Abstract
Some 200 different 5S rRNA sequences from eubacteria, chloroplasts, mitochondria, archaebacteria, and eukaryotes were analyzed for evolutionary kinship relationships and associated sequential features. Group-specific occupation schemes for the 149 positions of an overall alignment were established. Eubacterial, archaebacterial, and intermediate occupation schemes all yield a strongly biased base triplet pattern in one of the three possible reading frames strongest for eubacterial, chloroplastic, and archaebacterial, but still detectable for mitochondrial and eukaryotic cytoplasmic sequences. The frequency of triplets decays in the order RNY greater than RNR greater than YNY greater than YNR; R being a purine (guanine or adenine), Y is a pyrimidine (cytosine or uracil), and N is any base. A strong preference for guanine or cytosine was found in all triplet positions. The effects show no exceptions and are clearly above the level of statistical fluctuations.
Full text
PDF

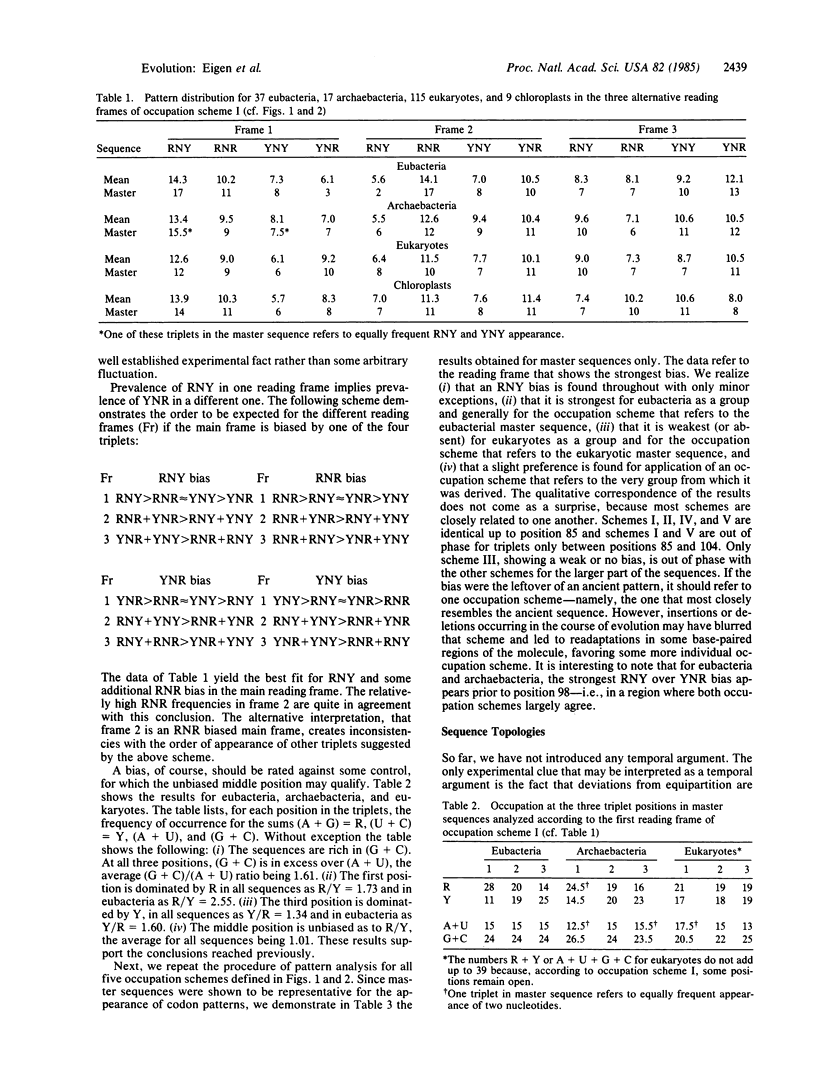
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Eigen M., Winkler-Oswatitsch R. Transfer-RNA, an early gene? Naturwissenschaften. 1981 Jun;68(6):282–292. doi: 10.1007/BF01047470. [DOI] [PubMed] [Google Scholar]
- Eigen M., Winkler-Oswatitsch R. Transfer-RNA: the early adaptor. Naturwissenschaften. 1981 May;68(5):217–228. doi: 10.1007/BF01047323. [DOI] [PubMed] [Google Scholar]
- Erdmann V. A., Wolters J., Huysmans E., Vandenberghe A., De Wachter R. Collection of published 5S and 5.8S ribosomal RNA sequences. Nucleic Acids Res. 1984;12 (Suppl):r133–r166. doi: 10.1093/nar/12.suppl.r133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klotz L. C., Komar N., Blanken R. L., Mitchell R. M. Calculation of evolutionary trees from sequence data. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4516–4520. doi: 10.1073/pnas.76.9.4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küntzel H., Heidrich M., Piechulla B. Phylogenetic tree derived from bacterial, cytosol and organelle 5S rRNA sequences. Nucleic Acids Res. 1981 Mar 25;9(6):1451–1461. doi: 10.1093/nar/9.6.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd J. C. Method to determine the reading frame of a protein from the purine/pyrimidine genome sequence and its possible evolutionary justification. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1596–1600. doi: 10.1073/pnas.78.3.1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd J. C. Periodic correlations in DNA sequences and evidence suggesting their evolutionary origin in a comma-less genetic code. J Mol Evol. 1981;17(2):94–102. doi: 10.1007/BF01732679. [DOI] [PubMed] [Google Scholar]