Abstract
To explain the frequency and distribution of heteromorphic sex chromosomes in the lizard genus Anolis we compared the relative roles of sex chromosome conservation vs. turnover of sex determining mechanisms. We used model based comparative methods to reconstruct karyotype evolution and the presence of heteromorphic sex chromosomes onto a newly generated Anolis phylogeny. We found that heteromorphic sex chromosomes evolved multiple times in the genus. Fluorescent in situ hybridization (FISH) of repetitive DNA showed variable rates of Y chromosome degeneration among Anolis species and identified previously undetected, homomorphic sex chromosomes in two species. We confirmed homology of sex chromosomes in the genus by performing FISH of an X-linked BAC and qPCR of X-linked genes in multiple Anolis species sampled across the phylogeny. Taken together, these results are consistent with long-term conservation of sex chromosomes in the group. Our results pave the way to address additional questions related to Anolis sex chromosome evolution and describe a conceptual framework that can be used to evaluate the origins and evolution of heteromorphic sex chromosomes in other clades.
Keywords: cytogenetics, homology, phylogeny, reptile, sex determination, X chromosome
Introduction
The discovery that the males and females of many species have morphologically distinct (heteromorphic) sex chromosomes provided some of the first direct evidence for the nature of inheritance and initiated more than a century of work on sex chromosome divergence and its consequences (Wilson 1905; Morgan 1911). Interest in how and why sex chromosomes diverge has intensified with the recognition that sex chromosomes play a central role in core evolutionary processes such as speciation, sex specific adaptation, and genetic conflict (Presgraves 2008; Bachtrog et al. 2009; Meiklejohn and Tao 2010; Zhou and Bachtrog 2012).
Theoretical and empirical work suggests that sex chromosomes can exhibit strikingly predictable divergence patterns over time. The prevailing model of sex chromosome evolution was developed primarily from empirical observations in a limited number of well-studied model taxa with heteromorphic sex chromosomes (e.g., Drosophila and mammals). In this model, degeneration of the heterogametic sex chromosome (the Y or W) follows evolution of a sex determining locus and reduced recombination in the vicinity of this locus (Muller 1914; Ohno 1967; Rice 1996; Charlesworth and Charlesworth 2000; Graves 2006). Among vertebrates, this model goes a long way toward explaining the presence of the heteromorphic sex chromosomes in most mammals, birds, and snakes. Most vertebrate species, however, including the vast majority of fish, amphibians, and lizards do not have heteromorphic sex chromosomes (Hillis and Green 1990; Devlin and Nagahama 2002; Ezaz et al. 2009b). The lack of morphologically distinct sex chromosomes in these species challenges the simple model of sex chromosome degeneration and suggests that frequent sex chromosome turnover or related processes can limit sex chromosome degeneration, potentially influencing the downstream evolutionary consequence of this differentiation on speciation and other evolutionary phenomena (Grossen et al. 2011; Stöck et al. 2011). Here we use phylogenetic, cytogenetic and molecular approaches to test alternative models of sex chromosome evolution in a species-rich lizard genus.
The classic model of sex chromosome evolution
Sex chromosomes evolve from autosomes (Muller 1914; Ohno 1967). The first step in their evolution is thought to be the appearance of a gene, via mutation or translocation, that controls the sex determination pathway (Rice 1996; Charlesworth and Charlesworth 2000; Graves 2006). Recombination is then restricted around the sex-determining locus and linked sexually antagonistic alleles (Bergero and Charlesworth 2008). Regions of reduced recombination then expand as the biased inheritance of sex chromosomes causes natural selection to favor the evolution or recruitment of sexually antagonistic loci (Rice 1987; Charlesworth et al. 2005; Bergero and Charlesworth 2008). Lack of recombination can ultimately lead to the degeneration of the heterogametic sex chromosome either by drift or the inability to purge deleterious alleles fixed by selection at linked loci (Rice 1987; Charlesworth et al. 2005; Bergero and Charlesworth 2008).
In general, heterogametic sex chromosomes show a net loss of genes, an accumulation of sex-specific alleles, and a gain of repetitive DNA sequences. This degeneration is expected to result in the morphological differences between the X and Y (or Z and W) chromosomes that are cytogenetically diagnosable (Graves 2006, 2008). Divergence in the gene content of X and Y chromosomes provides an opportunity for sex-specific and sexually antagonistic adaptation to occur on these chromosomes. The hemizygosity of the X (or Z) that results from this degeneration is also thought to explain the disproportionate role of sex chromosomes in speciation. Haldane’s rule, that heterogametic inter-specific hybrids suffer negative fitness consequences more often than the homogametic sex, is one of the most universal patterns in evolutionary biology (Haldane 1922). The prevailing explanation for this pattern, the dominance theory, states that if genetic incompatibilities between species are mostly recessive they will be masked if they occur on autosomes but will contribute to the reduced fitness of males if they occur on the X (Turelli and Orr 1995). Sex chromosome divergence may also lead to serious biological challenges, including dysfunction caused by dosage differences in the X-linked or W-linked genes between the two sexes. Although many taxa with strongly differentiated sex chromosomes have evolved efficient mechanisms for dosage compensation, the challenge of evolving such mechanisms may provide strong selective pressure against sex chromosome degeneration (Adolfsson and Ellegren 2013).
Applied broadly, the classic model of sex chromosome degeneration predicts that all species with genetic sex determination should eventually evolve heteromorphic sex chromosomes, or even lose the heterogametic chromosome (Y or W) altogether (Graves 2006, 2008). Support for this prediction can be found in several well-studied vertebrate groups including mammals, birds, and snakes. Species within each of these clades possess homologous sex chromosomes and exhibit varying degrees of sex chromosome heteromorphy. However, even some members of these well-studied groups possess homomorphic sex chromosomes that appear to have been maintained over relatively long periods of evolutionary time. For example, ratites (ostriches and their kin) and boas and pythons have homomorphic sex chromosomes while most other bird and snake species have heteromorphic sex chromosomes (Shetty et al. 1999; Matsubara et al. 2006; Nanda et al. 2008). Homomorphic sex chromosomes in these clades resemble the ancestral autosomes and contain fewer repetitive sequences and possess more functional genes than their heteromorphic orthologs in related species (Matsubara et al. 2006; O’Meally et al. 2010). Heteromorphic sex chromosomes are relatively rare in most other vertebrate lineages including fish, amphibians and lizards (Hillis and Green 1990; Devlin and Nagahama 2002; Ezaz et al. 2009b). In these clades a few species with heteromorphic sex chromosomes are scattered among a majority of species generally lacking chromosomal heteromorphism. Understanding sex chromosome evolution in these lineages is critical to understanding whether the classic model, and the evolutionary processes it explains, apply broadly to most vertebrates, or just to a few select lineages.
Explaining persistence of homomorphic sex chromosomes
Two hypotheses, which are not mutually exclusive, can explain the widespread occurrence of homomorphic sex chromosomes observed across most vertebrate species (Stöck et al. 2011). The first hypothesis, which we refer to as the “conservation” hypothesis, because it involves conservation of sex chromosome homology among related species, posits variable rates of degeneration of the heterogametic sex chromosomes. Under this hypothesis, variation in sex chromosome heteromorphism can be caused by a variety of processes that either change the size of sex chromosomes or slow the rate of degeneration. Occasional recombination between the X and Y in certain frog species, for example, can “refresh” the heterogametic sex chromosome and slow degeneration (Perrin 2009; Guerrero et al. 2012). In primates, meanwhile, sex chromosome degeneration may be slowed by intrachromosomal gene conversion among duplicated genes on the Y chromosome that counteracts degeneration due to a lack of homologous recombination (Rozen et al. 2003; Connallon and Clark 2010). The conservation hypothesis is the preferred model explaining the evolution of sex chromosomes in birds and snakes (Shetty et al. 1999; Matsubara et al. 2006).
An alternative to the conservation hypothesis is the “turnover” hypothesis, in which newly evolved sex chromosomes are homomorphic because they are too young for degeneration to have rendered them cytogenetically distinct (Charlesworth et al. 2005; Volff et al. 2007). Turnover has been observed among several closely related fish and amphibian species (Takehana et al. 2007; Tanaka et al. 2007; Volff et al. 2007; Henning et al. 2008; Ross et al. 2009; Miura et al. 2012) and inferred at higher taxonomic levels in lizards, amphibians and fish (Hillis and Green 1990; Mank et al. 2006; Ezaz et al. 2009b; Gamble 2010). Indeed, new sex determining mechanisms appear to have evolved repeatedly across the vertebrate phylogeny and the sex chromosomes of many vertebrate species are not homologous (Mank et al. 2006; O’Meally et al. 2012). It is less clear, however, whether turnover is broadly responsible for the commonly observed variation in sex chromosome heteromorphy among closely related species.
We investigate conservation and turnover of sex chromosomes in a diverse and well-studied lizard genus (Anolis) comprised mostly of species with homomorphic sex chromosomes. Anoles are ideally suited for this work for several reasons. First, all species studied to date are characterized by genetic sex determination (Gorman 1973; Viets et al. 1994). Second, preliminary evidence suggests a role for Anolis sex chromosomes in the evolution of reproductive isolation (Webster 1977). Third, karyotypes are available for one quarter of the nearly 400 recognized species of Anolis lizards; 2/3 of species have homomorphic sex chromosomes and those that do not have male heterogamety, either XY or XXY chromosomes (Gorman and Atkins 1966, 1968, 1969; Olmo 2005). Fourth, the recently completed genome of A. carolinensis recovered evidence for cryptic or homomorphic sex chromosomes and provides a foundation for identification of sex chromosome specific markers that are necessary to assess sex chromosome homology (Alföldi et al. 2011).
The conservation and turnover hypotheses make a number of specific predictions that can be tested with phylogenetic, cytogenetic, and genetic analyses of Anolis. First, the conservation hypothesis predicts that transitions will only occur from the homomorphic to the heteromorphic state, and not vice versa, whereas the turnover hypothesis predicts that transitions will occur in both directions. We test this hypothesis using phylogenetic comparative methods. Second, the conservation hypothesis predicts sex chromosome homology across species whereas the turnover hypothesis predicts that sex chromosomes will not be homologous. We test this hypothesis using cytogenetic and molecular methods. Finally, we test whether changes in chromosome number co-occur with changes in sex chromosome complement (e.g. homomorphic, XY or XXY). Our findings bear not only on the processes at play in Anolis sex chromosome evolution but also provide a conceptual framework to evaluate the origins and evolution of sex chromosomes in other clades. They also help explain the widespread variation in sex chromosome heteromorphy observed throughout vertebrates.
Materials and Methods
Phylogenetic Analyses
We generated a new phylogeny of Anolis species using ~1500bp of the mitochondrial genome including the entire NADH Dehydrogenase subunit 2 (ND2), 5 flanking tRNAs and the 5′ end of cytochrome c oxidase (COI) gene. By combining data from recently published Anolis datasets (Mahler et al. 2010; Rabosky and Glor 2010; Castañeda and deQueiroz 2011) with additional sequences from Genbank and newly generated sequences for ten species we obtain a dataset that includes 216 species. Previously unsampled species were amplified and sequenced following previously published protocols (Glor et al. 2001), and subsequently assembled and edited using GENEIOUS v5.4 (Drummond et al. 2011). Coding regions were aligned by eye and tRNAs were aligned using a secondary structural model (Kumazawa and Nishida 1993). In total, we compiled sequence data for 216 Anolis species plus three outgroup taxa, with one sequence for each species (Supplementary Table 1).
We estimated phylogenies using the Bayesian Metropolis-coupled Markov Chain Monte Carlo (MCMCMC) approach implemented in MrBayes v3.1.2 (Ronquist and Huelsenbeck 2003). We used a heating parameter of 0.01 and partitioned data by codon position, plus an additional partition for tRNAs. We selected models of molecular evolution for each partition by comparing Akaike information criterion (AIC) scores in JMODELTEST v0.1.1 (Posada 2008). We evaluated convergence using two strategies. First, we used TRACER v1.5 (Rambaut and Drummond 2007) to identify a plateau of likelihood scores and posterior estimates of model parameters for the two independent runs. Second, we used AWTY (Nylander et al. 2008), to examine the cumulative posterior probabilities of tree bipartitions using the “cumulative” utility, and compared bipartition posterior probabilities between independent analyses with the “compare” utility. We defined burn-in as the point at which posterior probabilities in both runs reached a stationary distribution. Final MrBayes analyses were run for 70 million generations, with the first 35 million discarded as burn-in and the remaining trees used to generate a consensus tree and posterior probabilities of nodes calculated as the frequency of a node among the posterior sample of trees.
To simultaneously estimate the topology and relative timing of the anole diversification, we conducted a Bayesian uncorrelated relaxed clock analysis in BEAST (Drummond et al. 2012). We constrained Anolis monophyly and set the root age to an arbitrary uniform distribution from 95 to 105. We used a Yule speciation topology prior with the MrBayes consensus tree as the starting tree. We performed four independent analyses each for 50 million generations, sampling trees every 10,000 generations. We assessed convergence and concordance among these four analyses using TRACER and AWTY, as described above. We removed trees from the first 25 million generations as burn-in, combined the output of independent runs and calculated the maximum clade credibility (MCC) chronogram for the remaining posterior sample (10,000 trees) using the LOGCOMBINER and TREEANNOTATOR utilities included in BEAST (Drummond et al. 2012).
Chromosomal evolution
We conducted comparative analyses on the posterior distribution of trees inferred by BEAST after pruning trees to include only the 98 taxa for which DNA sequence data, sex chromosome complement, and chromosome number were available (Supplementary Table 1).
We collected karyotypic data for anole species from ChromoRep, a database of karyotypic data for over 2,000 reptiles (Olmo 2005). We cross referenced data from ChromoRep with the original sources and in the two instances where they disagreed we used data from the original publication. We also supplemented this dataset with records from an additional unpublished account (Lieb 1981). For each species we recorded chromosome count as the haploid chromosome number of females. We coded sex chromosome complement as homomorphic or heteromorphic (XY or XXY). Species with morphologically indistinguishable sex chromosomes, so-called “cryptic” or homomorphic sex chromosomes (sensu Ezaz et al. 2005), such as the recently described A. carolinensis XY system (Alföldi et al. 2011), were coded as homomorphic.
We inferred ancestral states of sex chromosome complement both as a multistate character (homomorphic, XY and XXY) and as a binary character (heteromorphic or homomorphic) using a Markov Chain Monte Carlo approach and the BayesMultiState function in BAYESTRAITS v1.0 (Pagel et al. 2004). To account for uncertainty in estimates of topology and branch length we performed these analyses on a set of 10,000 trees drawn from the stationary posterior distribution of trees inferred by BEAST. Trees are archived in Dryad (doi:10.5061/dryad.dp848). We performed preliminary analyses to optimize three parameters (1) the per generation deviation in rates (ratedev), (2) the distribution shape and range of prior values for the reversible jump hyperprior (rjhp) and (3) the number of generations required for the MCMC estimates to achieve stationarity. From these preliminary analyses we determined a ratedev equal to 0.05 and rjhp priors drawn from an exponential distribution between 0 and 30 achieved an average proposal acceptance rate of 18% (range 0–42). Parameter estimates converged prior to 100,000 generations, therefore in our final analysis we conservatively discarded the first 200,000 generations as burn-in. We inferred ancestral states for all nodes and ran a final analysis using these parameters for 20 million generations.
Using binary state reconstructions we evaluated three models of sex chromosome evolution: (1) a model with equal rates of transition from heteromorphic to homomorphic sex chromosomes and from homomorphic to heteromorphic; (2) a model with independent rates for each type of change; and (3) a model with heteromorphic to homomorphic transitions fixed at zero. The first and second models, which include a non-zero rate of transition from heteromorphic to homomorphic sex chromosomes, are expected to be incompatible with the conservation hypothesis while the third model is incompatible with the turnover hypothesis. We evaluated the fit of these alternative models by calculating Bayes Factors: 2(log[harmonic mean likelihood model 1)] −log[harmonic mean likelihood model 2]).
Correlation Test
Heteromorphic sex chromosomes, including XY and XXY complements, occur in a variety of Anolis lineages. The presence of XXY sex chromosome complements suggests a possible role for the evolution of sex chromosome heteromorphism by fusion or fission with autosomes. We used the repeated independent evolution of sex chromosome heteromorphy in anoles to test for a relationship between chromosome number evolution and the evolution of sex chromosome heteromorphy. Specifically, we inferred ancestral states of chromosome number to test whether changes in sex chromosome heteromorphism and changes in chromosome number co-occur on the same branches of the anole phylogeny.
We used CHROMEVOL v1.3 (Mayrose et al. 2010), a maximum likelihood based approach specifically designed for ancestral state reconstruction of chromosomal data, to infer ancestral states for chromosome number. We evaluated two models of chromosome evolution. The first model estimates a constant but independent rate of chromosome loss and gain – model M0 sensu Mayrose et al. (2010); the second model estimates linear rates of chromosome loss and gain that are correlated with the total number chromosomes present – model M5 sensu Mayrose et al. (2010). The suitability of these models was evaluated using AIC scores and Akaike weights. We did not evaluate models that involve polyploidization as no genome duplication events appear to have occurred in any Anolis species.
We used a two-tailed Fisher’s Exact test to assess whether changes in sex chromosome complement, e,g, transitions between homomorphic and heteromorphic states or between XY and XXY, significantly coincide with changes in chromosome number. Using the maximum likelihood model inferred by CHROMEVOL, we filtered all branches for those with an inferred change in chromosome number greater than 0.5. To test for a correlation between change in sex chromosome complement and transitions in chromosome number we sorted branches into those with chromosome number transitions and those without, and branches that involve a change in sex chromosome complement and those that do not.
Cytogenetics
We examined chromosomes from five Anolis species to evaluate two distinct and related hypotheses. First, we tested whether repetitive sequences associated with Y chromosome degeneration in other vertebrate taxa occur on the Y chromosomes of anoles. Second, we assessed X chromosome homology among each of these five Anolis species using both fluorescent in situ hybridization (FISH) and quantitative PCR (qPCR).
We prepared chromosome spreads from both males and females of five Anolis species (A. carolinensis; A. sagrei; A. grahami; A. lineatopus; and A. distichus) from fibroblast cultures established from tail clips (Ezaz et al. 2008; Main et al. 2012). Cells were arrested in metaphase using vinblastine sulphate (1 mg/ml) for 2–3 hours. We collected cells after trypsin digestion and incubated them in a hypotonic solution (0.07 M KCl) for 20 min at 37°C. After hypotonic treatment, cells were centrifuged and fixed through a series of five washes in methanol:acetic acid (3:1). Cell suspensions were dropped onto clean glass slides, allowed to air dry, dehydrated in an ethanol series (70%, 95%, 100%) and stored at −80°C until used.
Testing Y Chromosome Degeneration
To test the hypothesis that some degree of Y chromosome degeneration occurs in anoles, we hybridized a GATA minisatellite repeat onto metaphase spreads. The GATA repeat, also called the Bkm satellite repeat, is one of several repetitive sequences that has been shown to accumulate on the heterogametic sex chromosomes of multiple animal species and is a good candidate marker for estimating the amount of molecular divergence between the X and Y chromosomes within species (Singh et al. 1980; Nanda et al. 1990; Singh et al. 1994; O’Meally et al. 2010). (GATA)n probes were generated by PCR in the absence of template DNA (Ijdo et al. 1991) using (GATA)7 and (TATC)7 primers. Probes were labeled via nick translation with Chromatide/Alexa Fluor fluorescently labeled dUTP 488-5 (Life Technologies); excitation 490nm, emission 520nm. Sizes of the nick translated fragments were checked by electrophoresis on a 1% TBE gel. Labeled DNA was ethanol precipitated and resuspended in 100uL hybridization buffer (Ezaz et al. 2005), denatured at 72°C for 10 minutes and snap-cooled on ice for five minutes. We added 20 uL of probe to each slide, affixed a cover slip using rubber cement, heated slides again to 72°C for 5 minutes and incubated overnight at 37°C. Slides were washed once at 60°C in 0.4%SSC, 0.3% Igepal CA-630 (Sigma Aldrich) for two minutes followed by a second two minute wash in 2%SSC, 0.1% Igepal CA-630 at room temperature. Slides were dehydrated in an ethanol series (70%, 95%, 100%) and air dried. We stained slides with 4,6-diamidino-2-phenylindole (DAPI) and mounted a cover slip using Permafluor (Lab Vision). Fluorescent signals were visualized on a Zeiss Imager Z1 microscope using a Zeiss MRm camera. Images were captured using Zeiss Axiovision software.
Testing X Chromosome Homology
We used two methods to test for homology of the X chromosome across Anolis. First, we used FISH to ask whether an X-linked bacterial artificial chromosome (BAC) identified by the A. carolinensis genome project maps to male and female metaphase spreads in five Anolis species in a pattern consistent with a single origin of male heterogamety in the genus. Second, we used qPCR of autosomal and X linked loci in males and females of nine species to test whether expected amounts of DNA generated by this procedure are consistent with the presence of targeted loci on autosomes or sex chromosomes. Male genomic DNA should have half the quantity of X-linked genes as measured by qPCR compared to females since males have only one copy of the X chromosome. Species with sex chromosomes lacking homology to A. carolinensis sex chromosomes should show no sex specific biases in the quantification of both of these X-linked genes.
FISH
The X-linked BAC, 206M13 - CHORI318, is localized to the Anolis carolinensis X chromosome, and FISH of this BAC to A. carolinensis metaphase spreads confirmed that it is present in two copies in females and one copy in males (Alföldi et al. 2011). In addition, the sequenced ends of 206M13 (NCBI Clone ID: 29314537) correspond to an unmapped linkage group, GL343282, which contains fragments of four protein coding genes: snap29, serpind1, pi4ka, and cltcl1. Orthologs of these genes occur on chicken chromosome 15, which is syntenic with the A. carolinensis X chromosome (Alföldi et al. 2011). All A. carolinensis genome data were referenced using draft assembly AnoCar2.0 on the UCSC Genome Browser (Meyer et al. 2013) and NCBI assembly database (Wheeler et al. 2007; Benson et al. 2010).
BAC DNA was labeled by nick translation (Nick Translation Kit - Abbott Molecular) using Orange - 552 dUTP (Enzo Life Science); excitation 552nm, emission 576nm. The labeled DNA was precipitated with COT-1 DNA, sheared A. carolinensis genomic DNA, sodium acetate and 95% ethanol, then dried and resuspended in 50% formamide hybridization buffer. The probe/hybridization buffer mix were denatured at 73°F and hybridized onto the slide for 24 hours at 37°. After hybridization, the FISH slides were washed in a 2xSSC solution at 72° for 15 seconds, and counterstained with DAPI stain. Fluorescent signals were visualized on an Olympus BX61 microscope workstation (Applied Spectral Imaging, Vista, CA) with DAPI and Texas Red filter sets. Images were captured using an interferometer-based CCD cooled camera (ASI) and BandView ASI software.
qPCR
FISH of an X chromosome BAC allowed us to determine sex chromosome homology among Anolis species, but we were limited with this approach to species where we had established cell lines for cytogenetics. Failure to sample the full breadth of Anolis diversity, particularly species from the basal Dactyloa clade, meant that any inferences made from cytogenetic results would not necessarily apply to the entire genus. We therefore used qPCR to expand our taxon sampling and screen additional Anolis species for genomic copy number of two X-linked genes in A. carolinensis.
We performed qPCR using two autosomal genes: kank1 (chromosome 2) and ngfb (chromosome 4); and two X-linked genes: pi4ka, and cltcl1. ngfb was used as the reference in all experiments. The two X-linked genes occur on the same BAC used to perform FISH experiments, 206M13 - CHORI318. Selecting genes from this BAC made results from the FISH experiments and qPCR results broadly comparable as they evaluate similar genic content.
We conducted qPCR on two to three individuals of each sex (see Supplementary Table 2) for nine Anolis species. This included the five species for which we had cytogenetic data to verify that qPCR could replicate the BAC FISH results. qPCR was performed using FastStart SYBR green (Roche) on an Eppendorf Realplex2 Mastercycler. Reactions were conducted in duplicate using 10 ng of genomic DNA in 12ul reactions. PCR primers are listed in Supplemetary Table 3. Cycle conditions involved an initial denaturation of 95°C for 10 minutes followed by 40 cycles of: 95°C for 20 seconds, 52°C for 30 seconds, 72°C for 40 seconds; and a final step to produce a melting curve going from 60°C to 95°C over 15 minutes with fluorescent data measured continuously. Reaction efficiencies were calculated for each gene and each species using a series of five serial dilutions (1/10) in duplicate. Data were analyzed using the comparative quantification approach, which measured the relative amount of a gene in a group of male samples compared to a group of female samples, in REST 2009 software (Pfaffl et al. 2002). Standard error and 95% confidence intervals of the normalized quantification values were calculated with 5,000 bootstrap replicates.
Results
Phylogenetic Analyses
MrBayes and BEAST analyses of our 216 species dataset converged by all metrics. The topology and node support estimated by both analyses are very similar to each other (Supplementary Figure 1) and consistent, at well-supported nodes, with other recently published Anolis phylogenies (Mahler et al. 2010; Rabosky and Glor 2010; Alföldi et al. 2011; Castañeda and deQueiroz 2011).
Sex Chromosome evolution
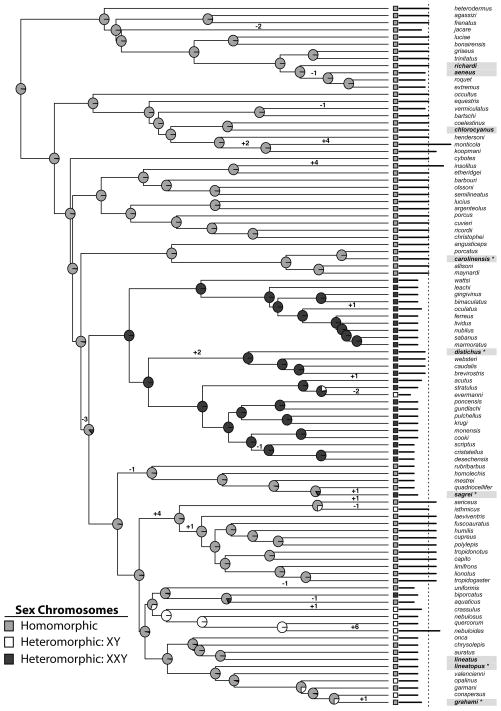
We used phylogenetic comparative methods to infer the ancestral sex chromosome states and test alternative models of chromosome evolution that are compatible with either the conservation or turnover hypotheses. The common ancestor to all anoles was inferred to have homomorphic sex chromosomes, a result suggested by previous studies (Gorman 1973). The ancestral state reconstruction inferred three transitions from homomorphic to XXY, five from homomorphic to XY, a single transition from XXY to XY, and no reversions to a homomorphic state.
Bayes factor analysis favored a model compatible with conservation, with the rate of transition from heteromorphic to homomorphic sex chromosome fixed at 0. The two turnover models: separate rates for heteromorphic to homomorphic and homomorphic to heteromorphic transitions, and a model where all transition rates were equal, were less well supported (Table 1). When both rates were estimated BayesTraits inferred a heteromorphic to homomorphic transition rate an order of magnitude lower than homomorphic to heteromorphic transitions (Table 1).
Table 1.
Comparison of full and constrained models of sex chromosome evolution and parameter estimates. “Het.” is an abbreviation of heteromorphic, “Hom.” is an abbreviation of homomorphic.
| Model | Rate: Het.→ Hom. | Rate: Hom.→ Het. | Harmonic Mean lnL | Bayes Factor (vs. best model) |
|---|---|---|---|---|
| Het→Hom = 0 | 0.0000 | 0.0037 | −34.2427 | 0.0000 |
| Rates different | 0.0004 | 0.0036 | −35.8609 | 3.2364 |
| Equal rates | 0.0032 | 0.0032 | −36.2698 | 4.0542 |
Correlation Test
Ancestral states for chromosome number were inferred using CHROMEVOL. AIC scores favored a constant rates model of chromosome evolution over linear rates. The maximum likelihood reconstruction inferred 1n=18 chromosomes (haploid number of chromosomes) in the ancestor of anoles with 13 gains and 10 losses (summarized in Figure 1, complete reconstruction Supplementary Figure 2). Changes in chromosome number were inferred to occur in terminal branches as well as branches nested deeply within the anole radiation. Three terminal branches are inferred to have undergone large gains in chromosome number (changes in haploid chromosome number between 4–6; Supplementary Figure 2). One of these is on the branch leading to Anolis monticola, which has four more chromosome pairss than its closest relatives and lacks any metacentric macrochromosomes. Webster and colleagues (1972) hypothesized this pattern was the result of Robertsonian fission of all of the ancestor’s macrochromosomes. We report here a similar scenario in Anolis nebuloides, which has 6 more chromosome pairs than its inferred ancestor, lacks metacentric macrochromosomes, and retains the same fundamental number (the number of chromosomal arms) as closely related species. The final instance of a large gain (on the branch leading to Anolis insolitus) cannot be assessed because the distribution of macro- and microchromosomes in this species is not known (Webster et al. 1972).
Figure 1.
Anolis phylogeny generated from ND2 data inferred using BEAST. The phylogeny is pruned to include only species with cytogenetic data. Sex chromosome complement is indicated by colored squares on the tips of the branches. Pie charts on each node indicate the posterior probability of that ancestor having homomorphic (gray), XY (white), or XXY (black) sex chromosomes. Species highlighted in grey were used in qPCR experiments. Species with asterisks (*) were used for cytogenetic experiments. Horizontal bars to the left of species names indicate the number of haploid chromosomes in each species. The vertical dashed line marks 1n=18, the inferred ancestral chromosome complement. Branches inferred to have undergone changes in female 1n chromosome number are labeled with the direction and magnitude of change.
Chromosome number changes occurred in a higher proportion of branches that also experienced a change in sex chromosome complement (3/9) compared to branches that did not experience a change in sex chromosome complement, although the difference was not statistically significant (20/185; p=0.0761; Table 2).
Table 2.
Counts of tree edges sorted into edges with inferred transitions in sex chromosome complement and for the presence of an inferred change in chromosome number on that edge.
| No chromosome number change | Chromosome number change | |
|---|---|---|
| Sex chromosome change | 6 | 3 |
| No sex chromosome change | 165 | 20 |
Cytogenetics
Chromosome number for the five Anolis species from which we obtained metaphase spreads was concordant with published karyotypes (Supplementary Table 1). Chromosome number in A. grahami varied from 2n=29–36. This intraspecific variation in chromosome number was due to disparities in the number of microchromosomes and is largely consistent with previous cytogenetic research on the species showing 2n=30–37 (Blake 1986).
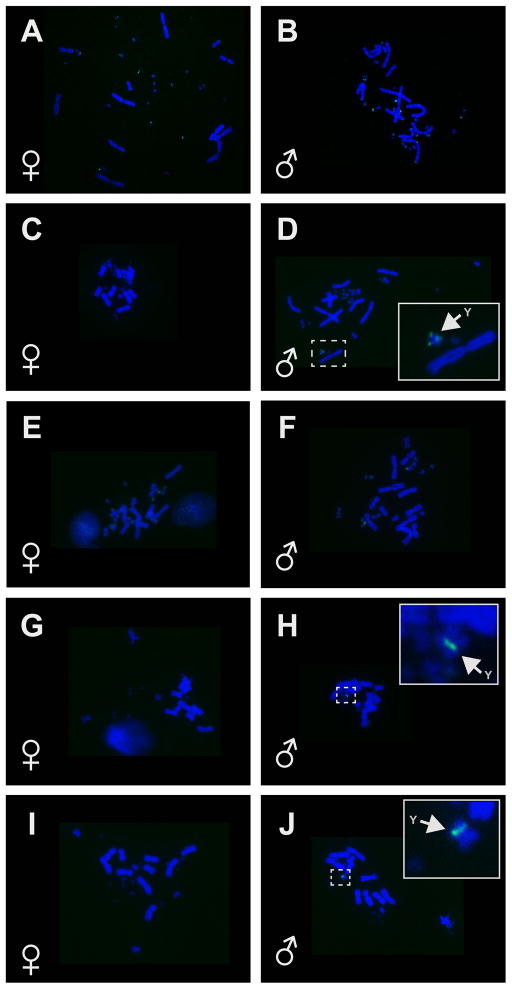
We identified previously unreported Y chromosomes in both A. grahami and A. lineatopus via sexually dimorphic accumulation of GATA repeats. We observed accumulation of GATA repeats in a discrete band on one of the small metacentric chromosomes in males and no GATA hybridization in females (Fig 2 H, J). GATA accumulation also appeared on the A. distichus Y chromosome (Fig 2 D). No sex-specific differences in GATA accumulation were apparent in A. carolinensis or A. sagrei (Fig 2 A, B, E, F).
Figure 2.
Fluorescent in situ hybridization (FISH) of the GATA minisatellite to chromosomes of males and females of 5 Anolis species. A. Anolis carolinensis female (TG1424); B. A. carolinensis male (TG1423); C. A. distichus female (TG1446); D. A. distichus male (TG1445); E. A. sagrei female (TG1463); F. A. sagrei male (TG1459); G. A. lineatopus female (TG1487); H. A. lineatopus male (TG1488); I. A. grahami female (TG1490); J. A. grahami male (TG1493). Sex specific hybridization on the Y chromosome, when identified, is indicated by an arrow. Solid lines indicate magnified views of areas in dashed lines.
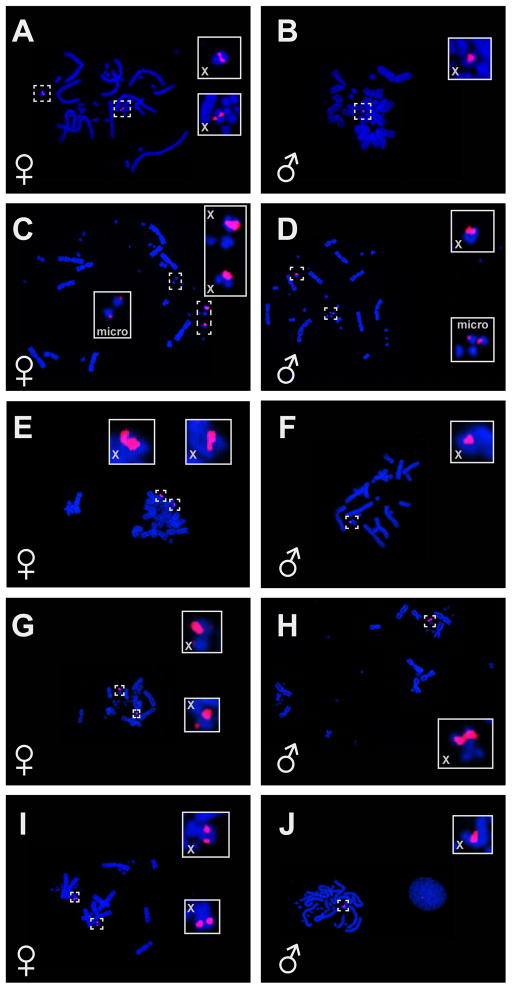
The X-specific BAC 206M13 hybridized to two microchromosomes in A. carolinensis females and one microchromosome in males (Fig 3) replicating previously published results (Alföldi et al. 2011). This sexually dimorphic hybridization pattern was also seen in the other four Anolis species we examined (Fig. 3). One of the XXY species examined, Anolis distichus, showed some additional BAC hybridization near the centromeres of two acrocentric microchromosomes (Fig 3 C, D).
Figure 3.
Fluorescent in situ hybridization (FISH) of an X-linked BAC (206M13) to chromosomes of males and females of five Anolis species. The BAC is found in two copies in females and one copy in males in all sampled species. A. Anolis carolinensis female (TG1424); B. A. carolinensis male (TG1423); C. A. distichus female (TG1446); D. A. distichus male (TG1445); E. A. sagrei female (TG1463); F. A. sagrei male (TG1459); G. A. lineatopus female (TG1487); H. A. lineatopus male (TG1488); I. A. grahami female (TG1491); J. A. grahami male (TG1493). Solid lines indicate magnified views of areas in dashed lines.
qPCR
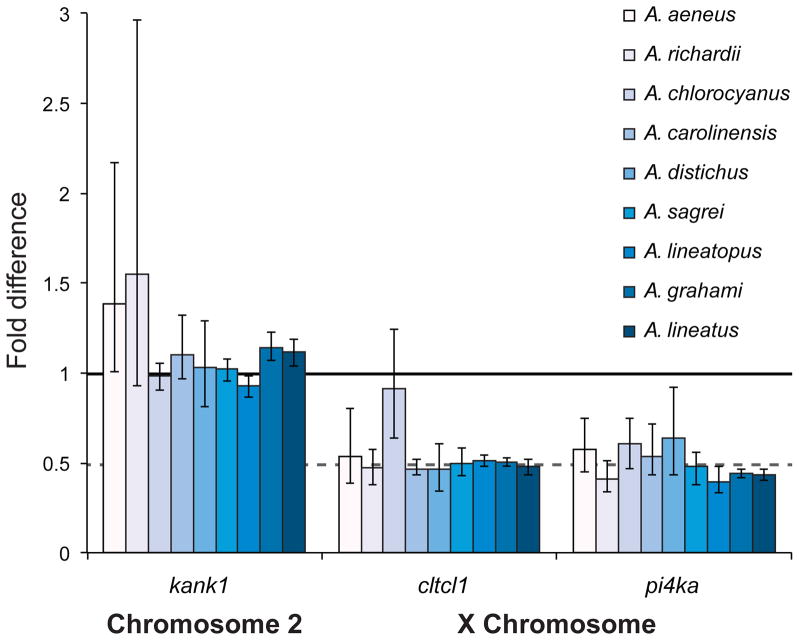
qPCR indicated that two genes known to be X-linked in A. carolinensis, pi4ka, and cltcl1, have quantification values in males that are half that of females for all Anolis species examined except A. chlorocyanus, which was hemizygous only for pi4ka (Figure 4). These X-linked genes also have quantification values in males that are half that of autosomal genes in both sexes. Standard error of quantification values did not encompass 1 for any X-linked genes in males with the exception of cltcl1in A. chlorocyanus (Supplementary Table 4). A value of 1 indicates equal quantification between males and females.
Figure 4.
Relative quantification and standard error of autosomal (kank1) and putative X-linked (cltcl1and pi4ka) genes in male Anolis compared to females using qPCR of genomic DNA. A value of one (solid horizontal line) is equivalent to standard diploid copy number, e.g. single copy autosomal genes and X-linked genes in females, a value of 0.5 (dashed horizontal line) is expected of hemizygous loci, e.g. X-linked genes in males. Species identification, from left to right: A. aeneus, A. richardii, A. chlorocyanus, A. carolinensis, A. distichus, A. sagrei, A. lineatopus, A. grahami, and A. lineatus.
Discussion
Independent origins of sex chromosome heteromorphism and variable rates of Y chromosome degeneration
Comparative phylogenetic analyses reject a turnover model in anoles yet indicate that sex chromosome heteromorphy has evolved repeatedly in multiple independent Anolis lineages. Our discovery of sex chromosome homology across sampled Anolis species further supports the conservation hypothesis in Anolis. The co-occurrence of heteromorphic and homomorphic sex chromosomes in anoles results from variable, lineage-specific rates of Y chromosome degeneration among species rather than repeated turnover of sex chromosomes or sex determining mechanisms. Rates of Y chromosome degeneration can vary due to several processes. Stochasticity related to variation in effective population size, mutation rate, and the fitness effects of individual mutations can affect rates of Y chromosome degeneration (Bachtrog 2008). In addition, Y chromosome degeneration does not appear to occur in a strict linear fashion meaning rates of gene loss may stabilize after an initial burst of degeneration (Bachtrog 2008; Hughes et al. 2012). Indeed, Y chromosome degeneration need not automatically translate to chromosomal heteromorphism at all as revealed by recent research on snake sex chromosomes (Vicoso et al. 2013).
The presence of heteromorphic sex chromosomes raises the question of how Anolis species that possess them mitigate the deleterious effects of Y chromosome degeneration. Turnover in sex chromosomes and sex determining mechanisms is one of several strategies that can remedy the adverse effects of recombination suppression and Y chromosome degeneration (Blaser et al. 2013). The accumulation of deleterious mutations on the nonrecombining portion of the Y chromosome eventually will result in the loss of function of some Y-linked alleles (Charlesworth 1978; Vicoso and Bachtrog 2009). This can reduce expression levels of affected genes by one half in males and presumably lead to a reduction in fitness. The best known strategy to deal with the loss of genes on the degenerating Y chromosome is to evolve some means of dosage compensation to balance expression levels of genes on the X chromosome with the expression levels of autosomal genes in both males and females (Charlesworth 1978; Vicoso and Bachtrog 2009). Global dosage compensation mechanisms are known from several groups with ancient sex chromosome systems and highly degenerate (or absent) Y chromosomes, e.g. Drosophila, mammals and Caenorhabditis elegans (Straub and Becker 2007). In these species dosage compensation operates differently on the X chromosome (increasing, reducing, or silencing expression from X) and involves different molecular mechanisms. Indirect evidence has also been used to suggest the presence of dosage compensation in at least one squamate reptile species (Quinn et al. 2007). Other clades, e.g. birds, snakes, and Lepidoptera, appear to have partial or incomplete dosage compensation where some, but not all, sex-linked genes are dosage compensated (Itoh et al. 2007; Harrison et al. 2012; Adolfsson and Ellegren 2013; Vicoso et al. 2013). Indeed, the failure to evolve an effective dosage compensation mechanism could help prevent sex chromosome degeneration (Adolfsson and Ellegren 2013). Another possible solution to the loss of alleles on the degenerating Y chromosome involves occasional recombination between homologous X and Y genes to “refresh” degenerating genes on the Y, a so-called “fountain of youth” (Perrin 2009; Stöck et al. 2011; Guerrero et al. 2012; Stöck et al. 2013). Fruitful areas for future research include determining whether Anolis have a full or partial dosage compensating mechanism and, if they meet the assumptions of the “fountain of youth” hypothesis, finding evidence for occasional recombination between the X and Y.
XXY systems have evolved three times in Anolis (Fig 1). We examined sex chromosome homology in two of those instances, i.e. the clade containing A. distichus and in A. sagrei. We observed BAC FISH signal on two chromosomes in the female and one chromosome in the male in both species. This pattern is consistent with either fusion of the Y chromosome to an autosome or fission of the X chromosomes (White 1973; Kitano and Peichel 2012). The coincident evolution of an XXY system with an increase in chromosome number in A. sagrei suggests evolution via a fission of the X chromosomes. We observed some X chromosome BAC hybridization near the centromere of two acrocentric microchromosomes in A. distichus. Two scenarios that could explain this pattern are the translocation of X-linked sequence to the microchromosomes or fission of ancestral X chromosomes to generate a new chromosome pair. Distinguishing between these two situations is not possible with our current data but FISH of additional autosomal and X-linked BACs could help test these hypotheses.
Chromosomal evolution
Chromosomal changes are often associated with major evolutionary transitions such as speciation events (Olmo 2005). Indeed this correlation has been used as evidence to suggest a mechanistic model of chromosomal speciation (King 1995). Although the sampling in this study is insufficient to properly analyze for any impact of chromosome number and sex chromosome complement on diversification rates, two general patterns are revealed by our analyses. First, changes in chromosome number (both increases and decreases) are distributed throughout the phylogeny of Anolis. This scenario contrasts with that of another squamate genus, Sceloporus, in which changes in chromosome number are concentrated on internal nodes followed by diversification of taxa all with similar or identical karyotypes (Leaché and Sites 2010). The distribution of losses and gains in chromosome number also contrasts with a previous study in mammals which found that lineages are biased for either fission or fusion (Pardo-Manuel de Villena and Sapienza 2001). Our reconstruction of chromosome evolution in Anolis reveals no evidence of a similar bias. Second, transitions from homomorphic to heteromorphic sex chromosomes occur throughout the Anolis phylogeny. Seven of the nine transitions in sex chromosome complement occur on terminal branches. Although this finding by itself might suggest reduced diversification rates among lineages with heteromorphic sex chromosomes, the remaining two transitions occur at deeply nested nodes with many descendant species. Furthermore, many of the taxa with terminal changes in sex chromosome complement have unsampled sister species that may share the same heteromorphic sex chromosomes. This suggests that in Anolis the evolution of heteromorphic sex chromosomes does not appear to be a direct path to increased or decreased diversification.
Valenzuela & Adams (2011) found increased rates of change in chromosome number in turtles were associated with changes in sex determining mechanism. Using a different approach we found a similar, though non-significant, trend in anoles. Whereas these authors suggest natural selection based mechanisms to explain correlated change in chromosome number and sex determining mechanisms, the association we recover in anoles may simply reflect that heteromorphism occasionally evolves by way of fissions of sex chromosomes thus simultaneously creating heteromorphic sex chromosomes and changing chromosome number but retaining sex chromosome homology.
Brandley et al. (2006) present the only other explicitly phylogenetic examination of chromosome number evolution in Anolis although their analysis was confined to the A. cristatellus series. Our results are concordant with theirs and we find changes in chromosome number occurring on the same branches within the A. cristatellus series. 2n=36 has been hypothesized as the ancestral number of chromosomes in Anolis (Gorman and Atkins 1969; Webster et al. 1972; Gorman 1973; Paull et al. 1976; Gorman et al. 1983), a result confirmed here.
Homology of Anolis sex chromosomes
Comparative BAC mapping and qPCR revealed X chromosome homology among sampled Anolis species, indicating a single origin of sex chromosomes present in their most recent common ancestor. This supports the conservation hypothesis and makes Anolis one of the most species-rich vertebrate clades to have sex chromosome systems that are homologous to each other and derived from the same ancestral XY chromosome pair. Other species-rich vertebrate clades with stable sex chromosome systems such as eutherian mammals, birds and snakes have proven important models in understanding the role of sex chromosomes in speciation, dosage compensation and genome evolution (Ohno 1967; Edwards et al. 2005; Graves 2006; Ellegren 2007; Wilson and Makova 2009; Vicoso et al. 2013) and Anolis provides an additional model for future comparative studies.
Homology of Anolis sex chromosomes indicates that Anolis species lacking heteromorphic sex chromosomes do indeed have sex chromosomes but these are homomorphic. This is illustrated by the recent finding of a homomorphic XX/XY system in Anolis carolinensis using BAC FISH (Alföldi et al. 2011). Further support is provided by our identification of homomorphic Y chromosomes in both A. grahami and A. lineatopus using FISH of repetitive DNA. qPCR results also bear this out. Our data confirm male heterogamety is more widespread in Anolis than published karyotypic data suggest and the lack of heteromorphic sex chromosomes cannot be used to dismiss male heterogamety in Anolis species. This finding likely applies to other vertebrate groups, e.g. garter snakes (Vicoso et al. 2013), and thus the observation of a few species with heteromorphic sex chromosomes nested within a larger clade of species lacking heteromorphic sex chromosomes should not automatically require an appeal to the turnover hypothesis.
Sex chromosomes have not previously been identified in A. grahami and A. lineatopus, although C-banding in several male A. grahami showed differential staining in one of the small pairs of metacentric chromosomes, with one chromosome being more heavily stained than the other (Blake 1983). Blake (1983) was hesitant to identify these as sex chromosomes because he only examined males, but it appears this is the same chromosomal pair that hybridized with the X-linked BAC and the sexually dimorphic accumulation of GATA repeats in our experiments.
Variation in the accumulation of repetitive GATA sequences among sampled Anolis species highlights the stochastic nature of Y chromosome evolution. The differential accumulation of the GATA probe clearly identified the Y chromosome in three of the five Anolis species sampled here: A. distichus; A. grahami; and A. lineatopus. Failure to identify the Y chromosome in A. sagrei and A. carolinensis using this technique may be due to the small Y chromosomes of both species, as the Y is a microchromosome in both (De Smet 1981; Alföldi et al. 2011), as well as the accumulation of GATA in both species on multiple microchromosomes further complicating identification of the putative Y chromosome. Additionally, the GATA minisatellite is just one of many repetitive DNA sequences that are known to accumulate on vertebrate Y chromosomes, and sex chromosomes in a variety of species can show very different patterns when hybridized with diverse repetitive DNA sequences (O’Meally et al. 2010; Pokorná et al. 2011; Cioffi et al. 2012). Performing FISH with several different repetitive DNA sequences might allow cytogenetic identification of the Y chromosome in both species.
The gene cltcl1 is X-linked in A. carolinesis but qPCR showed equal quantification in both male and female A. chlorocyanus. There are several possible explanations for this result. One possibility is that cltcl1 occurs in a pseudoautosomal region of the Y chromosome in A. chlorocyanus. An alternative is that cltcl1 has moved off of the X chromosome and is now autosomal. Distinguishing between these hypotheses is not possible given our data. Copy number in the X-linked pi4ka was half as much in males compared to females, similar to other sampled Anolis, and appears to remain X-linked in A. chlorocyanus confirming partial X chromosome homology with A. carolinensis.
Conclusions
Turnover and conservation have both been suggested as mechanisms to explain the lack of sex chromosome heteromorphism in most vertebrate species. Using cytogenetic and phylogenetic comparative analyses of anoles we find evidence supporting the conservation hypothesis of sex chromosome evolution as the primary mechanism in this group. Heteromorphic sex chromosomes have evolved multiple times throughout the genus but they are derived from a single ancestral pair. Furthermore, the degree of Y chromosome degeneration varies among lineages as measured both by heteromorphism and repeat accumulation. This study represents the first attempt to contrast turnover and conservation in a diverse and well-sampled clade. These results highlight the utility of combining phylogenetic comparative methods with cytogenetics and molecular genetics in understanding the origins and evolution of sex chromosomes. Only a handful of studies have used both approaches to fully explore the evolutionary origins of sex chromosomes in a clade, but none have been performed in a group where both turnover and conservation represent viable alternative hypotheses (Tanaka et al. 2007; Takehana et al. 2008; Ezaz et al. 2009a; Ross et al. 2009; Pokorná et al. 2012). Further use of these methods will undoubtedly prove useful in explaining the widespread variation is sex chromosome heteromorphy observed in other vertebrate groups.
Our results pave the way to using Anolis to address additional questions related to sex chromosome evolution. Anolis sex chromosomes provide an opportunity to investigate Y chromosome degeneration (or lack thereof) in a comparative framework. Much effort has been made to investigate the evolution of sex chromosomes because they have been implicated in a number of core evolutionary processes including dosage compensation, genetic conflict, adaptation, and speciation in a wide variety of taxa. For example, X-linked loci are related to hybrid male sterility in Drosophila (Presgraves 2008), mosquitoes (Slotman et al. 2004) and mice (Good et al. 2008; White et al. 2012). Similarly, Z-linked markers are associated with pre- and postzygotic isolation in birds (Edwards et al. 2005; Saether et al. 2007) and butterflies (Jiggins et al. 2001). Within anoles, sex chromosomes may be involved in sterility of hybrid F1 males derived from A. brevirostris and A. distichus (Webster 1977). Finally, there is evidence that dosage compensation is more prevalent in species with XY sex determining systems than in ZW systems (Mank In press). Anolis provides another exemplary XY vertebrate model to test these and other hypotheses.
Supplementary Material
Supplementary Figure 1. Majority rule phylogenetic tree inferred for 216 Anolis species using BEAST. Node support (posterior probability) for Beast and MrBayes analyses are indicated by color coded circles. Karyotype data were available for species indicated by black text.
Supplementary Figure 2. Ancestral state reconstruction of 1n female chromosome number onto the pruned phylogeny. Maximum likelihood estimate of ancestral chromosome number is indicated on each node. Female 1n chromosome number is indicated after each species name.
Supplementary Table 1. Samples used in the phylogenetic and comparative evolutionary analyses.
Supplementary Table 2. Samples used for qPCR.
Supplementary Table 3. Primers used for qPCR.
Supplementary Table 4. qPCR results showing mean fold change in quantification of genes in male samples compared to female samples with standard error and 95% confidence intervals.
Acknowledgments
Thanks to K. Krysko, J. Losos, and J. McGlothlin for access to specimens; L. Oseth for BAC FISH help; and two anonymous reviewers for comments that significantly improved the manuscript. BAC FISH was performed by the University of Minnesota Cytogenomics Shared Resource, supported by Masonic Comprehensive Cancer Center-NIH Grant #P30 CA077598-09. Funding was provided by NSF (IOS1146820 and DEB0920892); NIH (T32DE007288 from the National Institute of Dental & Craniofacial Research); University of Minnesota (Medical School and Office of the Vice President for Research).
Literature Cited
- Adolfsson S, Ellegren H. Lack of dosage compensation accompanies the arrested stage of sex chromosome evolution in ostriches. Mol Biol Evol. 2013;30:806–810. doi: 10.1093/molbev/mst009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alföldi J, Di Palma F, Grabherr M, Williams C, Kong L, Mauceli E, Russell P, Lowe CB, Glor RE, Jaffe JD, Ray DA, Boissinot S, Shedlock AM, Botka C, Castoe TA, Colbourne JK, Fujita MK, Moreno RG, ten Hallers BF, Haussler D, Heger A, Heiman D, Janes DE, Johnson J, de Jong PJ, Koriabine MY, Lara M, Novick PA, Organ CL, Peach SE, Poe S, Pollock DD, de Queiroz K, Sanger T, Searle S, Smith JD, Smith Z, Swofford R, Turner-Maier J, Wade J, Young S, Zadissa A, Edwards SV, Glenn TC, Schneider CJ, Losos JB, Lander ES, Breen M, Ponting CP, Lindblad-Toh K. The genome of the green anole lizard and a comparative analysis with birds and mammals. Nature. 2011;477:587–591. doi: 10.1038/nature10390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D. The temporal dynamics of processes underlying Y chromosome degeneration. Genetics. 2008;179:1513–1525. doi: 10.1534/genetics.107.084012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D, Jensen JD, Zhang Z. Accelerated adaptive evolution on a newly formed X chromosome. PLoS Biol. 2009;7:712–719. doi: 10.1371/journal.pbio.1000082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2010;38:D46–D51. doi: 10.1093/nar/gkp1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergero R, Charlesworth D. The evolution of restricted recombination in sex chromosomes. Trends Ecol Evol. 2008;24:94–102. doi: 10.1016/j.tree.2008.09.010. [DOI] [PubMed] [Google Scholar]
- Blake JA. Chromosomal C-banding in Anolis grahami. In: Rhodin AGJ, Miyata K, editors. Advances in Herpetology and Evolutionary Biology. Museum of Comparative Zoology; Cambridge, MA: 1983. pp. 621–625. [Google Scholar]
- Blake JA. Complex chromosomal variation in natural populations of the Jamaican lizard Anolis grahami. Genetica. 1986;69:3–17. [Google Scholar]
- Blaser O, Grossen C, Neuenschwander S, Perrin N. Sex-chromosome turnovers induced by deleterious mutation load. Evolution. 2013;67:635–645. doi: 10.1111/j.1558-5646.2012.01810.x. [DOI] [PubMed] [Google Scholar]
- Brandley MC, Wynn A, de Queiroz K. Karyotype and relationships of Anolis desechensis. J Herpetol. 2006;40:136–139. [Google Scholar]
- Castañeda MR, deQueiroz K. Phylogenetic relationships of the Dactyloa clade of Anolis lizards based on nuclear and mitochondrial DNA sequence data. Mol Phylogenet Evol. 2011;61:784–800. doi: 10.1016/j.ympev.2011.07.004. [DOI] [PubMed] [Google Scholar]
- Charlesworth B. Model for evolution of Y chromosomes and dosage compensation. Proc Natl Acad Sci U S A. 1978;75:5618–5622. doi: 10.1073/pnas.75.11.5618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D, Charlesworth B. The degeneration of Y chromosomes. Philos Trans R Soc Lond B Biol Sci. 2000;355:1563–1572. doi: 10.1098/rstb.2000.0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D, Charlesworth B, Marais G. Steps in the evolution of heteromorphic sex chromosomes. Heredity. 2005;95:118–128. doi: 10.1038/sj.hdy.6800697. [DOI] [PubMed] [Google Scholar]
- Cioffi MB, Kejnovsky E, Marquioni V, Poltronieri J, Molina WF, Diniz D, Bertollo LAC. The key role of repeated DNAs in sex chromosome evolution in two fish species with ZW sex chromosome system. Mol Cytogenet. 2012;5:28–35. doi: 10.1186/1755-8166-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connallon T, Clark AG. Gene duplication, gene conversion and the evolution of the Y chromosome. Genetics. 2010;186:277–286. doi: 10.1534/genetics.110.116756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet WHO. Description of the orcien stained karyotypes of 27 lizard species (Lacertilia: Reptilia) belonging to the families Iguanidae, Agamidae Chameleontidae and Gekkonidae (Ascalabota) Acta Zool Pathol Antverp. 1981;76:35–72. [Google Scholar]
- Devlin RH, Nagahama Y. Sex determination and sex differentiation in fish: An overview of genetic, physiological, and environmental influences. Aquaculture. 2002;208:191–364. [Google Scholar]
- Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A, Duran C, Field M, Heled J, Kearse M, Markowitz S, Moir R, Stones-Havas S, Sturrock S, Thierer T, Wilson A. Geneious. 2011;5.4 [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards SV, Kingan SB, Calkins JD, Balakrishnan CN, Jennings WB, Swanson WJ, Sorenson MD. Speciation in birds: Genes, geography, and sexual selection. Proc Natl Acad Sci U S A. 2005;102:6550–6557. doi: 10.1073/pnas.0501846102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H. Molecular evolutionary genomics of birds. Cytogenet Genome Res. 2007;117:120–130. doi: 10.1159/000103172. [DOI] [PubMed] [Google Scholar]
- Ezaz T, O’Meally D, Quinn AE, Sarre SD, Georges A, Graves JAM. A simple non-invasive protocol to establish primary cell lines from tail and toe explants for cytogenetic studies in Australian dragon lizards (Squamata: Agamidae) Cytotechnology. 2008;58:135–139. doi: 10.1007/s10616-009-9182-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezaz T, Quinn AE, Miura I, Sarre SD, Georges A, Marshall Graves JA. The dragon lizard Pogona vitticeps has ZZ/ZW micro-sex chromosomes. Chromosome Res. 2005;13:763. doi: 10.1007/s10577-005-1010-9. [DOI] [PubMed] [Google Scholar]
- Ezaz T, Quinn AE, Sarre SD, O’Meally D, Georges A, Graves JAM. Molecular marker suggests rapid changes of sex-determining mechanisms in Australian dragon lizards. Chromosome Res. 2009a;17:91–98. doi: 10.1007/s10577-008-9019-5. [DOI] [PubMed] [Google Scholar]
- Ezaz T, Sarre S, O’Meally D, Graves J, Georges A. Sex chromosome evolution in lizards: Independent origins and rapid transitions. Cytogenet Genome Res. 2009b;127:249–260. doi: 10.1159/000300507. [DOI] [PubMed] [Google Scholar]
- Gamble T. A review of sex determining mechanisms in geckos (Gekkota: Squamata) Sex Dev. 2010;4:88–103. doi: 10.1159/000289578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glor RE, Vitt LJ, Larson A. A molecular phylogenetic analysis of diversification in Amazonian Anolis lizards. Mol Ecol. 2001;10:2661–2668. doi: 10.1046/j.0962-1083.2001.01393.x. [DOI] [PubMed] [Google Scholar]
- Good JM, Dean MD, Nachman MW. A complex genetic basis to X-linked hybrid male sterility between two species of house mice. Genetics. 2008;179:2213–2228. doi: 10.1534/genetics.107.085340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman GC. The chromosomes of the Reptilia: A cytotaxonomic interpretation. In: Chiarelli B, Capanna E, editors. Cytotaxonomy and Vertebrate Evolution. Academic Press; London: 1973. pp. 349–424. [Google Scholar]
- Gorman GC, Atkins L. Chromosomal heteromorphism in some male lizards of the genus Anolis. Am Nat. 1966;100:579–583. [Google Scholar]
- Gorman GC, Atkins L. Confirmation of an X-Y sex determining mechanism in lizards (Anolis) Copeia. 1968;1968:159–160. [Google Scholar]
- Gorman GC, Atkins L. The zoogeography of Lesser Antillean Anolis lizards - An analysis based upon chromosomes and lactic dehydrogenases. Bull Mus Comp Zool. 1969;138:53–80. [Google Scholar]
- Gorman GC, Buth DG, Soule M, Yang SY. The relationships of the Puerto Rican Anolis: Electrophoretic and karyotypic studies. In: Rhodin AGJ, Miyata K, editors. Advances in Herpetology and Evolutionary Biology. Museum of Comparative Zoology; Cambridge, MA: 1983. pp. 626–642. [Google Scholar]
- Graves JAM. Sex chromosome specialization and degeneration in mammals. Cell. 2006;124:901–914. doi: 10.1016/j.cell.2006.02.024. [DOI] [PubMed] [Google Scholar]
- Graves JAM. Weird animal genomes and the evolution of vertebrate sex and sex chromosomes. Annu Rev Genet. 2008;42:565–586. doi: 10.1146/annurev.genet.42.110807.091714. [DOI] [PubMed] [Google Scholar]
- Grossen C, Neuenschwander S, Perrin N. Temperature-dependent turnovers in sex-determination mechanisms: A quantitative model. Evolution. 2011;65:64–78. doi: 10.1111/j.1558-5646.2010.01098.x. [DOI] [PubMed] [Google Scholar]
- Guerrero R, Kirkpatrick M, Perrin N. Cryptic recombination in the ever-young sex chromosomes of Hylid frogs. J Evol Biol. 2012;25:1947–1954. doi: 10.1111/j.1420-9101.2012.02591.x. [DOI] [PubMed] [Google Scholar]
- Haldane JB. Sex ratio and unisexual sterility in hybrid animals. J Genet. 1922;12:101–109. [Google Scholar]
- Harrison PW, Mank JE, Wedell N. Incomplete sex chromosome dosage compensation in the Indian meal moth, Plodia interpunctella, based on de novo transcriptome assembly. Genome Biol Evol. 2012;4:1118–1126. doi: 10.1093/gbe/evs086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henning F, Trifonov V, Ferguson-Smith MA, Almeida-Toledo LF. Non-homologous sex chromosomes in two species of the genus Eigenmannia (Teleostei: Gymnotiformes) Cytogenet Genome Res. 2008;121:55–58. doi: 10.1159/000124382. [DOI] [PubMed] [Google Scholar]
- Hillis DM, Green DM. Evolutionary changes of heterogametic sex in the phylogenetic history of amphibians. J Evol Biol. 1990;3:49–64. [Google Scholar]
- Hughes JF, Skaletsky H, Brown LG, Pyntikova T, Graves T, Fulton RS, Dugan S, Ding Y, Buhay CJ, Kremitzki C, Wang Q, Shen H, Holder M, Villasana D, Nazareth LV, Cree A, Courtney L, Veizer J, Kotkiewicz H, Cho TJ, Koutseva N, Rozen S, Muzny DM, Warren WC, Gibbs RA, Wilson RK, Page DC. Strict evolutionary conservation followed rapid gene loss on human and rhesus Y chromosomes. Nature. 2012;483:82–86. doi: 10.1038/nature10843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ijdo J, Wells R, Baldini A, Reeders S. Improved telomere detection using a telomere repeat probe (TTAGGG)n generated by PCR. Nucleic Acids Res. 1991;19:4780. doi: 10.1093/nar/19.17.4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y, Melamed E, Yang X, Kampf K, Wang S, Yehya N, Van Nas A, Replogle K, Band MR, Clayton DF. Dosage compensation is less effective in birds than in mammals. J Biol. 2007;6:2. doi: 10.1186/jbiol53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiggins CD, Linares M, Naisbit RE, Salazar C, Yang ZH, Mallet J. Sex-linked hybrid sterility in a butterfly. Evolution. 2001;55:1631–1638. doi: 10.1111/j.0014-3820.2001.tb00682.x. [DOI] [PubMed] [Google Scholar]
- King M. Species evolution: The role of chromosome change. Cambridge University Press; Cambridge: 1995. [Google Scholar]
- Kitano J, Peichel CL. Turnover of sex chromosomes and speciation in fishes. Environ Biol Fishes. 2012;94:549–558. doi: 10.1007/s10641-011-9853-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumazawa Y, Nishida M. Sequence evolution of mitochondrial tRNA genes and deep-branch animal phylogenetics. J Mol Evol. 1993;37:380–398. doi: 10.1007/BF00178868. [DOI] [PubMed] [Google Scholar]
- Leaché A, Sites J. Chromosome evolution and diversification in North American spiny lizards (genus Sceloporus) Cytogenet Genome Res. 2010;127:166–181. doi: 10.1159/000293285. [DOI] [PubMed] [Google Scholar]
- Lieb CS. Biochechemical and Karyological Systematics of the Mexican lizards of the Anolis gadovi and A. nebulosus species group (Reptilia: Iguanidae) University California; Los Angeles, Los Angeles, CA: 1981. p. 331. [Google Scholar]
- Mahler DL, Revell LJ, Glor RE, Losos JB. Ecological opportunity and the rate of morphological evolution in the diversification of Greater Antillean anoles. Evolution. 2010;64:2731–2745. doi: 10.1111/j.1558-5646.2010.01026.x. [DOI] [PubMed] [Google Scholar]
- Main H, Scantlebury DP, Zarkower D, Gamble T. Karyotypes of two species of Malagasy ground gecko (Paroedura: Gekkonidae) Afr J Herpetol. 2012;61:81–90. [Google Scholar]
- Mank JE. Sex chromosome dosage compensation: Definitely not for everyone. Trends Genet In press. [Google Scholar]
- Mank JE, Promislow DEL, Avise JC. Evolution of alternative sex-determining mechanisms in teleost fishes. Biol J Linn Soc. 2006;87:83–93. [Google Scholar]
- Matsubara K, Tarui H, Toriba M, Yamada K, Nishida-Umehara C, Agata K, Matsuda Y. Evidence for different origin of sex chromosomes in snakes, birds, and mammals and step-wise differentiation of snake sex chromosomes. Proc Natl Acad Sci U S A. 2006;103:18190–18195. doi: 10.1073/pnas.0605274103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayrose I, Barker MS, Otto SP. Probabilistic models of chromosome number evolution and the inference of polyploidy. Syst Biol. 2010;59:132–144. doi: 10.1093/sysbio/syp083. [DOI] [PubMed] [Google Scholar]
- Meiklejohn CD, Tao Y. Genetic conflict and sex chromosome evolution. Trends Ecol Evol. 2010;25:215–223. doi: 10.1016/j.tree.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer LR, Zweig AS, Hinrichs AS, Karolchik D, Kuhn RM, Wong M, Sloan CA, Rosenbloom KR, Roe G, Rhead B, Raney B, Pohl A, Malladi V, Li C, Lee B, Learned K, Kirkup V, Hsu F, Heitner S, Harte R, Haeussler M, Guruvadoo L, Goldman M, Giardine B, Fujita P, Dreszer T, Diekhans M, Cline M, Clawson H, Barber G, Haussler D, Kent W. The UCSC Genome Browser database: Extensions and updates 2013. Nucleic Acids Res. 2013;41:D64–D69. doi: 10.1093/nar/gks1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura I, Ohtani H, Ogata M. Independent degeneration of W and Y sex chromosomes in frog Rana rugosa. Chromosome Res. 2012;20:47–55. doi: 10.1007/s10577-011-9258-8. [DOI] [PubMed] [Google Scholar]
- Morgan TH. Sex-limited inheritance in Drosophila. Science. 1911;32:120–122. doi: 10.1126/science.32.812.120. [DOI] [PubMed] [Google Scholar]
- Muller HJ. A gene for the fourth chromosome of Drosophila. J Exp Zool. 1914;17:325–336. [Google Scholar]
- Nanda I, Feichtinger W, Schmid M, Schröder JH, Zischler H, Epplen JT. Simple repetitive sequences are associated with differentiation of the sex chromosomes in the guppy fish. J Mol Evol. 1990;30:456–462. [Google Scholar]
- Nanda I, Schlegelmilch K, Haaf T, Schartl M, Schmid M. Synteny conservation of the Z chromosome in 14 avian species (11 families) supports a role for Z dosage in avian sex determination. Cytogenet Genome Res. 2008;122:150–156. doi: 10.1159/000163092. [DOI] [PubMed] [Google Scholar]
- Nylander JAA, Wilgenbusch JC, Warren DL, Swofford DL. AWTY (are we there yet?): A system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics. 2008;24:581–583. doi: 10.1093/bioinformatics/btm388. [DOI] [PubMed] [Google Scholar]
- O’Meally D, Ezaz T, Georges A, Sarre SD, Graves JAM. Are some chromosomes particularly good at sex? Insights from amniotes. Chromosome Res. 2012;20:7–19. doi: 10.1007/s10577-011-9266-8. [DOI] [PubMed] [Google Scholar]
- O’Meally D, Patel HR, Stiglec R, Sarre SD, Georges A, Marshall Graves JA, Ezaz T. Non-homologous sex chromosomes of birds and snakes share repetitive sequences. Chromosome Res. 2010;18:787–800. doi: 10.1007/s10577-010-9152-9. [DOI] [PubMed] [Google Scholar]
- Ohno S. Sex Chromosomes and Sex-linked Genes. Springer Verlag; Berlin, Germany: 1967. [Google Scholar]
- Olmo E. Rate of chromosome changes and speciation in reptiles. Genetica. 2005;125:185–203. doi: 10.1007/s10709-005-8008-2. [DOI] [PubMed] [Google Scholar]
- Pagel M, Meade A, Barker D. Bayesian estimation of ancestral character states on phylogenies. Syst Biol. 2004;53:673–684. doi: 10.1080/10635150490522232. [DOI] [PubMed] [Google Scholar]
- Pardo-Manuel de Villena F, Sapienza C. Female meiosis drives karyotypic evolution in mammals. Genetics. 2001;159:1179–1189. doi: 10.1093/genetics/159.3.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull D, Williams EE, Hall WP. Lizard karyotypes from the Galapagos Islands: Chromosomes in phylogeny and evolution. Breviora. 1976:1–31. [Google Scholar]
- Perrin N. Sex reversal: A fountain of youth for sex chromosomes? Evolution. 2009;63:3043–3049. doi: 10.1111/j.1558-5646.2009.00837.x. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST©) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokorná M, Giovannotti M, Kratochvíl L, Caputo V, Olmo E, Ferguson-Smith MA, Rens W. Conservation of chromosomes syntenic with avian autosomes in squamate reptiles revealed by comparative chromosome painting. Chromosoma. 2012;121:409–418. doi: 10.1007/s00412-012-0371-z. [DOI] [PubMed] [Google Scholar]
- Pokorná M, Kratochvíl L, Kejnovsky E. Microsatellite distribution on sex chromosomes at different stages of heteromorphism and heterochromatinization in two lizard species (Squamata: Eublepharidae: Coleonyx elegans and Lacertidae: Eremias velox) BMC Genetics. 2011;12:90–97. doi: 10.1186/1471-2156-12-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D. jModelTest: Phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Presgraves DC. Sex chromosomes and speciation in Drosophila. Trends Genet. 2008;24:336–343. doi: 10.1016/j.tig.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn AE, Georges A, Sarre SD, Guarino F, Ezaz T, Graves JAM. Temperature sex reversal implies sex gene dosage in a reptile. Science. 2007;316:411. doi: 10.1126/science.1135925. [DOI] [PubMed] [Google Scholar]
- Rabosky DL, Glor RE. Equilibrium speciation dynamics in a model adaptive radiation of island lizards. Proc Natl Acad Sci U S A. 2010;107:22178–22183. doi: 10.1073/pnas.1007606107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A, Drummond AJ. Tracer. 2007. Distributed by authors. [Google Scholar]
- Rice WR. The accumulation of sexually antagonistic genes as a selective agent promoting the evolution of reduced recombination between primitive sex chromosomes. Evolution. 1987;41:911–914. doi: 10.1111/j.1558-5646.1987.tb05864.x. [DOI] [PubMed] [Google Scholar]
- Rice WR. Evolution of the Y sex chromosome in animals. Bioscience. 1996;46:331–343. [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Ross JA, Urton JR, Boland JE, Shapiro MD, Peichel CL. Turnover of sex chromosomes in the stickleback fishes (Gasterosteidae) PLoS Genet. 2009;5:1–12. doi: 10.1371/journal.pgen.1000391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H, Marszalek JD, Minx PJ, Cordum HS, Waterston RH, Wilson RK, Page DC. Abundant gene conversion between arms of palindromes in human and ape Y chromosomes. Nature. 2003;423:873–876. doi: 10.1038/nature01723. [DOI] [PubMed] [Google Scholar]
- Saether SA, Saetre GP, Borge T, Wiley C, Svedin N, Andersson G, Veen T, Haavie J, Servedio MR, Bures S, Kral M, Hjernquist MB, Gustafsson L, Traeff J, Qvarnstroem A. Sex chromosome-linked species recognition and evolution of reproductive isolation in flycatchers. Science. 2007;318:95–97. doi: 10.1126/science.1141506. [DOI] [PubMed] [Google Scholar]
- Shetty S, Griffin DK, Graves JAM. Comparative painting reveals strong chromosome homology over 80 million years of bird evolution. Chromosome Res. 1999;7:289–295. doi: 10.1023/a:1009278914829. [DOI] [PubMed] [Google Scholar]
- Singh L, Panicker SG, Nagaraj R, Majumdar KC. Banded krait minor-satellite (Bkm)-associated Y chromosome-specific repetitive DNA in mouse. Nucleic Acids Res. 1994;22:2289–2295. doi: 10.1093/nar/22.12.2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh L, Purdom I, Jones K. Sex chromosome associated satellite DNA: Evolution and conservation. Chromosoma. 1980;79:137–157. doi: 10.1007/BF01175181. [DOI] [PubMed] [Google Scholar]
- Slotman M, Della Torre A, Powell J. The genetics of inviability and male sterility in hybrids between Anopheles gambiae and An. arabiensis. Genetics. 2004;167:275–287. doi: 10.1534/genetics.167.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöck M, Horn A, Grossen C, Lindtke D, Sermier R, Betto-Colliard C, Dufresnes C, Bonjour E, Dumas Z, Luquet E, Maddalena T, Sousa HC, Martinez-Solano I, Perrin N. Ever-young sex chromosomes in European tree frogs. PLoS Biol. 2011;9:e1001062. doi: 10.1371/journal.pbio.1001062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöck M, Savary R, Betto-Colliard C, Biollay S, Jourdan-Pineau H, Perrin N. Low rates of X-Y recombination, not turnovers, account for homomorphic sex chromosomes in several diploid species of Palearctic green toads (Bufo viridis subgroup) J Evol Biol. 2013:674–682. doi: 10.1111/jeb.12086. [DOI] [PubMed] [Google Scholar]
- Straub T, Becker PB. Dosage compensation: The beginning and end of generalization. Nat Rev Genet. 2007;8:47–57. doi: 10.1038/nrg2013. [DOI] [PubMed] [Google Scholar]
- Takehana Y, Demiyah D, Naruse K, Hamaguchi S, Sakaizumi M. Evolution of different Y chromosomes in two medaka species, Oryzias dancena and O. latipes. Genetics. 2007;175:1335–1340. doi: 10.1534/genetics.106.068247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takehana Y, Hamaguchi S, Sakaizumi M. Different origins of ZZ/ZW sex chromosomes in closely related medaka shes, Oryzias javanicus and O. hubbsi. Chromosome Res. 2008;16:801–811. doi: 10.1007/s10577-008-1227-5. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Takehana Y, Naruse K, Hamaguchi S, Sakaizumi M. Evidence for different origins of sex chromosomes in closely related Oryzias fishes: Substitution of the master sex-determining gene. Genetics. 2007;177:2075–2081. doi: 10.1534/genetics.107.075598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli M, Orr HA. The dominance theory of Haldane’s rule. Genetics. 1995;140:389–402. doi: 10.1093/genetics/140.1.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela N, Adams DC. Chromosome number and sex determination co-evolve in turtles. Evolution. 2011;65:1808–1813. doi: 10.1111/j.1558-5646.2011.01258.x. [DOI] [PubMed] [Google Scholar]
- Vicoso B, Bachtrog D. Progress and prospects toward our understanding of the evolution of dosage compensation. Chromosome Res. 2009;17:585–602. doi: 10.1007/s10577-009-9053-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B, Emerson JJ, Zektser Y, Mahajan S, Bachtrog D. Comparative sex chromosome genomics in snakes: Differentiation, evolutionary strata, and lack of global dosage compensation. PLoS Biol. 2013;11:e1001643. doi: 10.1371/journal.pbio.1001643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viets BE, Ewert MA, Talent LG, Nelson CE. Sex–determining mechanisms in squamate reptiles. J Exp Zool. 1994;270:45–56. [Google Scholar]
- Volff JN, Nanda I, Schmid M, Schartl M. Governing sex determination in fish: Regulatory putsches and ephemeral dictators. Sex Dev. 2007;1:85–99. doi: 10.1159/000100030. [DOI] [PubMed] [Google Scholar]
- Webster TP. Hybridization of Hispaniolan lizards in the Anolis distichus species group. In: Williams EE, editor. The Third Anolis Newsletter. Museum of Comparative Zoology; Cambridge, MA: 1977. pp. 162–172. [Google Scholar]
- Webster TP, Williams EE, Hall WP. Fission in the evolution of a lizard karyotype. Science. 1972;177:611–613. doi: 10.1126/science.177.4049.611. [DOI] [PubMed] [Google Scholar]
- Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, Geer LY, Kapustin Y, Khovayko O, Landsman D, Lipman DJ, Madden TL, Maglott DR, Ostell J, Miller V, Pruitt KD, Schuler GD, Sequeira E, Sherry ST, Sirotkin K, Souvorov A, Starchenko G, Tatusov RL, Tatusova TA, Wagner L, Yaschenko E. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2007;35:D5–D12. doi: 10.1093/nar/gkl1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MA, Stubbings M, Dumont BL, Payseur BA. Genetics and evolution of hybrid male sterility in house mice. Genetics. 2012;191:917–934. doi: 10.1534/genetics.112.140251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MJD. Animal Cytology & Evolution. Cambridge University Press; Cambridge: 1973. [Google Scholar]
- Wilson EB. The chromosomes in relation to the determination of sex in insects. Science. 1905;22:500–502. doi: 10.1126/science.22.564.500. [DOI] [PubMed] [Google Scholar]
- Wilson MA, Makova KD. Genomic analyses of sex chromosome evolution. Annu Rev Genomics Hum Genet. 2009;10:333–354. doi: 10.1146/annurev-genom-082908-150105. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Bachtrog D. Sex-specific adaptation drives early sex chromosome evolution in Drosophila. Science. 2012;337:341–345. doi: 10.1126/science.1225385. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Majority rule phylogenetic tree inferred for 216 Anolis species using BEAST. Node support (posterior probability) for Beast and MrBayes analyses are indicated by color coded circles. Karyotype data were available for species indicated by black text.
Supplementary Figure 2. Ancestral state reconstruction of 1n female chromosome number onto the pruned phylogeny. Maximum likelihood estimate of ancestral chromosome number is indicated on each node. Female 1n chromosome number is indicated after each species name.
Supplementary Table 1. Samples used in the phylogenetic and comparative evolutionary analyses.
Supplementary Table 2. Samples used for qPCR.
Supplementary Table 3. Primers used for qPCR.
Supplementary Table 4. qPCR results showing mean fold change in quantification of genes in male samples compared to female samples with standard error and 95% confidence intervals.