Abstract
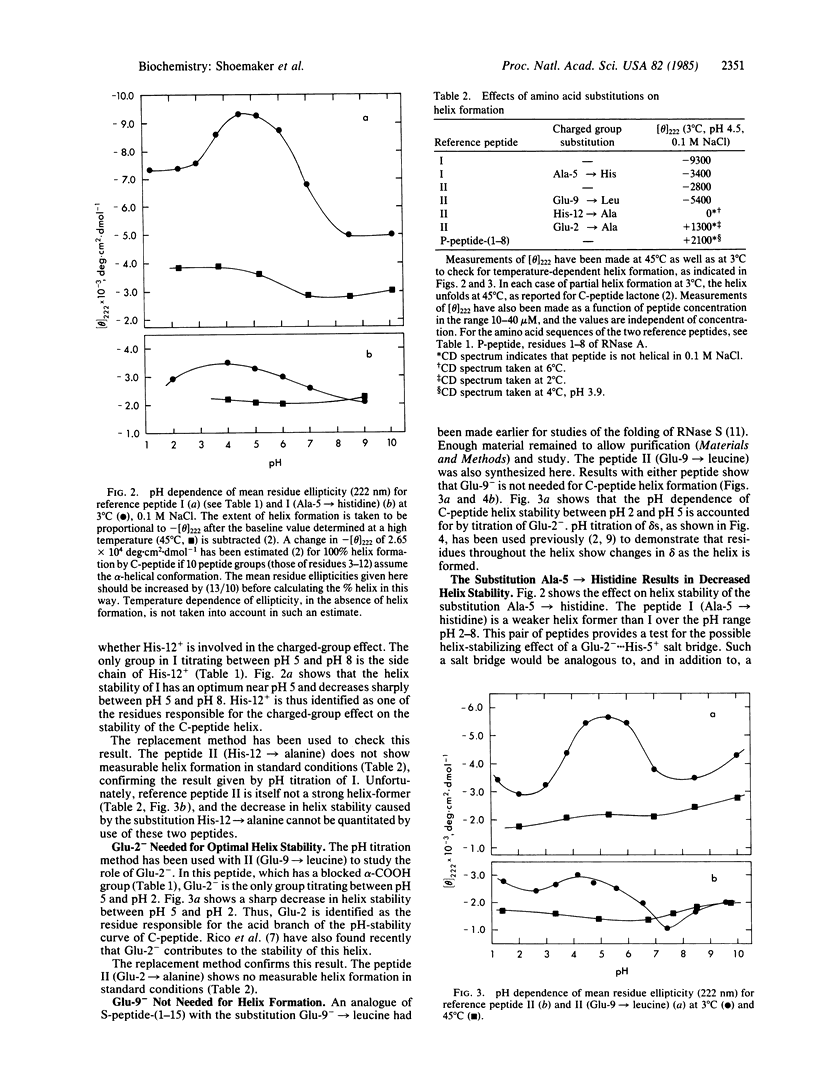
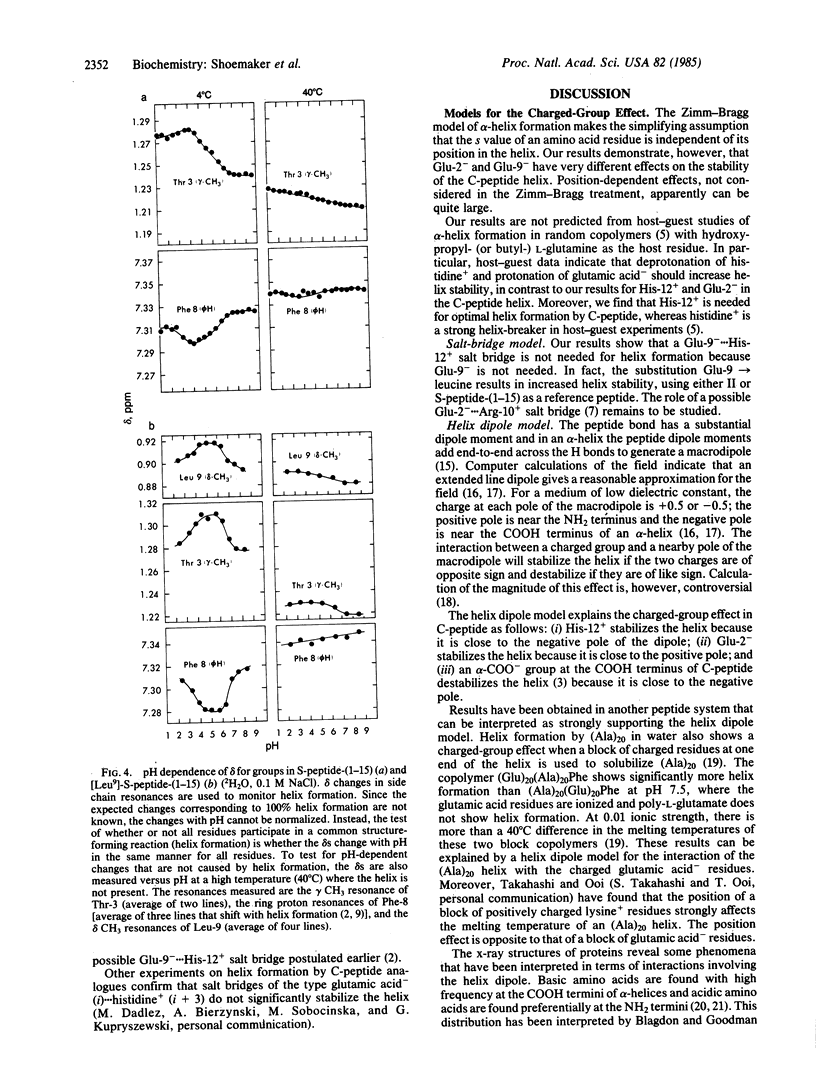
The residues responsible for the pH-dependent stability of the helix formed by the isolated C-peptide (residues 1-13 of ribonuclease A) have been identified by chemical synthesis of analogues and measurement of their helix-forming properties. Each of the residues ionizing between pH 2 and pH 8 has been replaced separately by an uncharged residue. Protonation of Glu-2- is responsible for the sharp decrease in helix stability between pH 5 and pH 2, and deprotonation of His-12+ causes a similar decrease between pH 5 and pH 8. Glu-9- is not needed for helix stability. The results cannot be explained by the Zimm-Bragg model and host-guest data for alpha-helix formation, which predict that the stability of the C-peptide helix should increase when Glu-2- is protonated or when His-12+ is deprotonated. Moreover, histidine+ is a strong helix-breaker in host-guest studies. In proteins, acidic and basic residues tend to occur at opposite ends of alpha-helices: acidic residues occur preferentially near the NH2-terminal end and basic residues near the COOH-terminal end. A possible explanation, based on a helix dipole model, has been given [Blagdon, D. E. & Goodman, M. (1975) Biopolymers 14, 241-245]. Our results are consistent with the helix dipole model and they support the suggestion that the distribution of charged residues in protein helices reflects the helix-stabilizing propensity of those residues. Because Glu-9 is not needed for helix stability, a possible Glu-9-...His-12+ salt bridge does not contribute significantly to helix stability. The role of a possible Glu-2-...Arg-10+ salt bridge has not yet been evaluated. A charged-group effect on alpha-helix stability in water has also been observed in a different peptide system [Ihara, S., Ooi, T. & Takahashi, S. (1982) Biopolymers 21, 131-145]: block copolymers containing (Ala)20 and (Glu)20 show partial helix formation at low temperatures, pH 7.5, where the glutamic acid residues are ionized. (Glu)20(Ala)20Phe forms a helix that is markedly more stable than (Ala)20(Glu)20Phe. The results are consistent with a helix dipole model.
Full text
PDF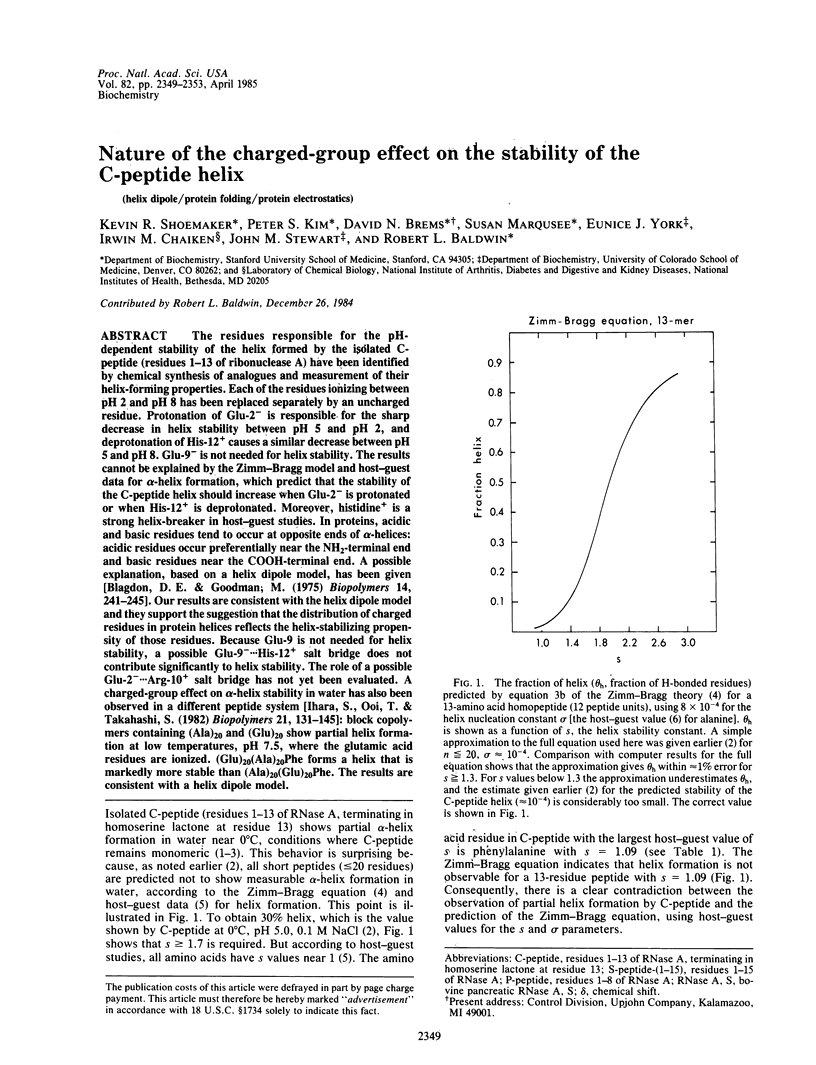


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bierzynski A., Kim P. S., Baldwin R. L. A salt bridge stabilizes the helix formed by isolated C-peptide of RNase A. Proc Natl Acad Sci U S A. 1982 Apr;79(8):2470–2474. doi: 10.1073/pnas.79.8.2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagdon D. E., Goodman M. Letter: Mechanisms of protein and polypeptide helix initiation. Biopolymers. 1975 Jan;14(1):241–245. doi: 10.1002/bip.1975.360140118. [DOI] [PubMed] [Google Scholar]
- Brown J. E., Klee W. A. Helix-coil transition of the isolated amino terminus of ribonuclease. Biochemistry. 1971 Feb 2;10(3):470–476. doi: 10.1021/bi00779a019. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Conformational parameters for amino acids in helical, beta-sheet, and random coil regions calculated from proteins. Biochemistry. 1974 Jan 15;13(2):211–222. doi: 10.1021/bi00699a001. [DOI] [PubMed] [Google Scholar]
- Dunn B. M., Chaiken I. M. Relationship between alpha-helical propensity and formation of the ribonuclease-S complex. J Mol Biol. 1975 Jul 15;95(4):497–511. doi: 10.1016/0022-2836(75)90313-7. [DOI] [PubMed] [Google Scholar]
- Hol W. G., Halie L. M., Sander C. Dipoles of the alpha-helix and beta-sheet: their role in protein folding. Nature. 1981 Dec 10;294(5841):532–536. doi: 10.1038/294532a0. [DOI] [PubMed] [Google Scholar]
- Hol W. G., van Duijnen P. T., Berendsen H. J. The alpha-helix dipole and the properties of proteins. Nature. 1978 Jun 8;273(5662):443–446. doi: 10.1038/273443a0. [DOI] [PubMed] [Google Scholar]
- Kim P. S., Baldwin R. L. A helix stop signal in the isolated S-peptide of ribonuclease A. 1984 Jan 26-Feb 1Nature. 307(5949):329–334. doi: 10.1038/307329a0. [DOI] [PubMed] [Google Scholar]
- Kim P. S., Bierzynski A., Baldwin R. L. A competing salt-bridge suppresses helix formation by the isolated C-peptide carboxylate of ribonuclease A. J Mol Biol. 1982 Nov 25;162(1):187–199. doi: 10.1016/0022-2836(82)90168-1. [DOI] [PubMed] [Google Scholar]
- Kuwajima K., Baldwin R. L. Nature and locations of the most slowly exchanging peptide NH protons in residues 1 to 19 of ribonuclease S. J Mol Biol. 1983 Sep 5;169(1):281–297. doi: 10.1016/s0022-2836(83)80184-3. [DOI] [PubMed] [Google Scholar]
- POTTS J. T., Jr, YOUNG D. M., ANFINSEN C. B. Reconstitution of fully active RNase S by carboxypeptidase-degraded RNase S-peptide. J Biol Chem. 1963 Jul;238:2593–2594. [PubMed] [Google Scholar]
- Rico M., Gallego E., Santoro J., Bermejo F. J., Nieto J. L., Herranz J. On the fundamental role of the Glu 2- ... Arg 10+ salt bridge in the folding of isolated ribonuclease A S-peptide. Biochem Biophys Res Commun. 1984 Sep 17;123(2):757–763. doi: 10.1016/0006-291x(84)90294-8. [DOI] [PubMed] [Google Scholar]
- Rico M., Nieto J. L., Santoro J., Bermejo F. J., Herranz J., Gallego E. Low-temperature 1H-NMR evidence of the folding of isolated ribonuclease S-peptide. FEBS Lett. 1983 Oct 17;162(2):314–319. doi: 10.1016/0014-5793(83)80779-0. [DOI] [PubMed] [Google Scholar]
- Rogers N. K., Sternberg M. J. Electrostatic interactions in globular proteins. Different dielectric models applied to the packing of alpha-helices. J Mol Biol. 1984 Apr 15;174(3):527–542. doi: 10.1016/0022-2836(84)90334-6. [DOI] [PubMed] [Google Scholar]
- Sheridan R. P., Levy R. M., Salemme F. R. alpha-Helix dipole model and electrostatic stabilization of 4-alpha-helical proteins. Proc Natl Acad Sci U S A. 1982 Aug;79(15):4545–4549. doi: 10.1073/pnas.79.15.4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada A. The alpha-helix as an electric macro-dipole. Adv Biophys. 1976:1–63. [PubMed] [Google Scholar]



