. After extensive washing with buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA and 0.5 M KCl), the bound proteins were eluted with elution buffer containing 10 mM Tris-HCl, pH 8.0, 1 mM EDTA and 0.5 mg/ml Flag peptide.
Protein expression and purification
All mutations were introduced using the QuikChange method (Stratagene). All proteins were expressed in E. coli BL21 (DE3)-codon Plus-RIPL using pET vectors (Stratagene). N-terminal His6-tagged fragments of fusion proteins of lnkH2B-H2A.Z and Anp32e and their mutants were first purified using Ni-NTA (Qiagen). The proteins were further purified using SP ion exchange columns (GE Healthcare). To prepare the lnkH2B-H2A.Z and Anp32e complexes, the fusion proteins were digested with Thrombin (Sigma) overnight at 30 °C. Samples were concentrated using Amicon Ultra (Millipore) devices and subjected to gel filtration on Superdex 200 10/300 GL columns (GE Healthcare). The collected fractions were exchanged to a final buffer of 20 mM MES, 0.5 M NaCl, pH 6.0.
Crystal structure determination
Native crystals of the Anp32e186-232-H2B-H2A.Z complex were grown at 16 °C using the hanging-drop vapor-diffusion method. Crystals were obtained with protein concentrations of 15 mg/ml after 15 days in 0.2 M sodium thiocyanate, 20% (w/v) polyethylene glycol 3350. X-ray diffraction data were collected at the Shanghai Synchrotron Radiation Facility (SSRF) on beamline BL17U equipped with a Quantum 315r CCD detector (ADSC). Details for data collection and processing are included in Supplementary information, Data S1 (also see Supplementary information, Figure S5).
RNAi
siRNAs were transfected into the cells using Oligofectamine RNAiMAX (Invitrogen), according to the manufacturer's protocols. The following list is the siRNAs (sense strands only) pooled for Anp32e knockdown:
Anp32e-1: 5′-TTGGAGCTTAGTGATAATATA-3′,
Anp32e-2: 5′-GTGGAATTCCCTAGCAATTTA-3′,
Anp32e-3: 5′-ATGGATTTGATCAGGAGGATA-3′.
ChIP-Seq
HeLa cells stably expressing Flag-Anp32e were used for ChIP-Seq experiments; the cells were crosslinked with 1% (v/v) formaldehyde for 10 min at room temperature, and the reaction was quenched by adding glycine at a final concentration of 0.15 M. The cells were washed with PBS and then resuspended in 20 mM Tris, pH 8.0, 300 mM NaCl and 0.1% SDS and sonicated to 200-to 500-bp chromatin fragments using a Bioruptor system (Diagenode). ChIP experiments were performed as previously reported41. Sequencing libraries were generated according to Illumina's recommendations using the NEBNext DNA Library Prep Master Mix Set for Illumina. The libraries were single-end sequenced using HiSeq 2000.
Additional methods for crystal structure determination, analytical ultracentrifugation, recombinant protein IP, and bioinformatics analysis for Chip-Seq are given as the Supplementary information, Data S1.
Accession numbers
The ID of the structural co-ordinates deposited in the Protein Data Bank is 4NFT. The ChIP-Seq data have been deposited in the Sequence Read Archive (SRA) under accession number SRA107538.
Acknowledgments
BZ is supported by the Howard Hughes Medical Institute International Early Career Scientist Program. ZZ and BZ are supported by grants from the Ministry of Science and Technology of China (2013CB910203 to ZZ, 2011CB812700 and 2011CB965300 to BZ). ZZ is supported by grants from the National Natural Science Foundation of China (31123003 and 31270833). ZZ is a CAS-Novo Nordisk Great Wall Professor. We thank SSRF beamline scientists for technical support during data collection.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Sequence alignment of human Anp32 family proteins.
Sequence alignment of human H2A.X, H2A and H2A.Z.
Superimposition of crystal structures of Anp32e-H2B-H2A.Z with nucleosomal H2A.Z-H2B in front view (left) and side view (right).
Structural comparison of the H2A.Z αC helix.
Solution stoichiometry and structure rigidity of Anp32e complex (A), Sedimentation experiments indicated that Anp32e186-232 and lnkH2B-H2A.Z formed a 1:1 complex.
(A-B) Stereo diagrams of the electron density map for the Anp32e-H2B-H2A.Z structure.
Supplementary Methods
Data collection and refinement statistics (molecular replacement)
References
- Henikoff S, Furuyama T, Ahmad K. Histone variants, nucleosome assembly and epigenetic inheritance. Trends Genet. 2004;20:320–326. doi: 10.1016/j.tig.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Sarma K, Reinberg D. Histone variants meet their match. Nat Rev Mol Cell Biology. 2005;6:139–149. doi: 10.1038/nrm1567. [DOI] [PubMed] [Google Scholar]
- Kusch T, Workman JL. Histone variants and complexes involved in their exchange. Subcel Biochem. 2007;41:91–109. [PubMed] [Google Scholar]
- Ray-Gallet D, Almouzni G. Nucleosome dynamics and histone variants. Essays Biochem. 2010;48:75–87. doi: 10.1042/bse0480075. [DOI] [PubMed] [Google Scholar]
- Banaszynski LA, Allis CD, Lewis PW. Histone variants in metazoan development. Dev Cell. 2010;19:662–674. doi: 10.1016/j.devcel.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan G, Zhu B. Histone variants and epigenetic inheritance. Biochim Biophys Acta. 2012;1819:222–229. doi: 10.1016/j.bbagrm.2011.06.007. [DOI] [PubMed] [Google Scholar]
- Zlatanova J, Thakar A. H2A.Z: view from the top. Structure. 2008;16:166–179. doi: 10.1016/j.str.2007.12.008. [DOI] [PubMed] [Google Scholar]
- Guillemette B, Bataille AR, Gévry N, et al. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Bio. 2005;3:e384. doi: 10.1371/journal.pbio.0030384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisner RM, Hartley PD, Meneghini MD, et al. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell. 2005;123:233–248. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Roberts DN, Cairns BR. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell. 2005;123:219–231. doi: 10.1016/j.cell.2005.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Pattenden SG, Lee D, et al. Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc Natl Acad Sci USA. 2005;102:18385–18390. doi: 10.1073/pnas.0507975102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert I, Mavrich TN, Tomsho LP, et al. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–576. doi: 10.1038/nature05632. [DOI] [PubMed] [Google Scholar]
- Mavrich TN, Jiang C, Ioshikhes IP, et al. Nucleosome organization in the Drosophila genome. Nature. 2008;453:358–362. doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D, Coleman-Derr D, Ballinger T, Henikoff S. Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature. 2008;456:125–129. doi: 10.1038/nature07324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Jin C, Zang C, Wei G, et al. H3.3/H2A.Z double variant-containing nucleosomes mark 'nucleosome-free regions' of active promoters and other regulatory regions. Nat Genet. 2009;41:941–945. doi: 10.1038/ng.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov M, Amrichova J, Parker BJ, et al. Histone H2A.Z inheritance during the cell cycle and its impact on promoter organization and dynamics. Nat Struct Mol Biol. 2012;19:1076–1083. doi: 10.1038/nsmb.2424. [DOI] [PubMed] [Google Scholar]
- Ku M, Jaffe JD, Koche RP, et al. H2A.Z landscapes and dual modifications in pluripotent and multipotent stem cells underlie complex genome regulatory functions. Genome Biol. 2012;13:R85. doi: 10.1186/gb-2012-13-10-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Cui K, Northrup D, et al. H2A.Z facilitates access of active and repressive complexes to chromatin in embryonic stem cell self-renewal and differentiation. Cell Stem Cell. 2013;12:180–192. doi: 10.1016/j.stem.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Markoulaki S, Levine SS, et al. H2AZ is enriched at polycomb complex target genes in ES cells and is necessary for lineage commitment. Cell. 2008;135:649–661. doi: 10.1016/j.cell.2008.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Keogh MC, Datta N, et al. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol Cell. 2003;12:1565–1576. doi: 10.1016/s1097-2765(03)00497-0. [DOI] [PubMed] [Google Scholar]
- Kobor MS, Venkatasubrahmanyam S, Meneghini MD, et al. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Bio. 2004;2:E131. doi: 10.1371/journal.pbio.0020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi G, Shen X, Landry J, et al. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- Ruhl DD, Jin J, Cai Y, et al. Purification of a human SRCAP complex that remodels chromatin by incorporating the histone variant H2A.Z into nucleosomes. Biochemistry. 2006;45:5671–5677. doi: 10.1021/bi060043d. [DOI] [PubMed] [Google Scholar]
- Choi J, Heo K, An W. Cooperative action of TIP48 and TIP49 in H2A.Z exchange catalyzed by acetylation of nucleosomal H2A. Nucleic Acids Res. 2009;37:5993–6007. doi: 10.1093/nar/gkp660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luk E, Vu ND, Patteson K, et al. Chz1, a nuclear chaperone for histone H2AZ. Mol Cell. 2007;25:357–368. doi: 10.1016/j.molcel.2006.12.015. [DOI] [PubMed] [Google Scholar]
- Radrizzani M, Vilá-Ortiz G, Cafferata EG, et al. Differential expression of CPD1 during postnatal development in the mouse cerebellum. Brain Res. 2001;907:162–174. doi: 10.1016/s0006-8993(01)02351-4. [DOI] [PubMed] [Google Scholar]
- Jiang M, Ma Y, Ni X, et al. Molecular cloning and characterization of a novel human gene ANP32E alias LANPL from human fetal brain. Cytogenet Genome Res. 2002;97:68–71. doi: 10.1159/000064058. [DOI] [PubMed] [Google Scholar]
- Seo SB, McNamara P, Heo S, et al. Regulation of histone acetylation and transcription by INHAT, a human cellular complex containing the set oncoprotein. Cell. 2001;104:119–130. doi: 10.1016/s0092-8674(01)00196-9. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Feng H, Hansen DF, et al. NMR structure of chaperone Chz1 complexed with histones H2A.Z-H2B. Nat Struct Mol Biol. 2008;15:868–869. doi: 10.1038/nsmb.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suto RK, Clarkson MJ, Tremethick DJ, Luger K. Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nat Struct Biol. 2000;7:1121–1124. doi: 10.1038/81971. [DOI] [PubMed] [Google Scholar]
- Andrews AJ, Chen X, Zevin A, Stargell LA, Luger K. The histone chaperone Nap1 promotes nucleosome assembly by eliminating nonnucleosomal histone DNA interactions. Mol Cell. 2010;37:834–842. doi: 10.1016/j.molcel.2010.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Feng H, Zhou BR, et al. Structural basis for recognition of centromere histone variant CenH3 by the chaperone Scm3. Nature. 2011;472:234–237. doi: 10.1038/nature09854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsasser SJ, Huang H, Lewis PW, et al. DAXX envelops a histone H3.3-H4 dimer for H3.3-specific recognition. Nature. 2012;491:560–565. doi: 10.1038/nature11608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CP, Xiong C, Wang M, et al. Structure of the variant histone H3.3-H4 heterodimer in complex with its chaperone DAXX. Nat Struct Mol Biol. 2012;19:1287–1292. doi: 10.1038/nsmb.2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Liu Y, Wang M, et al. Structure of a CENP-A-histone H4 heterodimer in complex with chaperone HJURP. Genes Dev. 2011;25:901–906. doi: 10.1101/gad.2045111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hondele M, Stuwe T, Hassler M, et al. Structural basis of histone H2A-H2B recognition by the essential chaperone FACT. Nature. 2013;499:111–114. doi: 10.1038/nature12242. [DOI] [PubMed] [Google Scholar]
- Wan Y, Chiang JH, Lin CH, et al. Histone chaperone Chz1p regulates H2B ubiquitination and subtelomeric anti-silencing. Nucleic Acids Res. 2010;38:1431–1440. doi: 10.1093/nar/gkp1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Wu T, Fu H, et al. Dense chromatin activates Polycomb repressive complex 2 to regulate H3 lysine 27 methylation. Science. 2012;337:971–975. doi: 10.1126/science.1225237. [DOI] [PubMed] [Google Scholar]
- Zhu B, Zheng Y, Pham AD, et al. Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Mol Cell. 2005;20:601–611. doi: 10.1016/j.molcel.2005.09.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence alignment of human Anp32 family proteins.
Sequence alignment of human H2A.X, H2A and H2A.Z.
Superimposition of crystal structures of Anp32e-H2B-H2A.Z with nucleosomal H2A.Z-H2B in front view (left) and side view (right).
Structural comparison of the H2A.Z αC helix.
Solution stoichiometry and structure rigidity of Anp32e complex (A), Sedimentation experiments indicated that Anp32e186-232 and lnkH2B-H2A.Z formed a 1:1 complex.
(A-B) Stereo diagrams of the electron density map for the Anp32e-H2B-H2A.Z structure.
Supplementary Methods
Data collection and refinement statistics (molecular replacement)