Abstract
Purpose
Metastatic urothelial carcinoma (UC) of the bladder is associated with multiple somatic copy number alterations (SCNAs). We evaluated SCNAs to identify predictors of poor survival in patients with metastatic UC treated with platinum-based chemotherapy.
Experimental Design
We obtained overall survival (OS) and array DNA copy number data from metastatic UC patients in two cohorts. Associations between recurrent SCNAs and OS were determined by a Cox proportional hazard model adjusting for performance status and visceral disease. mRNA expression was evaluated for potential candidate genes by Nanostring nCounter to identify transcripts from the region that are associated with copy number gain. In addition, expression data from an independent cohort was used to identify candidate genes.
Results
Multiple areas of recurrent significant gains and losses were identified. Gain of 1q23.3 was independently associated with a shortened OS in the both cohorts (adjusted HR 2.96; 95% CI, 1.35 to 6.48; P = 0.01 and adjusted HR 5.03; 95% CI 1.43-17.73; P < 0.001). The F11R, PFDN2, PPOX, USP21 and DEDD genes, all located on 1q23.3, were closely associated with poor outcome.
Conclusions
1q23.3 copy number gain displayed association with poor survival in two cohorts of metastatic UC. The identification of the target of this copy number gain is ongoing, and exploration of this finding in other disease states may be useful for the early identification of poor risk UC patients. Prospective validation of the survival association is necessary to demonstrate clinical relevance.
INTRODUCTION
Of the 75,000 new cases of urothelial carcinoma (UC) that are diagnosed annually in the US (1), approximately 40% develop invasive and/or metastatic UC. Metastatic UC remains incurable in the vast majority of patients, with a median survival of approximately six months without and 14 months with treatment (2). Unlike other solid tumors, targeted therapies thus far have failed to advance the standard of care for UC. Thus, there is an urgent need for new biomarkers and treatment approaches.
While multiple genetic alterations associated with UC development and progression have been identified, there is still a limited understanding of those changes that lead to poor prognosis in metastatic disease. For patients with metastatic UC, prognosis is primarily determined by clinical factors, such as performance status, visceral metastases, hemoglobin level, or liver metastases (3, 4). However, the variability of outcomes even within these groups is significant, and the ability to accurately predict survival and progression of disease is limited. Similarly, while multiple potential tissue-based biomarkers have been proposed in bladder cancer, none so far have been successfully validated.
Comparative genomic hybridization (CGH) studies of bladder carcinomas and cell lines have revealed recurrent genetic aberrations including gains on 8q22-24, 11q13, 17q21, and losses involving chromosomes 9, 8p22-23, and 17p (5–8). Some of these changes have since been associated with the pathological stage of and/or outcome from bladder cancer in diverse clinical cohorts (9, 10). The advent of high-resolution mapping array-based CGH (aCGH) has further facilitated the molecular delineation of gained and lost regions to the level of specific candidate genes (5, 7, 11). However, molecular markers of poor outcome in metastatic UC remain largely elusive. Furthermore, novel therapeutic approaches are needed, as there have been no major treatment advances for metastatic UC in 20 years.
We evaluated genomic copy number gains and losses using oligonucleotide aCGH in94 primary tumors from a clinically annotated cohort of UC patients who went on to develop metastatic disease. Using this unbiased approach, we screened for somatic gains and losses to identify loci strongly associated with poor survival. We identified copy number gain of 1q23.3 as being associated with poor survival. In an independent cohort we found similar survival associations with 1q23.3 copy number gain. We identified 1q23.3 candidate driver genes by mRNA expression in these patients and in a third independent cohort.
METHODS
Patients
Patients with metastatic bladder cancer were identified from two cohorts (Table 1). These cohorts, one from Spain (“Spanish cohort”) and the other from the Dana-Farber Cancer Institute (“DFCI cohort”) included patients who either presented with metastatic disease or subsequently developed metastatic disease after local therapy. The Spanish cohort consisted of primary urinary tract tumors, while the DFCI cohort both primary (n = 16) and metastatic (n = 18) tumors due to sample availability (see Table 1). All specimens were formalin-fixed and paraffin embedded (FFPE) blocks. Slides were evaluated by GU pathologists (DB and JAB) and tumor-bearing 0.6 mm cores were punched and processed for DNA and RNA extraction. All cases were collected under institutional review board-approved protocols at the different institutions, de-identified, and approved for use by the DFCI IRB.
Table 1.
Patient demographics and characteristics.
| Spanish aCGH | DFCI MIP | |||
|---|---|---|---|---|
| N | % | N | % | |
| 94 | 100% | 34 | 100% | |
| Pathological stage* | ||||
| Stage 0 (Ta) | 10 | 11% | 0 | 0% |
| Stage I (T1) | 5 | 5% | 1 | 3% |
| Stage II (T2) | 45 | 48% | 1 | 3% |
| Stage III (T3, T4) | 28 | 30% | 13 | 38% |
| Stage IV (L, M) | 5 | 5% | 10 | 29% |
| Missing | 1 | 1% | 9 | 26% |
| Metastatic sites | ||||
| Local | 16 | 17% | 9 | 26% |
| Bone | 13 | 14% | 5 | 15% |
| Liver | 8 | 9% | 9 | 26% |
| Lymph nodes | 36 | 38% | 13 | 38% |
| Lung | 7 | 7% | 13 | 38% |
| Others | 9 | 10% | 9 | 26% |
| Unknown | 5 | 5% | 0 | 0% |
| Visceral disease | ||||
| Yes | 34 | 36% | 19 | 56% |
| No | 60 | 64% | 15 | 44% |
| Unknown | ||||
| ECOG performance status | ||||
| 0 | 34 | 36% | 15 | 44% |
| > 0 | 60 | 63% | 17 | 50% |
| Unknown | 2 | 6% | ||
| Survival | ||||
| Died | 46 | 49% | 29 | 85% |
| Alive | 48 | 51% | 4 | 12% |
| Unknown | 1 | 3% | ||
Pathological stage at time of surgery. All patients either presented with metastatic disease or subsequently developed metastatic disease after local therapy.
Previously published gene expression data from patients who developed metastatic disease (Memorial Sloan-Kettering Cancer Center [MSKCC] cohort; n=37 (12)) was used to explore associations between the mRNA expression of genes located in SCNAs identified in the discovery and validation cohorts, and overall survival. We also obtained copy number data from the Cancer Genome Atlas (TCGA) bladder cancer analysis (TCGA cohort; n=99; included here with permission from TCGA), profiled with Affymetrix Genome-Wide Human SNP Arrays 6.0.
Array comparative genomic hybridization (aCGH)
aCGH was performed using genomic DNA isolated from primary tumors (Spanish cohort) and karyotypically normal reference genomic DNA (Promega, Madison, WI). The analysis was performed using Agilent Oligonucleotide Human Genome 180k CGH arrays. DNA was extracted from tumors using the QIAamp DNA FFPE Tissue Kit (Qiagen, Valencia, CA). The Genomic DNA ULS labeling kit for FFPE Samples (Agilent Technologies, Inc., Palo Alto, CA) was used to chemically label 500ng of DNA with either ULS-Cy5 (tumor) or ULS-Cy3 dye (normal/reference DNA) following the manufacturer’s protocol. Samples were hybridized to the Agilent SurePrint G3 Human CGH Microarray 4×180K. DNA samples were hybridized to the array using an Agilent microarray hybridization chamber for 40 hours in a Robbins Scientific oven with rotation at 20 rpm at 65°C. Post-hybridization, the slides were washed and scanned using an Agilent DNA microarray scanner. CGH Analytics software (version 3.4, Agilent Technologies, CA) was used to analyze the aCGH data.
Molecular inversion probe arrays (MIP)
DNA from primary tumor and metastatic specimens in the DFCI cohort was hybridized to Affymetrix OncoScanTM FFPE Express 2.0 SNP MIP arrays with 334,183 copy number and SNP probes (13). Matched normals were available for 23 samples. Copy numbers were estimated with the NEXUS software. Only samples that passed Affymetrix quality control metrics (median absolute pairwise difference [MAPD] value of ≤ 0.6) were considered.
Nanostring
mRNA was extracted from tumor specimens using standard protocols. Oligonucleotide probes for genes on 1q23.3 were synthesized by Nanostring Technologies, and transcripts were counted using the automated Nanostring nCounter system. Counts were normalized with the nSolver Analysis Software (v1.0) in which mRNA expression was compared to internal Nanostring controls, several housekeeping (ACTB, GAPHD, HPRT1, LDHA, PFKP, PGAM1, STAT1, TUBA4A, VIM) and invariant genes in bladder cancer (ANGEL1, DDX19A, NAGA, RPS10, RPS16, RPS24, RPS29). These invariant genes were identified by analyzing gene expression variances in several published datasets (12, 14, 15). After removing low quality samples flagged by the nSolver software, matched mRNA and copy number data was available for 79 patients in the Spanish cohort and 24 patients in the DFCI cohort.
Data availability
Raw and processed copy number data of the aCGH and MIP cohort is available at the Gene Expression Omnibus accession number GSE39282.
Clinical endpoints
To provide uniformity within the cohort, overall survival (OS) was determined from the start of first-line platinum-based chemotherapy for metastatic disease in the Spanish cohort. These dates were often unavailable for the DFCI cohort because of differences in patterns of care between the two cohorts. In the Spanish institutions, chemotherapy was administered at the center that contributed the specimens leading to improved quality of data regarding primary therapy and treatment start and end dates. At DFCI, the specimens were obtained from patients who were largely referred in for either surgical care or consideration of clinical trials. Due to the unavailability of initial chemotherapy treatment dates for most patients in the DFCI MIP and MSKCC cohorts, OS in these cohorts was defined as starting from the date of metastatic recurrence. The median time difference from diagnosis of metastasis to start of treatment was 1.2 months in the DFCI cohort for the 18 patients for which both dates were available, suggesting this difference is not clinically significant and that date of metastatic diagnosis was reasonable to use for these patients. Since overall survival and disease-specific survival are nearly identical in metastatic urothelial carcinoma, the former endpoint was chosen. Performance status was assessed at start of chemotherapy (Spanish) or at diagnosis of metastatic disease (DFCI).
Recurrent copy number alterations
Normalized copy number data was segmented using GLAD (16) with default parameters available in GenePattern version 3.3.3. Genomic Identification of Significant Targets in Cancer (GISTIC) software (17) (v2.0.12) was then used to identify regions of the genome that were significantly gained or deleted across a set of samples. The software estimated false discovery rates (q-values), as well as potential targets (drivers) of the aberrations. Threshold for copy number gain and loss was set so that approximately 99% of all segments in normal samples were below this threshold (+/− 0.15 for the Spanish and TCGA cohorts and +/− 0.25 for the DFCI cohort). GISTIC defines broad alterations as those spanning over more than a specified percentage of the chromosome arm, and we set this parameter the commonly used 50%.
Statistical analysis
Copy numbers of significantly gained or deleted regions (q-value < 0.25) were dichotomized based on the standard GISTIC cutoffs for amplifications or deletion (log base 2 ratio > 0.9 or < −1.3, respectively). Cox proportional hazard models were used to assess the associations of these regions with overall survival from either the time of initiation of chemotherapy for metastatic disease (Spanish cohort) or diagnosis of metastatic disease (DFCI cohort). Cox p-values in the Spanish discovery cohort were adjusted for multiple comparisons using the method by Benjamini and Hochberg (18). Regions that were associated with overall survival were further adjusted for Eastern Cooperative Oncology Group (ECOG) performance status (> 0) and presence of visceral metastasis, known prognostic factors for outcome in metastatic disease. Correlation with copy number and normalized Nanostring read counts was calculated using Spearman’s rank correlation. Statistical significance in the correlation analysis was estimated with the DRI package (19). To test for an association with survival, square root-transformed normalized read counts were used in univariate Cox regressions. MSKCC expression data cohort was obtained from GEO (GSE31684 (12)) and tested for OS association with univariate Cox regressions.
RESULTS
Clinical outcomes
For the Spanish and DFCI cohorts, median overall survival was 15.1 and 16.5 months, respectively. There were significant differences between the cohorts in the percentage of patients with visceral metastases and poor performance status. Table 1 summarizes patient characteristics for both cohorts.
Recurrent chromosomal gains and losses
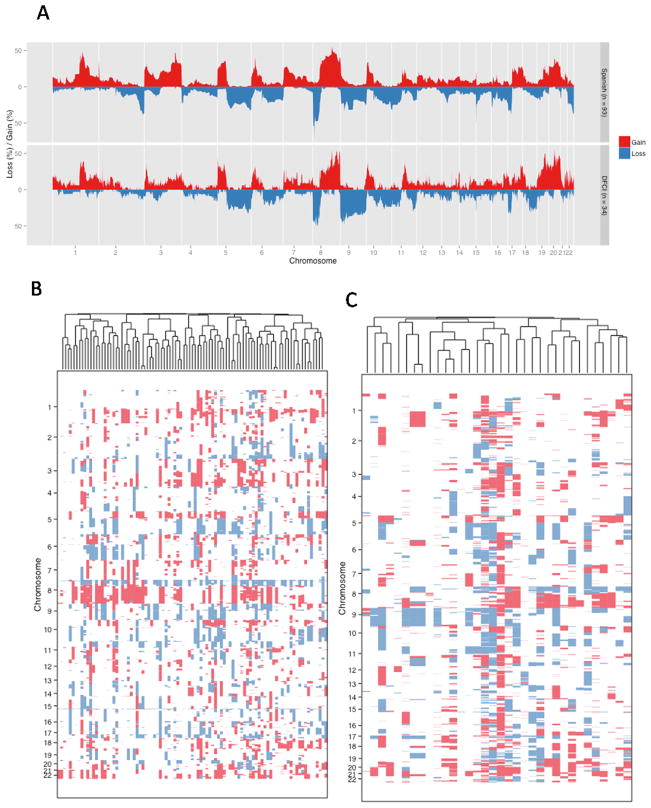
Using the GISTIC algorithm (17), we identified 96 focal (< 50% of a chromosome arm) and 22 broad (> 50% of a chromosome arm) events in the Spanish cohort and 39 focal and 13 broad events in the DFCI cohort (Figure 1A, Figures S1–5). Confirming and extending prior work (11, 20, 21), we found frequent losses of chromosomes 5q (29%), 8p (55%), 9 (p: 36%; q: 27%), 10q (31%), 11p (34%), 17p (36%), and 22q (32%) and recurrent gains of chromosomes 3q (32%), 5p (36%), 8q (41%), 19q (23%), and 20 (40%) (Tables S1–S2). The broad copy numbers were similar across the two cohorts, with only the 3q arm displaying borderline significant differences (FDR 0.05, Student t-test).
Figure 1. Recurrent copy number changes.
(a) The figure shows recurrent copy number gains and losses in the Spanish (top) and DFCI cohorts (bottom). In panels (b–c), we show hierarchical clustering of the (b) Spanish and (c) DFCI cohort.
For focal events, we focused on frequent and potentially targetable gains and amplifications. The most frequent focal gains found in the Spanish and DFCI cohorts included the YWHAZ (62%), MYC (52%), E2F3/SOX4 (47%), ERBB2 (40%), and PPARG (42%) loci (Table 2). Other alterations that are altered in a significant fraction of cases include known oncogenes such as FGFR1, MDM2, and AKT2. In contrast to previous reports (21–23), we could not indentify strong clusters in our cohorts (Figures 1B–C), likely due to the homogeneity of the cohorts, since all of these patients developed metastatic disease.
Table 2. Focal gains.
The table shows significant (q-value < 0.25) GISTIC focal gains in the Spanish and DFCI cohorts. Multiple peaks in one chromosome band were combined and the minimum q-value is shown.
| Chr | Spanish cohort | DFCI cohort | Possible Target Genes* | ||
|---|---|---|---|---|---|
| Freq. | q-value | Freq. | q-value | ||
| 1p34.2 | 14 (14.9%) | < 0.001 | 14 (41.2%) | 0.006 | MYCL1 |
| 1q23.3 | 53 (56.4%) | < 0.001 | 18 (52.9%) | 0.011 | F11R, ARHGAP30, PFDN2, PPOX, USP21, DEDD, PVRL4 |
| 3p25.1 | 40 (42.6%) | < 0.001 | 16 (47.1%) | < 0.001 |
PPARG SDHA, SLC9A3, TERT, TRIP13, PDCD6, SLC12A7, TPPP, EXOC3, CEP72, AHRR, BRD9, ZDHHC11, CLPTM1L, NKD2, LOC116349, CCDC127, PLEKHG4B, SLC6A19, |
| 5p15.33 | 32 (34%) | 0.036 | 20 (58.8%) | 0.066 | SLC6A18, LOC389257 |
| 6p22.3 | 44 (46.8%) | < 0.001 | 13 (38.2%) | < 0.001 | E2F3, SOX4 |
| 6p23 | 30 (31.9%) | 0.016 | 12 (35.3%) | 0.024 | CD83, CCDC90A, RNF182 |
| 7p21.1 | 35 (37.2%) | 0.02 | 12 (35.3%) | < 0.001 | AHR |
| 8p11.23 | 63 (67%) | < 0.001 | 12 (35.3%) | 0.005 | ADAM18, HOOK3 |
| 8p12 | 29 (30.9%) | < 0.001 | 11 (32.4%) | 0.001 | FGFR1, WHSC1L1 |
| 8q22.3 | 58 (61.7%) | < 0.001 | 21 (61.8%) | < 0.001 | YWHAZ, PABPC1, SNX31 |
| 8q24.13 | 49 (52.1%) | 0.008 | 20 (58.8%) | 0.045 | MYC |
| 10p14 | 37 (39.4%) | < 0.001 | 15 (44.1%) | 0.024 | GATA3 |
| 11q13.2 | 30 (31.9%) | < 0.001 | 14 (41.2%) | < 0.001 | CCND1 |
| 12q15 | 17 (18.1%) | < 0.001 | 11 (32.4%) | 0.003 | MDM2 |
| 17q12 | 38 (40.4%) | < 0.001 | 11 (32.4%) | 0.019 | ERBB2 |
| 18p11.31 | 31 (33%) | 0.026 | 10 (29.4%) | 0.005 | TGIF1, MYOM1, DLGAP1, MRCL3, MRLC2, FLJ35776 |
| 19q13.2 | 32 (34%) | < 0.001 | 18 (52.9%) | 0.002 | AKT2, ZNF331 |
Either known cancer genes (50) within the region or GISTIC predictions
Associations of SCNAs with OS
No broad copy number event was significantly associated with OS in the Spanish cohort (Tables S1–S2). Chromosomal instability as measured by the fraction of genome altered (21) was neither associated with initial pathologic stage, tumor site (primary vs metastasis) nor OS (Figure S6).
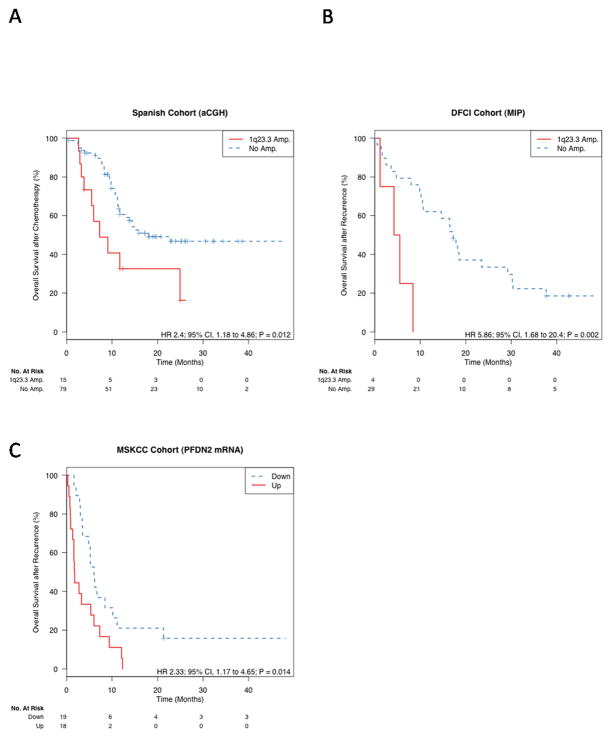
With regard to focal events, we found that only the MCL1 locus 1q21.2 and the 1q23.3 region (chromosome 1:147645-159407Mb; human genome build 18, Figure S7) were independently associated with OS after adjusting for the relevant confounding characteristics ECOG performance status and visceral metastasis (false discovery rate [FDR] 0.11, Table S3). The locus 1q23.3 (adjusted HR 2.96; 95% CI, 1.35 to 6.48; P = 0.007) represented the GISTIC peak region with the lowest, most significant q-value and largest number of copy number amplifications across all patients, and thus we focused further analysis on this region. This locus was gained (log base 2 ratio > 0.15) in 40% of all patients and amplified in 16% (log base 2 ratio > 0.9, Table S4). Amplification of 1q23.3 was not associated with initial pathologic stage at time of surgery (odds ratio [OR] 1.17; 95% CI 0.22–12.1 for pT >=2 versus pT < 2). Validation of the 1q23.3 amplification showed it was significantly associated with overall survival after recurrence in the DFCI cohort (n = 33; adj. HR 5.03; 95% CI, 1.43–17.73; P = 0.012; Figure 2). In the 100 available matched normal samples from all cohorts, we found no evidence of germline alterations of 1q23.3.
Figure 2. Prognostic utility of 1q23.3 amplification.
The figure shows the Kaplan-Meier plot for the Spanish cohort (a), DFCI MIP cohort (b). In panel (c), patients of an independent cohort were stratified by the expression median of the PFDN2 gene, located at the peak of the 1q23.3 amplification.
Gain of 1q23.3 is often accompanied by gain of the MCL1 locus 1q21.2, one of the most frequent copy number alterations across all cancer types (25). Gains and amplifications were more frequent at 1q23.3 than at 1q21.2 in the cohorts presented here and in other studies (e.g. (10, 22)), which demonstrated a driver role of 1q23.3 in UC progression independent of MCL1.
1q23.3 analysis
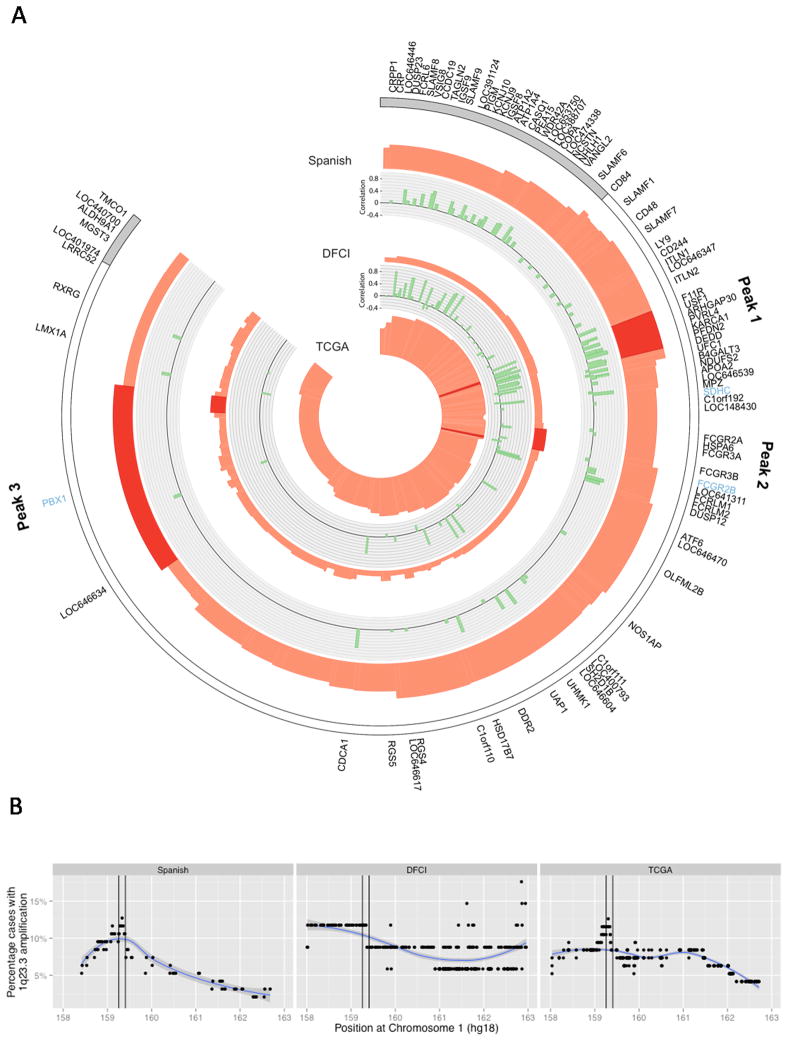
To identify potential targets of 1q23.3 copy number gain, we examined the GISTIC wide peak regions in three independent UC cohorts: the Spanish aCGH, DFCI, and TCGA cohorts (Figure 3, Supplemental Figure S8). We identified three different peak regions, each of which was observed in at least two patient cohorts (Figure 3, Table S4). Peak 1 was the highest GISTIC peak in the Spanish and TCGA cohorts, indicating the region in 1q23.3 with highest likelihood of harboring driver genes in these two cohorts according to the GISTIC algorithm. Peaks 2 and 3 had weaker associations with OS (Tables S5–6, Figures S9–10). Patients with 1q23.3 amplification often had high copy numbers in more than one peak (Figure S11). We then tested for correlation of mRNA expression and copy number in the Spanish and DFCI cohorts and observed similar results (Figure 3, Table S8).
Figure 3. Potential drivers of the 1q23.3 amplification.
(a) The figure displays the wide peak amplification regions in three bladder cancer cohorts (Spanish, DFCI and TCGA). Red peak heights visualize the statistical significance (GISTIC q-values). The q-value provides an estimate of the likelihood of the observed copy numbers at the corresponding locus in the cohort; the higher the peak, the higher the probability is that this SCNA has a driver role. The DFCI q-values are lower due to the smaller sample size, not due to a lower prevalence of this peak in this cohort. The GISTIC wide peak regions, defined by the algorithm as regions that likely contain driver genes, are colored in dark red. For the Spanish and DFCI cohorts, the correlation coefficients of gene expression and copy number are plotted in green. Genes with very low or even anti-correlation are unlikely driver genes of 1q23.3 amplification. Blue gene names are known cancer genes (50). Figure S8 provides heatmaps of this region for the Spanish and DFCI cohort. Panel (b) shows the frequency of 1q23.3 amplification in all three cohorts. Peak 1 is marked in black. This plot demonstrates that the frequency of 1q23.3 amplification (log 2 copy number ratio > 0.9) is highest in peak 1.
Copy numbers of all genes in peak 1 were highly correlated with expression (FDR < 0.001). While expression correlation alone is not sufficient to indentify driver genes (24), quantitative measures of expression such qRT-PCR and NanoString can accurately identify genes for which copy number gains and amplification elevate expression levels significantly. PVRL4 and F11R genes displayed the most profound expression changes of all peak 1 genes when we compared patients with normal and high copy numbers (Figure S12–S13). Genes in peaks 2 and 3 had a weaker copy number association with expression than genes in peak 1. Similarly, elevated mRNA expression of peak 1 genes, and genes close to this peak, was highly associated with OS (Table S7). For 64 genes in the 1q23.3 region, NanoString and copy number data was available (Table S7, Figure 3a) and expression of 24 of these genes was associated with survival (FDR < 0.2). While F11R expression displayed strong association with survival, PVRL4 did not. We then tested PVRL4 survival association on the protein level via immunohistochemistry staining in the Spanish cohort and similarly found only a weak survival association, confirming that PVRL4 is unlikely the main driver of 1q23.3 amplification (Figure S14). Finally, amplification of 1q23.3 was most frequent in peak 1 for the two large cohorts Spanish and TCGA (Figure 3b).
We then tested whether expression of 1q23.3 genes was associated with survival in an independent cohort of patients who developed metastatic UC from MSKCC (12). Median survival after recurrence in this MSKCC cohort was 4.9 months and data on follow-up until death was available for all but one patient. Copy number data was not available for this cohort, but the PFDN2, PPOX, USP21, and DEDD genes, all located in the 1q23.3 region of the first peak, showed highly correlated patterns of gene expression (pair wise ρ > 0.62, Figure S15a). These high correlations of gene expression were only observed in Spanish and DFCI cohort patients with 1q23.3 copy number gain (Figure S16), implying that those MSKCC samples with correlated over-expression of all of these genes also had copy number gain of the 1q23.3 locus. Over-expression of all four genes was associated with shorter survival after metastatic recurrence, but not with survival after cystectomy (Figure 2, Table S8). This is consistent with the findings from the Spanish and DFCI cohorts. Expression of MCL1, the FCGR family genes and PBX1, the genes found at the other peaks identified in 1q21.2 and 1q23.3, was not associated with OS in the MSKCC cohort (Table S9).
DISCUSSION
Through an unbiased genomic screen of DNA copy number in two distinct cohorts, we have shown that focal copy number gain of chromosome 1q23.3 is associated with poor overall survival in patients with metastatic urothelial carcinoma. Gain of 1q23.3 is one of the most frequent copy number alterations in UC, is most prevalent in invasive tumors (21, 22) and is further observed in multiple other cancer types (25). This work presents to our knowledge the first attempt in characterizing this major alteration in detail. We found evidence that 1q23.3 amplification could predict outcome independently of previously established prognostic variables, further strengthening the importance of these results. The supporting data from the bladder cancer cohort of the TCGA identifies the same region showing significant copy number gains.
The precise underlying target for 1q23.3 amplification remains unclear, although target prediction (17) in three cohorts with available aCGH copy number data (Spanish, DFCI and TCGA) yielded remarkably consistent results, identifying three small peak regions in this large, gene-rich 1q23.3 amplicon (Figure 3). Several candidates were further identified through expression profiling of the two cohorts presented in this study (Spanish and DFCI) and of a clinically characterized MSKCC cohort of metastatic bladder cancer patients. These data suggest that candidate genes associated with this phenotype include PVRL4, PFDN2, PPOX, USP21, F11R and DEDD. Further experimental validation is needed to demonstrate an effect of one or more of these genes in bladder cancer.
In addition to the 1q23.3 findings, this work confirms the presence of focal copy number gains of potentially druggable oncogenes in metastatic UC. ERBB2 gains are present in a subset of patients, and may highlight a patient population for which FDA-approved agents such as trastuzumab and lapatinib may be evaluated. FGFR1 and Akt are both targets of ongoing clinical investigation, with agents targeting these pathways in clinical trials. Similarly, CDKN2A deletion, and amplification of CDK4, cyclin D1, and E2F3 are potentially targetable by novel agents inhibiting CDK4.
There are several limitations to this study. While the association of 1q23.3 copy number gain with poor outcomes has been shown in unrelated cohorts in this study, the sample sizes for these validation cohorts were relatively small, treatments were non-uniform, and the tissue of origin (primary vs. metastatic) in the DFCI cohort was heterogeneous. Larger validation cohorts are necessary for a demonstration of a potential clinical utility of 1q23.3 as a biomarker. Even then, it is unlikely that this biomarker on its own will provide sufficient accuracy for the clinic, and will need to be complemented with other predictors of outcome in metastatic UC.
While the genomic alterations occurring in 1q23.3 appear complex, GISTIC peak 1 appears to be the most consistently altered region and the alteration most strongly associated with poor outcomes. This peak harbors the DEDD, PPOX, USP21, PFDN2 and F11R genes. DEDD associates with caspase-8 and -10, and plays a role in death receptor-mediated apoptosis (27). In addition, it may inhibit Cdk1/cyclin B1 signaling and play a role in cell growth (28). Over-expression of DEDD has further been shown to decrease rates of apoptosis in vitro (29). However, in breast and colon cancer, increased immunohistochemical staining was associated with improved survival, and breast cancer cell lines with overexpression demonstrated decreased metastatic potential (30). PPOX, the protoporphyrinogen oxidase, is involved in heme biosynthesis, and its inactivation causes variegate porphyria (31). USP21 is an ubiquitin-specific protease acting as regulator of centrosome and microtubule structure and is critical for development of radial microtubule formation, and primary cilia formation (32)(33). USP21 is also a major deubiquitinase of histone H2A leading to increased gene transcription (34). PFDN2 is a nuclear chaperone protein that forms a subunit of the prefoldin complex, which stabilizes newly synthesized peptides to facilitate appropriate protein folding, and may be critically important for tubulin folding (36). Another study (37) reported that PFND2 overexpression was strongly associated with disease-specific survival in a cohort of 181 high-grade bladder cancer patients with mixed pathological stages. F11R, otherwise known as Junctional Adhesion Molecule A (JAM-A), is involved in the regulation of tight junction development between epithelial cells. Downregulation of JAM-A has been associated with increased invasion, metastasis, poor prognosis, and progression in certain cancers (38, 39)(41, 42), while other studies suggest an inverse relationship between JAM-A expression and cancer cell invasiveness in vitro (40).
Another gene in within the peak is PVRL4. Intriguingly, the PVRL4 gene encodes for Nectin-4, a cell adhesion molecule involved in E-cadherin-based junctions. It is expressed in embryogenesis as well as in tumors of bladder, ovarian, breast, and lung cancer (43). Somatic missense mutations of this gene have been observed in colon, ovarian and squamous cell lung cancer (44, 45). While PVRL4 was not strongly associated with bladder cancer survival on the mRNA and protein level, suggesting that PVRL4 is not a primary target of 1q23.3 amplification, copy number gains and amplifications increased mRNA levels significantly (Figure S12). Data from other cancers suggest that Nectin-4 is associated with poor outcomes. High levels of this protein have been shown to be associated with poor survival in breast and lung cancer (46, 47). Inhibition of Nectin-4 expression by RNA interference results in decreased lung adenocarcinoma growth, and overexpression results in Rac1 activation which is postulated to lead to increased cellular invasiveness (46). In vitro evaluation will be necessary to further characterize the role of Nectin-4.
The second GISTIC peak contains an immunomodulatory gene cluster encoding low affinity Fc fragment of IgG receptors (FCGR proteins). These genes also modulate apoptotic responses and are frequently over-expressed as result of 1q23 chromosomal alterations in hematological malignancies (48). The third GISTIC peak includes the PBX1 pre-B-cell leukemia homeobox transcription factor. Over-expression of PBX1 is associated with reduced metastasis-free survival in ERα-positive breast cancer (49). We tested peaks 2 and 3 for association with survival and found a weaker relationship than with the first peak; mRNA expression showed no association with survival (Tables S5–7; Figures S9–10).
Metastatic UC remains a fatal disease in the majority of patients, and identification of novel genomic drivers of this disease may yield new therapeutic opportunities. Based on the results of this analysis, genes within 1q23.3 may lead to poor outcomes in a subset of patients with metastatic UC, and further exploration of this chromosomal region is warranted.
Supplementary Material
Table 3. Association between 1q23.3 amplification and survival.
The table shows the hazard ratios for 1q23.3 amplification in all cohorts.
| Model | Characteristic | Hazard Ratio (95% CI) | ||
|---|---|---|---|---|
| Spanish aCGH (n=94) | DFCI MIP (n=33) | |||
| Unadjusted | 1q23.3 Amp | Yes vs No | 2.4 (1.18–4.86) | 5.86 (1.68–20.39) |
|
| ||||
| Adjusted | 1q23.3 Amp | Yes vs No | 2.96 (1.35–6.48) | 5.03 (1.43–17.73) |
| ECOG PS | >1 vs 0 | 1.95 (1–3.81) | 1.6 (0.73–3.5) | |
| Visceral disease | Yes vs No | 2.39 (1.26–4.51) | 1.32 (0.62–2.82 ) | |
Translational Relevance.
No validated molecular biomarkers exist for predicting prognosis in patients with metastatic urothelial carcinoma (UC). To identify genomic predictors of poor outcome in metastatic disease, we evaluated DNA copy number alterations of primary tumors of patients who developed metastatic urothelial carcinoma. While many regions showed copy number gain and loss, gain of a short segment of chromosome 1q23.3, one of the most frequent alterations in UC, was determined to confer a poor prognosis independent of known prognostic factors, and externally validated in a cohort of primary and metastatic tumors. To elucidate the mechanism underlying this finding, an integrated analysis of this region was undertaken, which suggests that F11R, PVRL4, PFDN2, PPOX, USP21 and DEDD may play a pathogenic role in the aggressiveness of UC. Further studies into the underlying biology are ongoing.
Acknowledgments
Supported by NCI R21 CA164613-01. Spanish tissue collection supported by biobank grants from Instituto de Salud Carlos III FEDER, RD09/0076/00101. JR acknowledges support from the DFCI PART Fellowship, and MR and FM support from the DFCI Physical Sciences-Oncology Center (U54CA1437980).
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Loehrer PJ, Sr, Einhorn LH, Elson PJ, Crawford ED, Kuebler P, Tannock I, et al. A randomized comparison of cisplatin alone or in combination with methotrexate, vinblastine, and doxorubicin in patients with metastatic urothelial carcinoma: a cooperative group study. J Clin Oncol. 1992;10:1066–73. doi: 10.1200/JCO.1992.10.7.1066. [DOI] [PubMed] [Google Scholar]
- 3.Bajorin DF, Dodd PM, Mazumdar M, Fazzari M, McCaffrey JA, Scher HI, et al. Long-term survival in metastatic transitional-cell carcinoma and prognostic factors predicting outcome of therapy. J Clin Oncol. 1999;17:3173–81. doi: 10.1200/JCO.1999.17.10.3173. [DOI] [PubMed] [Google Scholar]
- 4.Bellmunt J, Choueiri TK, Fougeray R, Schutz FA, Salhi Y, Winquist E, et al. Prognostic factors in patients with advanced transitional cell carcinoma of the urothelial tract experiencing treatment failure with platinum-containing regimens. J Clin Oncol. 2010;28:1850–5. doi: 10.1200/JCO.2009.25.4599. [DOI] [PubMed] [Google Scholar]
- 5.Hurst CD, Fiegler H, Carr P, Williams S, Carter NP, Knowles MA. High-resolution analysis of genomic copy number alterations in bladder cancer by microarray-based comparative genomic hybridization. Oncogene. 2004;23:2250–63. doi: 10.1038/sj.onc.1207260. [DOI] [PubMed] [Google Scholar]
- 6.Kallioniemi A, Kallioniemi OP, Citro G, Sauter G, DeVries S, Kerschmann R, et al. Identification of gains and losses of DNA sequences in primary bladder cancer by comparative genomic hybridization. Genes, chromosomes & cancer. 1995;12:213–9. doi: 10.1002/gcc.2870120309. [DOI] [PubMed] [Google Scholar]
- 7.Richter J, Beffa L, Wagner U, Schraml P, Gasser TC, Moch H, et al. Patterns of chromosomal imbalances in advanced urinary bladder cancer detected by comparative genomic hybridization. Am J Pathol. 1998;153:1615–21. doi: 10.1016/S0002-9440(10)65750-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sandberg AA, Berger CS. Review of chromosome studies in urological tumors. II. Cytogenetics and molecular genetics of bladder cancer. The Journal of urology. 1994;151:545–60. doi: 10.1016/s0022-5347(17)35014-0. [DOI] [PubMed] [Google Scholar]
- 9.Hoglund M, Sall T, Heim S, Mitelman F, Mandahl N, Fadl-Elmula I. Identification of cytogenetic subgroups and karyotypic pathways in transitional cell carcinoma. Cancer research. 2001;61:8241–6. [PubMed] [Google Scholar]
- 10.Hurst CD, Platt FM, Taylor CF, Knowles MA. Novel tumor subgroups of urothelial carcinoma of the bladder defined by integrated genomic analysis. Clin Cancer Res. 2012;18:5865–77. doi: 10.1158/1078-0432.CCR-12-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blaveri E, Brewer JL, Roydasgupta R, Fridlyand J, DeVries S, Koppie T, et al. Bladder cancer stage and outcome by array-based comparative genomic hybridization. Clin Cancer Res. 2005;11:7012–22. doi: 10.1158/1078-0432.CCR-05-0177. [DOI] [PubMed] [Google Scholar]
- 12.Riester M, Taylor JM, Feifer A, Koppie T, Rosenberg JE, Downey RJ, et al. Combination of a novel gene expression signature with a clinical nomogram improves the prediction of survival in high-risk bladder cancer. Clin Cancer Res. 2012;18:1323–33. doi: 10.1158/1078-0432.CCR-11-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang R, Morris DS, Tomlins SA, Lonigro RJ, Tsodikov A, Mehra R, et al. Development of a multiplex quantitative PCR signature to predict progression in non-muscle-invasive bladder cancer. Cancer research. 2009;69:3810–8. doi: 10.1158/0008-5472.CAN-08-4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim WJ, Kim EJ, Kim SK, Kim YJ, Ha YS, Jeong P, et al. Predictive value of progression-related gene classifier in primary non-muscle invasive bladder cancer. Mol Cancer. 2010;9:3. doi: 10.1186/1476-4598-9-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanchez-Carbayo MSN, Lozano J, Saint F, Cordon-Cardo C. Defining molecular profiles of poor outcome in patients with invasive bladder cancer using oligonucleotide microarrays. J Clin Oncol. 2006;24:778–89. doi: 10.1200/JCO.2005.03.2375. [DOI] [PubMed] [Google Scholar]
- 16.Hupe P, Stransky N, Thiery JP, Radvanyi F, Barillot E. Analysis of array CGH data: from signal ratio to gain and loss of DNA regions. Bioinformatics. 2004;20:3413–22. doi: 10.1093/bioinformatics/bth418. [DOI] [PubMed] [Google Scholar]
- 17.Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benjamini YHY. Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. 1995;B:289–300. [Google Scholar]
- 19.Salari K, Tibshirani R, Pollack JR. DR-Integrator: a new analytic tool for integrating DNA copy number and gene expression data. Bioinformatics. 2010;26:414–6. doi: 10.1093/bioinformatics/btp702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fadl-Elmula I. Chromosomal changes in uroepithelial carcinomas. Cell & chromosome. 2005;4:1. doi: 10.1186/1475-9268-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knowles MA, Elder PA, Williamson M, Cairns JP, Shaw ME, Law MG. Allelotype of human bladder cancer. Cancer research. 1994;54:531–8. [PubMed] [Google Scholar]
- 22.Lindgren D, Sjodahl G, Lauss M, Staaf J, Chebil G, Lovgren K, et al. Integrated genomic and gene expression profiling identifies two major genomic circuits in urothelial carcinoma. PLoS One. 2012;7:e38863. doi: 10.1371/journal.pone.0038863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindgren DFA, Gudjonsson S, Sjodahl G, Hallden C, Chebil G, Veerla S, Ryden T, Mansson W, Liedberg F, Hoglund M. Combined gene expression and genomic profiling define two intrinsic molecular subtypes of urothelial carcinoma and gene signatures for molecular grading and outcome. Cancer research. 2010;70:3463–72. doi: 10.1158/0008-5472.CAN-09-4213. [DOI] [PubMed] [Google Scholar]
- 24.Akavia UD, Litvin O, Kim J, Sanchez-Garcia F, Kotliar D, Causton HC, et al. An integrated approach to uncover drivers of cancer. Cell. 2010;143:1005–17. doi: 10.1016/j.cell.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei G, Margolin AA, Haery L, Brown E, Cucolo L, Julian B, et al. Chemical genomics identifies small-molecule MCL1 repressors and BCL-xL as a predictor of MCL1 dependency. Cancer Cell. 2012;21:547–62. doi: 10.1016/j.ccr.2012.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alcivar A, Hu S, Tang J, Yang X. DEDD and DEDD2 associate with caspase-8/10 and signal cell death. Oncogene. 2003;22:291–7. doi: 10.1038/sj.onc.1206099. [DOI] [PubMed] [Google Scholar]
- 28.Arai S, Miyake K, Voit R, Nemoto S, Wakeland EK, Grummt I, et al. Death-effector domain-containing protein DEDD is an inhibitor of mitotic Cdk1/cyclin B1. Proc Natl Acad Sci U S A. 2007;104:2289–94. doi: 10.1073/pnas.0611167104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stegh AH, Schickling O, Ehret A, Scaffidi C, Peterhansel C, Hofmann TG, et al. DEDD, a novel death effector domain-containing protein, targeted to the nucleolus. EMBO J. 1998;17:5974–86. doi: 10.1093/emboj/17.20.5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lv Q, Wang W, Xue J, Hua F, Mu R, Lin H, et al. DEDD interacts with PI3KC3 to activate autophagy and attenuate epithelial-mesenchymal transition in human breast cancer. Cancer research. 2012;72:3238–50. doi: 10.1158/0008-5472.CAN-11-3832. [DOI] [PubMed] [Google Scholar]
- 31.Deybach JC, Puy H, Robreau AM, Lamoril J, Da Silva V, Grandchamp B, et al. Mutations in the protoporphyrinogen oxidase gene in patients with variegate porphyria. Hum Mol Genet. 1996;5:407–10. doi: 10.1093/hmg/5.3.407. [DOI] [PubMed] [Google Scholar]
- 32.Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hassounah NB, Bunch TA, McDermott KM. Molecular pathways: the role of primary cilia in cancer progression and therapeutics with a focus on Hedgehog signaling. Clin Cancer Res. 2012;18:2429–35. doi: 10.1158/1078-0432.CCR-11-0755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakagawa T, Kajitani T, Togo S, Masuko N, Ohdan H, Hishikawa Y, et al. Deubiquitylation of histone H2A activates transcriptional initiation via trans-histone cross-talk with H3K4 di- and trimethylation. Genes Dev. 2008;22:37–49. doi: 10.1101/gad.1609708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu G, Tan X, Wang H, Sun W, Shi Y, Burlingame S, et al. Ubiquitin-specific peptidase 21 inhibits tumor necrosis factor alpha-induced nuclear factor kappaB activation via binding to and deubiquitinating receptor-interacting protein 1. J Biol Chem. 2010;285:969–78. doi: 10.1074/jbc.M109.042689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vainberg IE, Lewis SA, Rommelaere H, Ampe C, Vandekerckhove J, Klein HL, et al. Prefoldin, a chaperone that delivers unfolded proteins to cytosolic chaperonin. Cell. 1998;93:863–73. doi: 10.1016/s0092-8674(00)81446-4. [DOI] [PubMed] [Google Scholar]
- 37.Lopez V, Gonzalez-Peramato P, Suela J, Serrano A, Algaba F, Cigudosa JC, et al. Identification of prefoldin amplification (1q23.3-q24.1) in bladder cancer using comparative genomic hybridization (CGH) arrays of urinary DNA. J Transl Med. 2013;11:182. doi: 10.1186/1479-5876-11-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fong D, Spizzo G, Mitterer M, Seeber A, Steurer M, Gastl G, et al. Low expression of junctional adhesion molecule A is associated with metastasis and poor survival in pancreatic cancer. Ann Surg Oncol. 2012;19:4330–6. doi: 10.1245/s10434-012-2381-8. [DOI] [PubMed] [Google Scholar]
- 39.Gutwein P, Schramme A, Voss B, Abdel-Bakky MS, Doberstein K, Ludwig A, et al. Downregulation of junctional adhesion molecule-A is involved in the progression of clear cell renal cell carcinoma. Biochem Biophys Res Commun. 2009;380:387–91. doi: 10.1016/j.bbrc.2009.01.100. [DOI] [PubMed] [Google Scholar]
- 40.Wang Y, Lui WY. Transforming growth factor-beta1 attenuates junctional adhesion molecule-A and contributes to breast cancer cell invasion. Eur J Cancer. 2012;48:3475–87. doi: 10.1016/j.ejca.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 41.McSherry EA, Brennan K, Hudson L, Hill AD, Hopkins AM. Breast cancer cell migration is regulated through junctional adhesion molecule-A-mediated activation of Rap1 GTPase. Breast Cancer Res. 2011;13:R31. doi: 10.1186/bcr2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gotte M, Mohr C, Koo CY, Stock C, Vaske AK, Viola M, et al. miR-145-dependent targeting of junctional adhesion molecule A and modulation of fascin expression are associated with reduced breast cancer cell motility and invasiveness. Oncogene. 2010;29:6569–80. doi: 10.1038/onc.2010.386. [DOI] [PubMed] [Google Scholar]
- 43.Fabre-Lafay S, Monville F, Garrido-Urbani S, Berruyer-Pouyet C, Ginestier C, Reymond N, et al. Nectin-4 is a new histological and serological tumor associated marker for breast cancer. BMC Cancer. 2007;7:73. doi: 10.1186/1471-2407-7-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–50. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hammerman PS, Hayes DN, Wilkerson MD, Schultz N, Bose R, Chu A, et al. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–25. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takano A, Ishikawa N, Nishino R, Masuda K, Yasui W, Inai K, et al. Identification of nectin-4 oncoprotein as a diagnostic and therapeutic target for lung cancer. Cancer research. 2009;69:6694–703. doi: 10.1158/0008-5472.CAN-09-0016. [DOI] [PubMed] [Google Scholar]
- 47.Athanassiadou AM, Patsouris E, Tsipis A, Gonidi M, Athanassiadou P. The significance of Survivin and Nectin-4 expression in the prognosis of breast carcinoma. Folia Histochem Cytobiol. 2011;49:26–33. doi: 10.5603/fhc.2011.0005. [DOI] [PubMed] [Google Scholar]
- 48.Chen W, Palanisamy N, Schmidt H, Teruya-Feldstein J, Jhanwar SC, Zelenetz AD, et al. Deregulation of FCGR2B expression by 1q21 rearrangements in follicular lymphomas. Oncogene. 2001;20:7686–93. doi: 10.1038/sj.onc.1204989. [DOI] [PubMed] [Google Scholar]
- 49.Magnani L, Ballantyne EB, Zhang X, Lupien M. PBX1 genomic pioneer function drives ERalpha signaling underlying progression in breast cancer. PLoS Genet. 2011;7:e1002368. doi: 10.1371/journal.pgen.1002368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Futreal PACL, Marshall M, Down T, Hubbard T, Wooster R, Rahman N, Stratton MR. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–83. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw and processed copy number data of the aCGH and MIP cohort is available at the Gene Expression Omnibus accession number GSE39282.