Abstract
One can interpret fragmentation spectra stemming from peptides in mass spectrometry-based proteomics experiments using so called database search engines. Frequently, one also runs post-processors such as Percolator to assess the confidence, infer unique peptides and increase the number of identifications. A recent search engine, MS-GF+, has shown promising results, due to a new and efficient scoring algorithm. However, MS-GF+ provides few statistical estimates about the peptide-spectrum matches, hence limiting the biological interpretation. Here, we enabled Percolator-processing for MS-GF+ output, and observed an increased number of identified peptides for a wide variety of datasets. In addition, Percolator directly reports p values and false discovery rate estimates, such as q values and posterior error probabilities, for peptide-spectrum matches, peptides and proteins, functions that are useful for the whole proteomics community.
Introduction
A critical component of mass spectrometry-based proteomics is the database searching, where search engines are used to match fragmentation spectra to theoretical spectra of peptides in a database.1 While the most common examples of the search engines are Sequest,2 Mascot3 and X!Tandem,4 a newer alternative, named MS-GF+,5,6 is discussed here. These search engines all produce peptide-spectrum matches (PSMs) from which the researcher can infer the peptides and the proteins present in the analyzed sample. The biological interpretation is typically confounded by the relatively large proportion of spectra that are matched incorrectly by the search engines, i.e. matched to peptides that were not actually in the mass spectrometer and undergoing fragmentation. Such mismatching is likely the result of various effects, such as unusual peptide fragmentation,7 unaccounted-for post-translation modifications (PTMs)8,9 and incomplete databases.10
To help discriminate between correct and incorrect PSMs, the search engines assign scores to each PSM, as a measure of how well the peptide matches the spectrum. The scoring algorithms often make up the most fundamental difference between search engines, and although the scores do not necessarily have a direct probabilistic interpretation, they indicate the quality of the match. In the end, the researcher typically chooses a score threshold associated with a certain confidence level, above which the PSMs are accepted as predominantly correct matches. Regardless of how we measure the confidence level, the actual discrimination is performed by the scores, hence the various search engines will produce different sets, and numbers, of PSMs for a certain confidence level.
The standard procedure for inferring identifications from high-throughput experiments is to control the false discovery rate (FDR).11–13 This is the expected fraction of incorrect identifications among the set of identifications accepted as correct. Here, the FDR is represented by the q value, the minimal FDR required to call an identification significant, which has the desirable property of being monotonically increasing with the number of identifications.13,14 In the field of mass spectrometry-based proteomics, the target-decoy analysis15 is arguably the most commonly used approach for estimating the q value. The method requires matching the spectra against a shuffled or reversed decoy database, in addition to the target database of the studied organism. The matches to the decoy database are true negatives and serve to model the incorrect matches to the target database.
An advantage of a properly performed target-decoy analysis is that the results from different search engines can be compared directly, with the FDR as the common denominator. Furthermore, as the decoy PSMs are assumed to be good models of incorrect target PSMs, they can be used to train a machine learning algorithm to produce scores to improve the separation between correct and incorrect target PSMs. This idea is embodied in Percolator, a post-processing tool that accepts target and decoy PSMs from a search engine, and trains a linear support vector machine (SVM) to improve the classification of correct target PSMs.16 Percolator considers a set of features that describes each PSM, and combines these into a new score, tailored for the dataset at hand. This score routinely increases the number of confident identifications, as the typical original search engine scores fail to address the specific characteristics of each individual experiment.
So far, the improvements made by Percolator have been seen for the classical search engines Sequest, Mascot and X!Tandem, as their inherent, general, scoring scheme is not fully adjusted for each individual dataset. However, the recently developed MS-GF+ has been demonstrated to perform well for a wide range of different datasets, due to its highly sophisticated scoring algorithm. MS-GF+ uses a dynamic programming algorithm to match all peptides, not restricted to the ones in the searched database, against each spectrum. The rendered score distribution can then be utilized to calibrate the scoring function for each individual spectrum.5 With this information, the score assigned to the best matching peptide in the searched database becomes more interpretable.
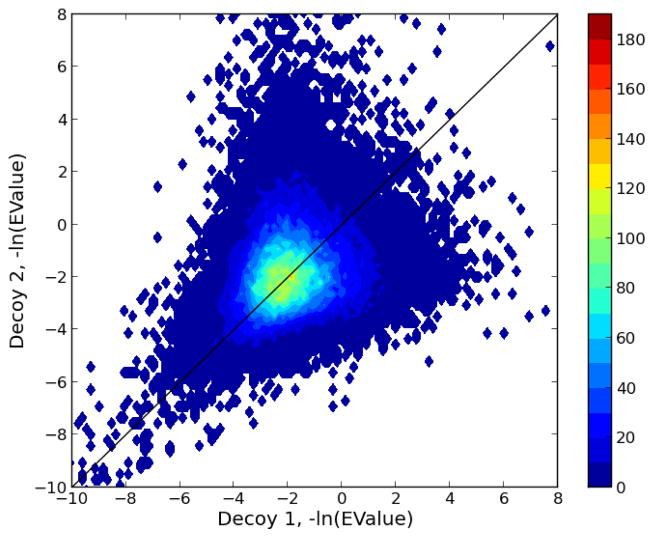
However, despite the impressive performance of MS-GF+, a closer look at the scores of incorrect PSMs reveals some room for improvement. Figure 1 shows the relationship between scores of the same set of spectra searched against two different decoy databases. Low scoring PSMs are clearly correlated between the two runs. Ideally, the scores of these pairs of incorrect PSMs should not be correlated, to facilitate the discrimination of correct PSMs. In practice, however, most search engines exhibit this type of correlation as a result of confounding features among the spectra. An example of such a confounding variable, that influences the scores without providing evidence for the match quality, is the number of peptide candidates to a spectrum.17 A solution is offered by post-processors that can decrease the influences of confounding variables by accounting for them while learning how to optimally separate between correct and incorrect matches.18
Figure 1. The covariation of two MS-GF+ decoy searches.
MS-GF+ was run against a reversed decoy database (Decoy 1) and a shuffled decoy database (Decoy 2) generated with Mimic. For each PSM, the negative logarithm of the E value reported by MS-GF+ was plotted.
Here, we present a joint effort to combine the sophisticated scoring algorithm of MS-GF+ and the machine learning of Percolator. The new tool, denoted by MS-GF+Percolator and implemented as a converter of the output format from MS-GF+ to the input format for Percolator, increases the number of identifications over MS-GF+ alone. In addition, post-processing by Percolator enables direct statistical interpretation of the results, as q values, posterior error probabilities (PEP, also known as “local FDR”) and p values are reported for PSMs, unique peptides, and proteins. Previously, these estimates required additional analyses subsequent to MS-GF+. In this study, we first validate the statistical assumptions made by Percolator, and then present the observed improvements of MS-GF+Percolator for a wide variety of datasets.
Experimental procedures
Updates to MS-GF+
Percolator’s performance relies on characteristics, called features, of the PSMs. The features help to tell correct from incorrect matches. By default, MS-GF+ outputs a set of scores, called RawScore and DeNovoScore, as well as the statistical estimates E values and spectral E values, for each PSM. RawScore is the match quality score of MS-GF+, and DeNovoScore is the maximum possible RawScore to the given spectrum, considering all possible peptide sequences.
To further facilitate Percolator’s discriminative performance, MS-GF+ was amended with an option (-addFeatures) to output an additional set of features. These values include the fragmentation spectrum ion current, and the fractions of it that can be explained by theoretical fragment ions from the peptide sequence. Moreover, the number of theoretical fragment ions that were matched to fragment peaks is also reported, as well as the average mass errors and standard deviation of the top seven fragment peaks.
In Percolator, the default MS-GF+ output and these additional features make up the basis for about half of the features used for learning and discrimination. The other half of the features comes from the peptide mass, charge state and other quantities relating to the identified peptide. The feature generation is explained in more detail in the Results section.
The latest version of MS-GF+ can be obtained freely for research and non-profit institutions from http://proteomics.ucsd.edu/Software/MSGFPlus.html.
Percolator and file format converter
To enable Percolator to post-process results from MS-GF+, its mzIdentML format19 output is converted to the input file format (pin) of Percolator. A converter was implemented in C++ and added to the Percolator package, which is freely available online at http://per-colator.com/. The converter uses the CodeSynthesis XSD library (http://www.codesynthesis.com/products/xsd/) to parse and write the two formats, both written in the Extensible Markup Language (XML).
The converter, named msgf2pin, takes as input a target and a decoy mzIdentML format file from MS-GF+ run with the addFeatures option set to 1. The generated pin-file contains the feature information for each PSM, and can be directly interpreted and post-processed by Percolator.
Experimental data
To evaluate the MS-GF+Percolator algorithm, we analyzed previously published mass spectrometry-based proteomics data from one purified protein sample and four complex samples. The purified sample consisted of Orbitrap data from mixture 7 of the ISB18 dataset, obtained online at https://regis-web.systemsbiology.net/PublicDatasets/.20
As for complex samples, first, a human dataset of LTQ-Orbitrap spectra from a prostate cancer cell line was used.21 Second, LTQ-Orbitrap spectra from mouse brain tissue published by Huttlin et al.22 were used as an example of phosphorylation enriched datasets. Third, data of human peripheral blood mononuclear cells from the Proteome Informatics Research Group (iPRG) 2013 study was used as an example of data with high accuracy fragmentation spectra.23 The sample is labeled with 6plex Tandem mass tags (TMT)24 for quantification, and has been used to benchmark a variety of tools and approaches. All the above samples were digested with trypsin. Fourth, yeast datasets digested with various enzymes (trypsin, Lys-C, Arg-C, Glu-C and Asp-N) published by Swaney et al.25 were used to evaluate MS-GF+Percolator’s applicability to different cleavage specificities. From the study, the CID spectra were used here.
Database searching
For all datasets, we used MS-GF+ version 9540, followed by post-processing by Percolator, version 2.05. In addition, we used the search engines Crux26 version 1.40 in the sequest-search mode, and X!Tandem4 version Sledgehammer 2013.09.01.1.
The ISB18 mix spectra, downloaded in mzXML file format, were searched against a database of the 18 purified proteins as well as common contaminants. Appended to the database was a larger (25x) proportion of shuffled sequences, as entrapment sequences for incorrect matches.27 In addition to the target database, a decoy database containing the reversed target database sequences was searched separately to estimate the statistical confidence. When a second decoy database was used, Mimic (https://github.com/percolator/mimic) was used to shuffle the amino acid sequences while retaining the same level of homology as in the target databases. We used a ± 10 ppm mass tolerance window, with no enzyme specificity.
All remaining datasets were converted from their native raw file format to the mgf, ms2 or mzXML file format using msconvert of ProteoWizard version 2.2 to 3.0.28 Subsequently, they were searched against Ensembl29 (release 68) databases of the appropriate organism and, separately, against the corresponding reversed decoy databases. A ± 10 ppm precursor mass tolerance was used in all cases, and appropriate settings for activation technique and fragmentation spectrum mass accuracy. The enzymatic parameters were set to allow one non-enzymatic termini for each peptide, for the respective enzyme. Crux was run in non-enzymatic mode for enzymes other than trypsin. Cysteine carbamidomethylation was accounted for by a fixed modification of 57.02 u on C, and methionine oxidation by a variable modification of 15.99 u on M. For the phosphorylated mouse dataset, an additional 79.96 u mass shift was allowed on amino acids S, T and Y. In the TMT labeled sample, a fixed modification of 229.16 u on the N-terminal and K was used.
The target and decoy outputs from MS-GF+ were converted from the mzIdentML format to the Percolator-in format by the program msgf2pin supplied in Percolator’s converter package. Subsequent Percolator processes were run with peptide termini-features for the respective enzyme, and using the MS-GF+ E value as the initial feature direction for MS-GF+. The same analysis was done for the Crux and X!Tandem output, but converting the file format using the programs sqt2pin and tandem2pin, also supplied in Percolator’s converter package.
To estimate the confidence of the identified unique peptides, the canonical amino acid sequence was considered, disregarding charge states and PTMs. As MS-GF+ reports only confidence estimates for PSMs, proper comparisons to MS-GF+Percolator on the level of unique peptides require an additional “Weed-out then estimate” (WOTE) procedure, described and evaluated previously.30 This includes discarding all but the best scoring duplicate PSMs matching to a given target or decoy peptide, followed by statistical evaluation of the remaining unique peptides. Here, a target-decoy confidence estimation was performed using the MS-GF+ E values, by the software qvality31 downloaded on April 23, 2013. Percolator performs a target-decoy analysis for unique peptides by default.
Results
Selection of Percolator features
For learning, Percolator obtains a set of features from the MS-GF+ output. These features are listed and described in Table 1. However, classification and prediction using machine learning algorithms always include risks of overfitting and biased learning. Percolator uses a nested cross-validation scheme to avoid overfitting,32 but its statistical accuracy still relies on the assumption that matches to the decoy database are good models of incorrect target matches. This assumption must hold for all features used by Percolator during learning. Otherwise, the algorithm would easily discriminate between matches from the two databases, but not necessarily between correct and incorrect target matches.
Table 1. MS-GF+ features in Percolator.
List of the features from MS-GF+ used by Percolator.
| Feature name | Feature description |
|---|---|
| RawScore | Raw match score of MS-GF+ |
| DeNovoScore | Maximum possible raw match score to this spectrum |
| ScoreRatio | RawScore divided by DeNovoScore |
| Energy | Difference between RawScore and DeNovoScore |
| lnEValue | Negative MS-GF+ E Value, logged |
| lnSpecEValue | Negative MS-GF+ Spectral E Value, logged |
| IsotopeError | Number of additional neutrons in peptide |
| lnExplainedIonCurrent | Summed intensity of identified fragment ions, divided by that of all fragment ions, logged |
| lnNTermIonCurrentRatio | Summed intensity of identified N-terminal fragments, divided by that of all identified fragments, logged |
| lnCTermIonCurrectRatio | Summed intensity of identified N-terminal fragments, divided by that of all identified fragments, logged |
| lnMS2IonCurrent | Summed intensity of all observed fragment ions, logged |
| Mass | Peptide mass |
| PepLen | Peptide length |
| dM | Difference between theoretical and experimental mass |
| absdM | Absolute value of the difference between theoretical and experimental mass |
| MeanErrorTop7 | Mean of mass errors of the seven fragment ion peaks with the highest intensities |
| sqMeanErrorTop7 | Squared MeanErrorTop7 |
| StdevErrorTop7 | Standard deviation of mass errors of the seven fragment ion peaks with the highest intensities |
| ChargeN | Boolean, peptide charge is N |
| enzN | Boolean, N-terminal agrees with enzymatic cleavage rules |
| enzC | Boolean, C-terminal agrees with enzymatic cleavage rules |
| enzInt | Number of internal cleavage sites |
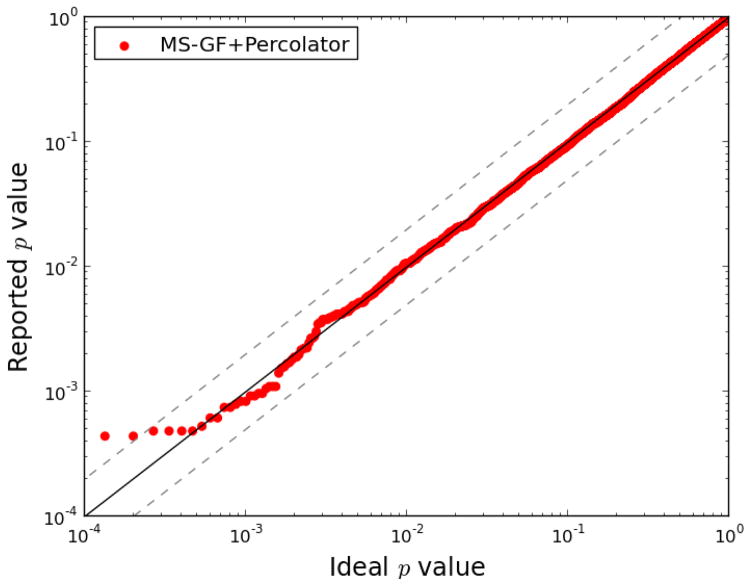
To validate that the decoy model is suitable for all features used by Percolator on the MS-GF+ output, a previously described semi-labeled calibration protocol was applied using the ISB18 dataset.27 Accordingly, only the incorrect matches of spectra from a purified mixtures of known proteins were analyzed. With this dataset, such incorrect matches are easily identified as the PSMs that match parts of the database that are not made up of sequences from the 18 ISB proteins (or common contaminants). This portion of the database is larger, and made up of shuffled sequences, or proteins from an evolutionary distant organism. Furthermore, p values reported for incorrect PSMs should be uniformly distributed, according to their definition. Percolator assigns p values to PSMs based on their final Percolator score. Hence, we isolated the surely incorrect PSMs of the ISB18 mix sample, and plotted their p values against a rank from 0 to 1 to illustrate their uniformity, see Figure 2. Based on this quantile-quantile plot we can see the deviation of the p values from uniformity, i.e. biases in the Percolator output, as p values diverging from the x = y diagonal.
Figure 2. Statistical calibration of null p values from MS-GF+Percolator.
Using only incorrect matches from the Orbitrap spectra of ISB18 mix 7, p values were estimated from the Percolator score, after post-processing the MS-GF+ results. The y axis represents the p values reported by Percolator, and the x axis a uniform rank from 0 to 1. The black line marks the x = y diagonal, and the gray dashed lines x = 2y and x = y/2. A calculated Kolomogorov-Smirnov test D value of 0.014 between the reported and ideal p values indicates a small difference between the two distributions.
Performance of MS-GF+Percolator
In the previous section, we validated the accuracy of the statistical estimates reported from MS-GF+Percolator. With this confirmation, the performance of the algorithm can be benchmarked more safely against other approaches for analyzing shotgun proteomics data. To measure the performance, we compared the numbers of confidently identified unique peptides from the different algorithms. The main benefit of using unique peptides instead of PSMs is that they represent a biologically more relevant entity. Note, however, that the number of unique peptides is lower than the number of PSMs, as many PSMs can map to a single peptide sequence.30 We tested MS-GF+Percolator on a wide range of datasets, and compared it to MS-GF+ alone, as well as to Crux+Percolator and X!Tandem+Percolator. Crux is an open source alternative to Sequest, that represents the traditional method for running Percolator. X!Tandem is also an open source search engine that have been associated with Percolator previously.33,34
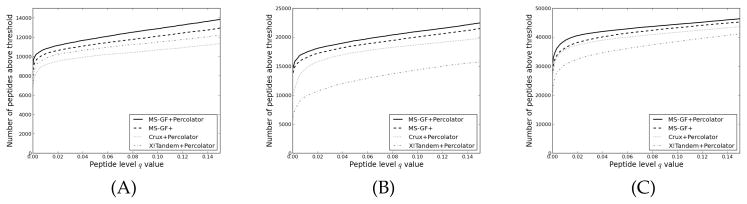
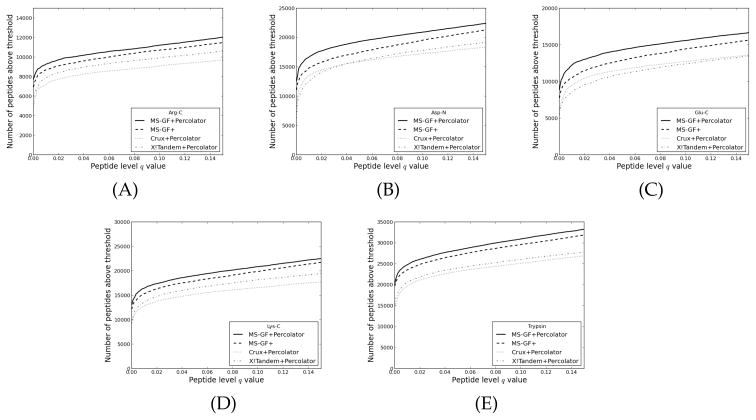
Figure 3 (A) shows the performance of MS-GF+Percolator on the prostate cancer cell line dataset, measured by the number of identified peptides at different peptide level q value thresholds. The complete analysis took less than an hour on a desktop computer with an Intel Core 2 Quad 2.66 GHz processor, about half of which is the Percolator analysis. At a peptide level q value of 0.01, the increase of MS-GF+Percolator corresponds to 5.1% more peptides over the native MS-GF+. Similar results were seen for a highly phosphorylated mouse dataset (Figure 3 (B), 5.0% increase), and the iPRG data (Figure 3 (C), 7.1% increase). The yeast sample digested with five different enzymes showed that Percolator’s performance varies substantially depending on the enzyme (Figure 4 A–E). The improvements in the number of confidently identified unique peptides over native MS-GF+ were 6.4%, 12.9%, 16.2%, 7.5% and 5.1% for Arg-C, Asp-N, Glu-C, Lys-C and trypsin, respectively. The results suggest that Percolator increases the number of confident identifications from MS-GF+ on a wide variety of data, especially for enzymes other than trypsin. As discussed in more detail later, a likely explanation is that the scoring algorithm of MS-GF+ is better optimized for tryptic peptides.
Figure 3. MS-GF+Percolator performance on tryptic datasets.
The numbers of accepted unique peptides are plotted as a function of the peptide level q value threshold. Panel (A) shows the results for the human prostate cancer data, (B) the phosphorylated mouse data and (C) the TMT labeled iPRG data. Peptide level confidence estimates from MS-GF+ were obtained by using qvality as described in Experimental procedures.
Figure 4. The performance of MS-GF+Percolator on the yeast data digested by multiple enzymes.
The number of accepted unique peptides for different peptide level q values thresholds, for several different enzymes. The enzymes used for digestions are (A) Arg-C, (B) Asp-N, (C) Glu-C, (D) Lys-C and (E) trypsin.) Peptide level confidence estimates from MS-GF+ were obtained by using qvality as described in Experimental procedures.
For completeness we also compared the performance of the different methods on the PSM level. As expected, the same trends as for unique peptides were seen for the number of confidently identified PSMs. At a PSM level q value of 0.01, MS-GF+ and MS-GF+Percolator identified 22,712 and 23,877 PSMs, respectively, for the prostate cancer set. The corresponding numbers for the phosphorylated mouse dataset were 58,734 and 59,235 PSMs and for the iPRG dataset 65,260 and 70,303 PSMs. In the yeast samples digested with multiple enzymes, the following numbers of PSMs were identified with MS-GF+ and MS-GF+Percolator, respectively: Arg-C; 23,140 and 24,947, Asp-N; 59,456 and 67,631, Glu-C; 19,279 and 23,037, Lys-C; 65,045 and 68,870, trypsin; 123,049 and 129,833.
Feature analysis
To estimate the importance of the different groups of features for MS-GF+Percolator, we repeatedly removed groups of related features from the analysis. The features were grouped based on whether they described MS-GF+ raw scores, E values (as well as these first two groups combined), descriptions of the fragment ions, the precursor ions, the peptide or the enzymatic digestion. By removing one of these groups at the time, the contribution of the corresponding features can be inferred from the change in MS-GF+Percolator performance, computed by counting the number of confident unique peptides at a q value of 1%.
The human prostate cancer and the iPRG data are examples of datasets with low and high accuracy fragmentation spectra, respectively. Table 2 and Table 3 show the results of the feature analyses for these datasets. Included in the tables are the corresponding numbers for Crux. The features used by Percolator for Crux differ somewhat from those in MS-GF+Percolator, but they generally represent the same types of information and can, to some extent, be grouped similarly.
Table 2. Feature analysis for human prostate data.
The performance of Percolator using reduced sets of features, on a dataset with low accuracy fragmentation spectra. The performance is estimated by counting the number of unique peptides with a peptide level q value lower than or equal to 0.01. The first number represents the number of identified peptides, for MS-GF+Percolator or Crux+Percolator. The number in parenthesis shows the corresponding percental change in performance relative the full feature set, for the respective approach.
| Removed features | MS-GF+Percolator | Crux+Percolator |
|---|---|---|
| None | 10791 | 9123 |
| RawScores: RawScore, DeNovoScore, ScoreRatio, Energy | 10684 (−0.99%) | |
| E values: lnEValue, lnSpecEValue | 10677 (−1.06%) | |
| Scores: RawScores and E values | 5260 (−51.26%) | 7693 (−15.67%) |
| Precursor: IsotopeError, dM, absdM | 10709 (−0.76%) | 8992 (−1.44%) |
| Fragment ions: lnExplainedIonCurrentRatio, lnNTermIonCurrentRatio, lnCTermIonCurrentRatio, lnMS2IonCurrentRatio, MeanErrorTop7, sqMeanErrorTop7, StdevErrorTop7 | 10749 (−0.39%) | 9096 (−0.30%) |
| Peptide: Mass, PepLen, ChargeN | 10725 (−0.61%) | 9069 (−0.59%) |
| Enzyme: enzN, enzC, enzInt | 10646 (−1.34%) | 7745 (−15.10%) |
Table 3. Feature analysis for iPRG data.
The performance of Percolator using reduced sets of features, on a dataset with high accuracy fragmentation spectra. The performance is estimated by counting the number of unique peptides with a peptide level q value lower than or equal to 0.01. The first number represents the number of identified peptides, for MS-GF+Percolator or Crux+Percolator. The number in parenthesis shows the corresponding percental change in performance relative the full feature set, for the respective approach.
| Removed features | MS-GF+Percolator | Crux+Percolator |
|---|---|---|
| None | 38813 | 35587 |
| RawScores: RawScore, DeNovoScore, ScoreRatio, Energy | 38050 (−1.97%) | |
| E values: lnEValue, lnSpecEValue | 35363 (−8.89%) | |
| Scores: RawScores and E values | 27225 (−29.86%) | 4131 (−88.39%) |
| Precursor: IsotopeError, dM, absdM | 38737 (−0.20%) | 35421 (−0.47%) |
| Fragment ions: lnExplainedIonCurrentRatio, lnNTermIonCurrentRatio, lnCTermIonCurrentRatio, lnMS2IonCurrentRatio, MeanErrorTop7, sqMeanErrorTop7, StdevErrorTop7 | 38043 (−1.98%) | 35478 (−0.31%) |
| Peptide: Mass, PepLen, ChargeN | 38372 (−1.14%) | 35571 (−0.04%) |
| Enzyme: enzN, enzC, enzInt | 38246 (−1.46%) | 31831 (−10.55%) |
Discussion
This study shows that post-processing PSMs obtained from MS-GF+ by Percolator increases the number of confidently identified peptides. Using a sample of known protein content, p values assigned to absent peptides were demonstrated to be uniformly distributed. We interpret the uniformity as an indication of that the statistical estimates from MS-GF+Percolator are accurate. In turn, this is a consequence of that the features used for training the classifier do not behave differently between the target and decoy databases, but rather between correct and incorrect PSMs.
Users of MS-GF+Percolator can expect an increased number of confidently identified peptides compared to the native MS-GF+. In agreement with previous studies,16 the improvements from Percolator are generally more pronounced for enzymes alternative to trypsin. Most likely, scoring algorithms tend to be designed and optimized with an emphasis on trypsin, while leaving some room for improvement for other enzymes. Percolator, on the other hand, attempts to optimally weigh the enzymatic cleavage features for the dataset at hand, needing only the cleavage specificity rules of the enzyme. Thus, Percolator works to adjust the imperfections of the scoring function of the search engine.
Regardless of the performance improvements over MS-GF+ alone, however, perhaps the most attractive attribute of MS-GF+Percolator is its rigorous statistical estimates. Percolator directly outputs p values, q values and posterior error probabilities on the level of PSMs, peptides and proteins (using the inbuilt Fido algorithm35). This greatly facilitates the downstream interpretation of the results. With MS-GF+Percolator, the versatility and accuracy of MS-GF+ are made more accessible to proteomics researchers.
The feature analysis clearly shows that the removal of some groups of features have a large impact on the performance of MS-GF+Percolator, while the removal of other groups is less detrimental. However, there are clear differences between the two datasets. The fragment ion features, for example, have a more positive effect for high accuracy than low accuracy fragment data, as expected. Despite these differences, the combination of RawScores and E values generally tends to be the most informative group of features. Still, the feature analysis highlights that correct and incorrect identifications can be discriminated without the use of any search engine scores, although this becomes harder with low accuracy fragmentation spectra. Note, however, that the search engines scores are still taken into account implicitly as they were used to rank the PSMs to identify the top scoring match for each spectrum.
The feature analysis also shows some differences between choosing Sequest (in its Crux implementation) or MS-GF+ as the search engine. The most striking difference, with respect to the influence of the features, is the large contribution from the enzymatic cleavage features in Crux+Percolator. An explanation is that in MS-GF+, cleavage information from the C and N terminal ends of the peptide sequence is incorporated into the RawScore, which is not done in Sequest’s cross correlation score (XCorr). The results also show that removing the raw scores and E values leads to a large performance decrease when removed both at the same time, however when removing the groups individually only a smaller performance decrease is observed. The reason is that these features are dependent, and encode almost the same information.
Critical evaluation
In this study, the performance of MS-GF+Percolator was compared to that of MS-GF+, Crux+Percolator and X!Tandem+Percolator. In contrast, many published methods for analyzing proteomics experiments are benchmarked against a wider set of algorithms. In our case, however, the message is simple; to demonstrate how Percolator improves the results of MS-GF+. We apply the principle of ceteris paribus, meaning to only alter one variable at the time. Showing the performance of MS-GF+ before and after the Percolator post-processing gives a clear picture of the gain from using Percolator. The comparisons to Crux+Percolator are motivated by that Crux, and Sequest, traditionally has been the default search engines for the stand alone version of Percolator, similarly, X!Tandem has been increasingly associated with Percolator.
In the performance tests we have focused on the number of identified unique peptides, in contrast to the number of PSMs. Like many have done previously,30,36,37 we would like to emphasize the distinction between PSMs and unique peptides. In short, the statistical confidence estimated for a PSM is not valid for the corresponding peptide. The reason is that the hypotheses underlying the confidence measures are different in the two cases. The null hypothesis, for instance, could be formulated for a PSM as; “no appropriate match was found for the spectrum”, but differently for a peptide; “no evidence was found for the peptide”. As a consequence, PSM and peptide FDRs differ, and can not be directly compared. We argue that the peptide level accuracy measurements have a more direct interpretation to the end user for most biological applications.
Although Percolator is flexible and accepts virtually any feature, we implemented a limited number of features in this study. A more rigorous attempt to produce and test the performance of different features could boost the performance of MS-GF+Percolator further, and compose an interesting direction for the future. Some promising candidates could be predictions of the iso-electric points (pI) and retention times of peptides, as well as features describing the potential co-fragmentation of multiple peptides species.
Acknowledgments
This work was supported by grants from the Swedish Research Council, the Swedish Foundation for Strategic Research, the Lawski Foundation. S.K. and R.D.S. were supported by the National Institute of General Medical Sciences Proteomics Research Center (P41 GM 103493-10) and by the Department of Energy Office of Biological and Environmental Research Genome Sciences Program under the Pan-omics project. Work was partially performed in the Environmental Molecular Science Laboratory, a U.S. Department of Energy (DOE) national scientific user facility at Pacific Northwest National Laboratory (PNNL) in Richland, WA. Battelle operates PNNL for the DOE under contract DE-AC05-76RLO01830.
Footnotes
Notes
The authors declare no conflicting interests.
References
- 1.Käll L, Vitek O. Computational Mass Spectrometry–Based Proteomics. PLoS computational biology. 2011;7:e1002277. doi: 10.1371/journal.pcbi.1002277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eng J, McCormack A, Yates J, et al. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. Journal of the American Society for Mass Spectrometry. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 3.Perkins D, Pappin D, Creasy D, Cottrell J. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 4.Craig R, Beavis R. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004:921. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 5.Kim S, Gupta N, Pevzner P. Spectral probabilities and generating functions of tandem mass spectra: a strike against decoy databases. J Proteome Res. 2008;7:3354–3363. doi: 10.1021/pr8001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim S, Mischerikow N, Bandeira N, Navarro J, Wich L, Mohammed S, Heck A, Pevzner P. The generating function of CID, ETD and CID/ETD pairs of tandem mass spectra: Applications to database search. Molecular & Cellular Proteomics. 2010 doi: 10.1074/mcp.M110.003731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paizs B, Suhai S. Fragmentation pathways of protonated peptides. Mass spectrometry reviews. 2005;24:508–548. doi: 10.1002/mas.20024. [DOI] [PubMed] [Google Scholar]
- 8.Tsur D, Tanner S, Zandi E, Bafna V, Pevzner P. Identification of post-translational modifications via blind search of mass-spectra. Computational Systems Bioinformatics Conference, 2005. Proceedings. 2005 IEEE; 2005. pp. 157–166. [DOI] [PubMed] [Google Scholar]
- 9.Dasari S, Chambers M, Codreanu S, Liebler D, Collins B, Pennington S, Gallagher W, Tabb D, Dasari S, Chambers M, et al. Sequence tagging reveals unexpected modifications in toxicoproteomics. Chemical research in toxicology. 2011;24:204. doi: 10.1021/tx100275t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dasari S, Chambers M, Slebos R, Zimmerman L, Ham A, Tabb D. TagRecon: high-throughput mutation identification through sequence tagging. Journal of proteome research. 2010;9:1716–1726. doi: 10.1021/pr900850m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sorić B. Statistical “discoveries” and effect-size estimation. Journal of the American Statistical Association. 1989;84:608–610. [Google Scholar]
- 12.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 13.Storey J, Tibshirani R. Statistical significance for genomewide studies. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:9440. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Käll L, Storey J, MacCoss M, Noble W. Assigning significance to peptides identified by tandem mass spectrometry using decoy databases. Journal of Proteome Research. 2008;7:29–34. doi: 10.1021/pr700600n. [DOI] [PubMed] [Google Scholar]
- 15.Moore R, Young M, Lee T. Qscore: an algorithm for evaluating SEQUEST database search results. Journal of the American Society for Mass Spectrometry. 2002;13:378–386. doi: 10.1016/S1044-0305(02)00352-5. [DOI] [PubMed] [Google Scholar]
- 16.Käll L, Canterbury J, Weston J, Noble W, MacCoss M. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nature Methods. 2007;4:923–925. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- 17.Klammer AA, Park CY, Noble WS. Statistical calibration of the SEQUEST XCorr function. Journal of proteome research. 2009;8:2106. doi: 10.1021/pr8011107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Granholm V, Käll L. Quality assessments of peptide–spectrum matches in shotgun proteomics. Proteomics. 2011;11:1086–1093. doi: 10.1002/pmic.201000432. [DOI] [PubMed] [Google Scholar]
- 19.Jones AR, Eisenacher M, Mayer G, Kohlbacher O, Siepen J, Hubbard SJ, Selley JN, Searle BC, Shofstahl J, Seymour SL, et al. The mzIdentML data standard for mass spectrometry-based proteomics results. Molecular & Cellular Proteomics. 2012;11 doi: 10.1074/mcp.M111.014381. year. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klimek J, Eddes J, Hohmann L, Jackson J, Peterson A, Letarte S, Gafken P, Katz J, Mallick P, Lee H, et al. The standard protein mix database: a diverse data set to assist in the production of improved peptide and protein identification software tools. The Journal of Proteome Research. 2007;7:96–103. doi: 10.1021/pr070244j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Serang O, Moruz L, Hoopmann M, Käll L. Recognizing uncertainty increases robustness and reproducibility of mass spectrometry-based protein inferences. Journal of Proteome Research. 2012 doi: 10.1021/pr300426s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huttlin E, Jedrychowski M, Elias J, Goswami T, Rad R, Beausoleil S, Villén J, Haas W, Sowa M, Gygi S. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143:1174–1189. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen R, Mias GI, Li-Pook-Than J, Jiang L, Lam HY, Chen R, Miriami E, Karczewski KJ, Hariharan M, Dewey FE, et al. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell. 2012;148:1293–1307. doi: 10.1016/j.cell.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson A, Schäfer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, Neumann T, Hamon C. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Analytical chemistry. 2003;75:1895–1904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- 25.Swaney D, Wenger C, Coon J. Value of using multiple proteases for large-scale mass spectrometry-based proteomics. Journal of proteome research. 2010;9:1323–1329. doi: 10.1021/pr900863u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park C, Käll L, Klammer A, MacCoss M, Noble W. Rapid and accurate peptide identification from tandem mass spectra. Journal of proteome research. 2008;7:3022. doi: 10.1021/pr800127y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Granholm V, Noble W, Käll L. On using samples of known protein content to assess the statistical calibration of scores assigned to peptide-spectrum matches in shotgun proteomics. Journal of proteome research. 2011 doi: 10.1021/pr1012619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chambers MC, Maclean B, Burke R, Amodei D, Ruderman DL, Neumann S, Gatto L, Fischer B, Pratt B, Egertson J, et al. A cross-platform toolkit for mass spectrometry and proteomics. Nature biotechnology. 2012;30:918–920. doi: 10.1038/nbt.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flicek P, Amode M, Barrell D, Beal K, Brent S, Chen Y, Clapham P, Coates G, Fairley S, Fitzgerald S, et al. Ensembl 2011. Nucleic acids research. 2011;39:D800–D806. doi: 10.1093/nar/gkq1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Granholm V, Navarro JC, Noble WS, Käll L. Determining the calibration of confidence estimation procedures for unique peptides in shotgun proteomics. Journal of proteomics. 2012 doi: 10.1016/j.jprot.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kall L, Storey J, Noble W. QVALITY: non-parametric estimation of q-values and posterior error probabilities. Bioinformatics. 2009;25:964. doi: 10.1093/bioinformatics/btp021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Granholm V, Noble W, Käll L. A cross-validation scheme for machine learning algorithms in shotgun proteomics. BMC Bioinformatics. 2012;13:S3. doi: 10.1186/1471-2105-13-S16-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang P, Ma J, Wang P, Zhu Y, Zhou BB, Yang YH. Improving X! Tandem on peptide identification from mass spectrometry by self-boosted Percolator. Computational Biology and Bioinformatics, IEEE/ACM Transactions on. 2012;9:1273–1280. doi: 10.1109/TCBB.2012.86. [DOI] [PubMed] [Google Scholar]
- 34.Xu M, Li Z, Li L. Combining Percolator with X! Tandem for Accurate and Sensitive Peptide Identification. Journal of proteome research. 2013;12:3026–3033. doi: 10.1021/pr4001256. [DOI] [PubMed] [Google Scholar]
- 35.Serang O, MacCoss MJ, Noble WS. Efficient marginalization to compute protein posterior probabilities from shotgun mass spectrometry data. Journal of proteome research. 2010;9:5346. doi: 10.1021/pr100594k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bern M, Goldberg D. Improved ranking functions for protein and modification-site identifications. Journal of Computational Biology. 2008;15:705–719. doi: 10.1089/cmb.2007.0119. [DOI] [PubMed] [Google Scholar]
- 37.Shteynberg D, Deutsch EW, Lam H, Eng JK, Sun Z, Tasman N, Mendoza L, Moritz RL, Aebersold R, Nesvizhskii AI. iProphet: multi-level integrative analysis of shotgun proteomic data improves peptide and protein identification rates and error estimates. Molecular & Cellular Proteomics. 2011;10 doi: 10.1074/mcp.M111.007690. year. [DOI] [PMC free article] [PubMed] [Google Scholar]