Abstract
Techniques enabling precise genome modifications enhance the safety of gene-based therapy. DLC1 is a hot spot for phiC31 integrase-mediated transgene integration in vitro and in vivo. Here we show that integration of a coagulation factor VIII transgene into intron 7 of DLC1 supports durable expression of factor VIII in primary human umbilical cord-lining epithelial cells. Oligoclonal cells with factor VIII transgene integrated in DLC1 did not have altered expression of DLC1 or neighbouring genes within a 1-Mb interval. Only 1.9% of all expressed genes were transcriptionally altered; most were downregulated and mapped to cell cycle and DNA repair pathways. DLC1-integrated cells were not tumourigenic in vivo and were normal by high-resolution genomic DNA copy number analysis. Our data identify DLC1 as a locus for durable transgene expression that does not incur features of insertional oncogenesis, thus expanding options for developing ex vivo cell therapy mediated by site-specific integration methods.
Introduction
One of the fundamental requirements for developing gene-based cell therapy for disorders caused by deficient production of specific proteins, such as the haemophilias, is durable expression of the corrective transgene product. This can be optimally achieved by stably integrating the appropriate transgene into the genome. Several clinical trials that used integrating viral vectors have provided proof-of-principle by successfully correcting the disease phenotype of several monogenic disorders.1, 2, 3, 4, 5, 6 However, serious adverse outcomes that later emerged in some of these studies brought to light oncogenic complications incurred by random integration of transgenes. It is now understood that transactivation of neighbouring oncogenes such as LMO2, CCND2, BMI1 and the MDS-EVI1 complex following retroviral vector integration can lead to clonal cell expansion, myelodysplasia and overt leukaemia.7, 8, 9 These oncogenic complications motivate the continuing search for non-viral methods that integrate therapeutic transgenes in safe genomic regions.10
The non-viral phiC31 integrase system induces stable expression of transgenes that are integrated into endogenous pseudo attP sites in mammalian genomes.11, 12, 13, 14 It has been successfully used to correct deficiencies of fumarylacetoacetate hydrolase,15 factor IX,16 factor VIII17, 18 and dystrophin19 in murine disease models. Bioinformatic analysis predicted that phiC31 integrase could potentially mediate integrations into about 370 different genomic sites.11 In practice, however, integrations have been found experimentally in only a small subset of these sites. Along with others, we have identified a few sites in the human genome where transgenes integrate with high frequency. Among these hot spots are hitherto uncharacterized loci in 8p22(refs 12, 13, 17, 20) and 19q13.31.11
We have reported that phiC31 integrase-modified primary human cord-lining epithelial cells (CLECs) durably expressed a human coagulation factor FVIII (FVIII) transgene and corrected the disease phenotype when implanted in FVIII-deficient mice.17 Up to 40% of FVIII transgene integrations in a mixed population of CLECs occurred in 8p22, and clonal cultures of genome-modified CLECs showed no clear markers of genotoxic risk. This drew our attention to the 8p22 locus that appeared to be both permissive for durable transgene expression and potentially safe against oncogenic risk. Here, we show that transgene integration into intron 7 of DLC1 in 8p22 leaves a minimal and benign footprint in the genome and transcriptome, and does not induce tumourigenic behaviour in genome-modified cells. Our data identify this site as a likely safe harbour for gene-based cell therapies that require integration of transgenes.
Results
A high expression human–porcine FVIII transgene
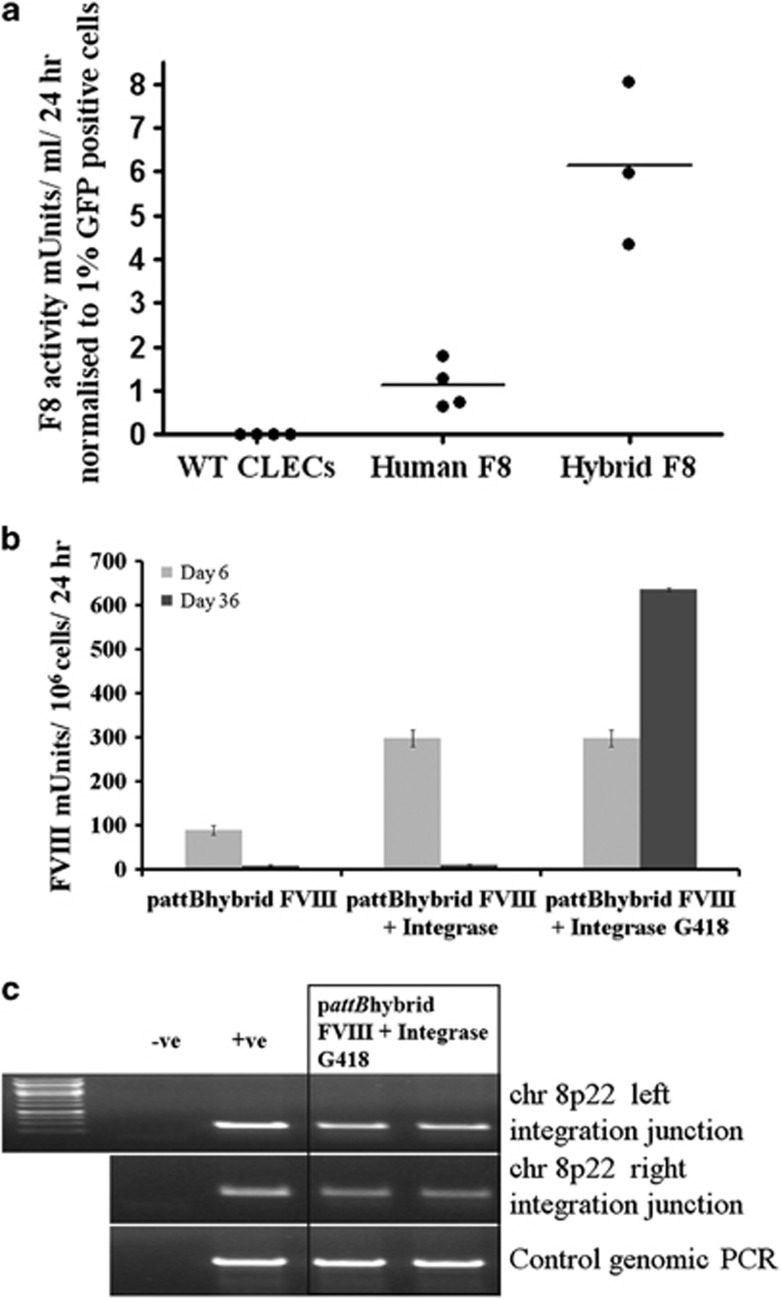
We designed and assembled a hybrid human–porcine FVIII complementary DNA (cDNA) that was similar to a construct reported to induce 10-fold higher expression than human FVIII cDNA.21 A plasmid encoding B domain-deleted (BDD) hybrid FVIII cDNA comprising porcine A1 and A3 domains, a residual human B domain retaining eight glycosylation sites, and human A2, C1 and C2 domains. To compare the efficacy of the human–porcine hybrid BDD FVIII cDNA with BDD human FVIII cDNA F309S,17 CLECs were co-electroporated with plasmids expressing enhanced green fluorescent protein and either human or human–porcine FVIII. Both FVIII cDNAs were expressed from the human ferritin light chain promoter. After normalizing for transfection efficiency, assays of FVIII activity secreted by transiently transfected CLECs showed approximately 5.5-fold higher secretion of human–porcine FVIII (6.13±1.07 mU ml–1 per 24 h) compared with human FVIII (1.12±0.26 mU ml–1 per 24 h; P=0.003; Figure 1a).
Figure 1.
Enhanced secretion of human–porcine hybrid BDD-FVIII and phiC31 integrase-mediated integration by transfected CLECs. (a) FVIII activity (Coamatic FVIII assay) in conditioned media of naive CLECs or CLECs co-electroporated with an enhanced green fluorescent protein reporter gene and either human BDD-FVIII F309S cDNA or human–porcine hybrid BDD-FVIII cDNA. Data are expressed as mU ml–1 per 24 h normalized to transfection efficiency. Each data point shows the mean of triplicate assays of a single experiment. An average 5.5-fold increase in FVIII levels (P=0.003; Student's unpaired t-test) was secreted by the hybrid FVIII cDNA compared with human FVIII cDNA. (b) FVIII activity of overnight conditioned media of three groups of electroporated CLECs: (i) pattBhybrid FVIII, no G418 selection; (ii) pattBhybrid FVIII+Integrase, no G418 selection; and (iii) pattBhybrid FVIII+Integrase with G418 selection. Conditioned media was collected on day 6 (before G418 selection) and day 36 (after G418 selection; where indicated) post-electroporation. Data are mean±s.e.m.; n=3 per group. (c) Genomic DNA (200 ng) extracted from a bulk population of genome-modified CLECs was amplified with primer pairs specific for the vector and chromosome 8p22 to detect the presence of left and right integration junctions indicative of correct integration at the pseudo attP site in 8p22 (done in duplicates). ‘–ve' denotes minus template PCR amplification while ‘+ve' denotes amplification from genomic DNA isolated from a clonal line of CLEC previously identified to have integration at 8p22. Control PCR amplified a 900-bp region in 19q13.42 (AAVS1 locus). Amplified products were resolved by 1% agarose gel electrophoresis and imaged using BioRad Gel Doc 2000 transilluminator and QuantityOne software (Bio-Rad Laboratories).
PhiC31 integrase-mediated integration of hybrid FVIII cDNA in CLECs
To determine durability of FVIII transgene expression by phiC31 integrase-mediated genomic integration, CLECs were electroporated only with pattBhybrid FVIII (an attB-bearing plasmid encoding the neomycin resistance gene and hybrid FVIII cDNA expressed from human light chain ferritin promoter; Figure 1b: pattBhybrid FVIII) or co-electroporated with pCMV-Int (a plasmid encoding CMV promoter-phiC31 integrase)22 using the Amaxa Nucleofector I device (Lonza, Basel, Switzerland). Electroporated CLECs were either unselected (Figure 1b: pattBhybrid FVIII and pattBhybrid FVIII+Integrase) or subjected to G418 selection (Figure 1b: pattBhybrid FVIII+Integrase G418) to derive cultures stably integrated with the pattBhybrid FVIII construct. FVIII activity assay (Figure 1b) of conditioned media showed durable FVIII expression from G418-selected CLECs that were co-electroporated with pattBhybrid FVIII and phiC31 integrase (day 6: 298.30±19.01 mU FVIII/106 cells per 24 h; day 36: 635.6±3.40 mU FVIII/106 cells per 24 h) but not from CLECs that were similarly transfected without G418 selection (day 6: 298.30±19.01 mU FVIII/106 cells per 24 h; day 36: 8.97±2.51 mU FVIII/106 cells per 24 h), indicating that G418 selection enriched for cells with stable genomic integration of the FVIII transgene. CLECs that were transfected only with pattBhybrid FVIII and not G418-selected also showed a steep decline in FVIII secretion from day 6 to day 36 (day 6: 89.02±9.77 mU FVIII/106 cells per 24 h; day 36: 7.16±2.40 mU FVIII/106 cells per 24 h), consistent with loss of episomal FVIII plasmid with successive cell divisions and negligible genomic integration in the absence of the integrase.
In our earlier study, transgene integrations into 8p22 accounted for about 40% of all recovered integration events in a mixed population of CLECs.17 In order to characterize only CLECs with 8p22 integration at high resolution, we screened stable G418-resistant CLECs derived by co-electroporating pattBhybrid FVIII and phiC31 integrase by junction PCR using primers specific for sequences within the FVIII vector and a previously identified pseudo attP site in 8p22.11, 20 PCR amplification of both left and right integration junctions was evidence for site-specific integration of FVIII transgene cassette at 8p22 (Figure 1c). Sequencing the integration junction PCR products confirmed transgene integration at the 8p22 hot spot. Sequence analysis showed a 6-bp deletion in the vector sequence at the left integration junction and a 7-bp deletion in the genomic region at the right integration junction (Supplementary Figure S1).
Screening oligoclonal CLECs with 8p22 integration
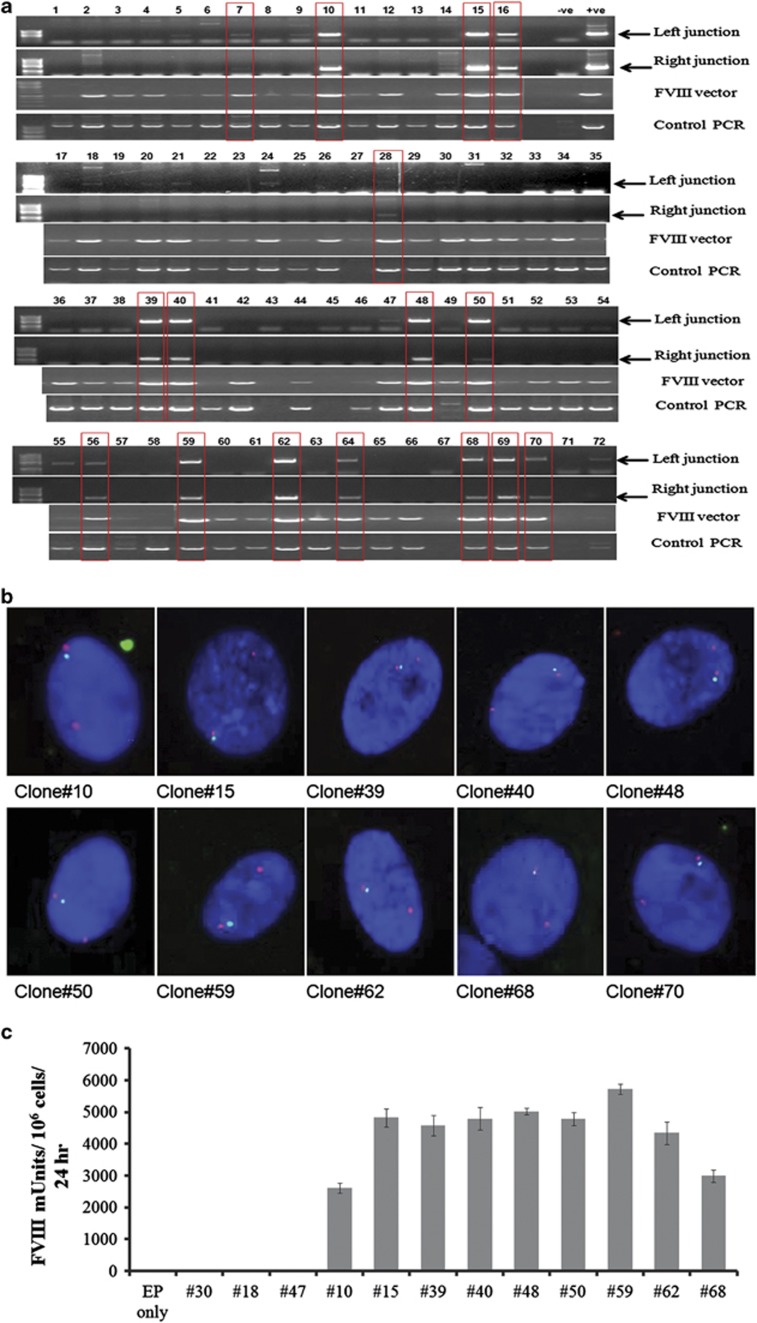
Evidence of 8p22 integrations in the bulk-transfected population prompted us to derive clonal populations of genome-modified CLECs having this specific integration. We examined oligoclonal CLEC cultures obtained by flow-sorting four cells into each well of 96-well plates for evidence of 8p22-specific transgene integration by direct in situ PCR. Using two primer pairs specific to the vector sequence and sequence at the 8p22 hot spot, genomic DNA from in situ lysed cells were screened by PCR for the presence of left and right integration junctions, indicative of complete transgene integration. Intron-spanning primers specific to C1-domain of FVIII cDNA were used to amplify and detect the presence of the integrated transgene. A control PCR that amplified an adjacent genomic region in 19q13.42 served both to verify integrity of genomic DNA and to act as a positive control for PCR among all samples. Sixty-six of 72 oligoclonal populations were positive for the control PCR amplification. Of these positive oligoclones, 16 (24%) amplified both left and right integration junctions at 8p22 (Figure 2a).
Figure 2.
Identification of oligoclonal CLECs with transgene integration at 8p22 and FVIII secretion. (a) Oligoclonal cells (four flow-sorted cells per well) from a bulk population of G418-selected CLECs electroporated with pattBhybrid FVIII and phiC31 integrase were investigated by direct in situ PCR for transgene integration at the 8p22 hot spot. In situ lysed cells were screened using Phusion human specimen direct PCR kit (Thermo Scientific) and primers specific for vector and genomic sequences at the 8p22 integration site to detect the presence of left and right integration junctions. Intron-spanning vector-specific primers were used to amplify C1 domain of the integrated FVIII cDNA (FVIII vector). Control genomic PCR amplified a 900-bp region in 19q13.42 (AAVS1 locus). ‘–ve' denotes minus template PCR amplification while ‘+ve' denotes amplification from genomic DNA isolated from a previously identified clonal CLEC with integration at 8p22. Amplified products were electrophoresed on 1% agarose gels and imaged using BioRadGel Doc 2000 transilluminator and QuantityOne software. Red box highlights samples, which were positive for left and right integration junctions, FVIII vector and control PCR. (b) Genome-modified oligoclonal CLECs identified by junction PCR to be positive for transgene integration at chromosome 8p22 were screened by FISH. Fluorescence images of 4,6-diamino-2-phenylindole (DAPI)-stained cells hybridized with fluorescein isothiocyanate (FITC)-labelled probes specific to integrated vector (green signal) and Texas Red-labelled centromeric probes specific to chromosome 8 (red signal) are shown (original magnification × 600). Integration of transgene at chromosome 8 is indicated by the close proximity of the vector-specific green signal and chromosome 8 centromere red signal. (c) CLECs that were electroporated without any plasmid DNA (EP only) and oligoclonal CLECs that were positive for transgene integration at 8p22 (10, 15, 16, 28, 39, 40, 50, 59, 62, 68 and 69) and three oligoclonal populations that were negative for 8p22 transgene integration (18, 30 and 47) were evaluated for FVIII secretion (day 40 post-electroporation). Overnight conditioned media was assayed for FVIII activity using a Coamatic FVIII kit (Chromogenix). Data are mean±s.e.m.; n=3 per group.
Transgene integration in chromosome 8 in oligoclonal CLECs was confirmed by fluorescence in situ hybridization (FISH) data, which showed close proximity of signals specific to probes for chromosome 8 centromere (red signal) and the integrated transgene (green signal; Figure 2b). This was consistent with results of junction PCR (Figure 2a) and amplicon sequencing (Supplementary Figures S1c and d). Oligoclonal CLECs with transgene integration at 8p22 were further characterized by FISH using transgene-specific probes to determine transgene copy number (Supplementary Table 1). The majority of cells had a single copy of the integrated transgene. However, a small proportion of cells (0.6–7%) had two copies suggesting transgene integration into other loci besides 8p22. We screened 13 oligoclonal CLECs derived from the mixed population of G418-selected CLECs treated with pattBhybrid FVIII+Integrase for FVIII expression 40 days post-electroporation. Nine oligoclones (Figure 2a; oligoclones 10, 15, 39, 40, 48, 50, 59, 62 and 68) with evidence of transgene integration at 8p22 by junction PCR were selected, whereas three oligoclones (Figure 2a; oligoclones 18, 30 and 47) were negative by junction PCR. All nine positive oligoclones secreted FVIII at high levels (2610–5724 mU FVIII/106 cells per 24 h), whereas the three negative oligoclones did not secrete FVIII (Figure 2c), showing that transgene integration at 8p22 sustained durable FVIII secretion by genome-modified cells.
We proceeded to characterize eight 8p22-modified oligoclones for oncogenic and genotoxic alterations, compared with unmodified CLECs.
DLC1 expression
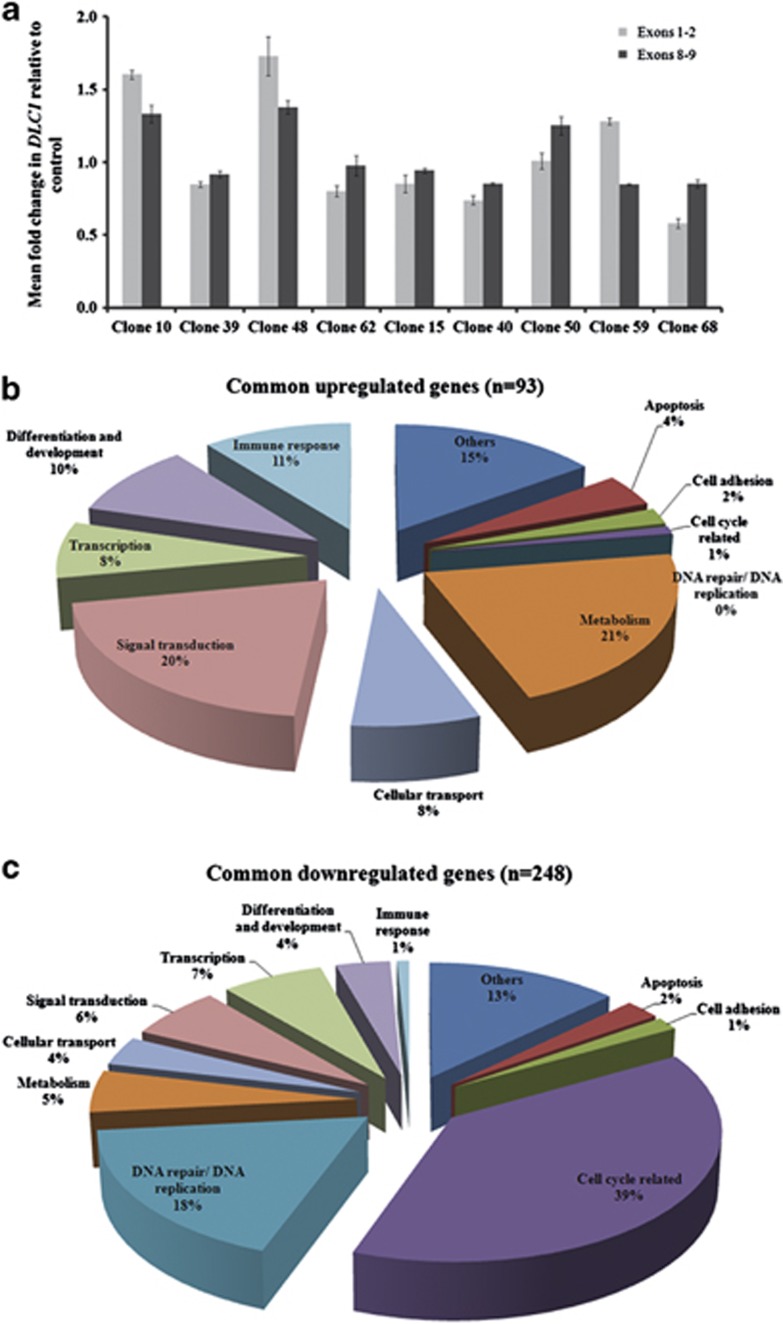
As the transgene integration site in 8p22 is within intron 7 of a tumour-suppressor gene, DLC1,23 it was of interest to determine its expression in genome-modified CLECs. We performed quantitative reverse transcription (RT)-PCR to compare DLC1 transcript levels (exons 1–2 and exons 8–9) in nine 8p22-integrated oligoclones and in unmodified control CLECs from the same donor (that is, electroporated without plasmid DNA but having the same number of population doublings in culture). DLC1 transcript levels were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression in all reactions. The ratio of DLC1 transcript levels in CLECs with transgene integration in 8p22 relative to DLC1 transcripts levels of control CLECs ranged between 0.58–1.603 (exons 1–2) and 0.851–1.38 (exons 8–9), showing minimal perturbations to DLC1 expression caused by transgene integration at 8p22 (Figure 3a).
Figure 3.
RT-PCR and transcriptome analysis of 8p22 oligoclonal CLECs. (a) Quantitative RT-PCR was performed on control CLECs that had been electroporated without any DNA and oligoclonal CLECs with transgene integration at 8p22 to quantify DLC1 transcript levels (exons 1–2 and exons 8–9) and GAPDH expression. The mean fold change in GAPDH-normalized DLC1 expression levels in oligoclonal samples (exons 1–2 and exons 8–9) are shown relative to that of control CLECs as determined by the 2-ΔΔCt method. Data are mean±s.e.m.; n=3 experiments per group. (b, c) Categorization of genes that differed in their expression by ⩾2.5-fold in all eight oligoclonal CLECs relative to control CLECs. The percentage of (b) significantly upregulated and (c) significantly downregulated genes categorized according to their biological functions are depicted.
Transcriptome and genome copy number analyses of DLC1-integrated oligoclonal CLECs
The extent to which transgene integration in DLC1 altered global gene expression of oligoclonal CLECs was investigated by transcriptome profiling on Affymetrix PrimeView expression arrays (Affymetrix, Santa Clara, CA, USA). Transcripts that differed by ⩾2.5-fold compared with unmodified control CLECs were identified.
Transcriptome analysis identified a total of 341 genes out of a total of 17 719 expressed genes (1.9%) that were commonly altered in expression in all 8 oligoclonal CLECs with DLC1 integration compared with control CLECs (Supplementary Table 2). Of these, 93 genes (0.5%) were upregulated and 248 genes (1.4%) were downregulated. No changes were detected in DLC1 expression or genes that mapped within a 1-Mb window (LONRF1, KIAA456 and C8orf48) centred on the 8p22 integration site for all eight oligoclonal CLECs. Genes whose expressions were altered in oligoclonal CLECs were classified and clustered according to their biological functions and their proportional categorizations are depicted in Figures 3b and c. It was noteworthy that up to 40% of underexpressed genes were involved in cell cycle regulation, suggesting that oligoclonal CLECs with DLC1 integration could be less proliferative compared with control CLECs. A possible reason for downregulated cell cycle genes was the increased metabolic burden from artificially induced overexpression of a transgenic protein.24 Given that oligoclonal CLECs had been cultured continuously in vitro for at least 2 months, downregulation of cell cycle genes could also reflect onset of senescence of these primary cells. In either case, downregulation of cell cycle genes was contrary to the transcriptional profile of transformed cells. Further characterization of oligoclonal CLECs by the MTS assay revealed decreases in proliferation in five of six oligoclonal CLECs evaluated (15, 39, 48, 50, 59 and 62) by 28%−47% (Supplementary Figure S3). This was in accord with the observed downregulated transcription of cell cycle genes that caused a slowdown in proliferation of these cells.
The list of genes with altered expression was submitted to DAVID pathway mapping analysis and compared against a list of known and potential oncogenes.25 This clustered downregulated genes (46 genes) mainly to cell cycle and DNA repair pathways (KEGG pathway database; Supplementary Figure S2). Genes with upregulated expression were not significantly clustered to any pathway. Referencing the altered genes to a database of known and potential oncogenes identified 9 and 31 oncogenes that were up- and downregulated, respectively (Supplementary Table 2). In summary, transcriptome analysis of oligoclonal CLECs with transgene integration in intron 7 of DLC1 did not display signatures of transformed cells.
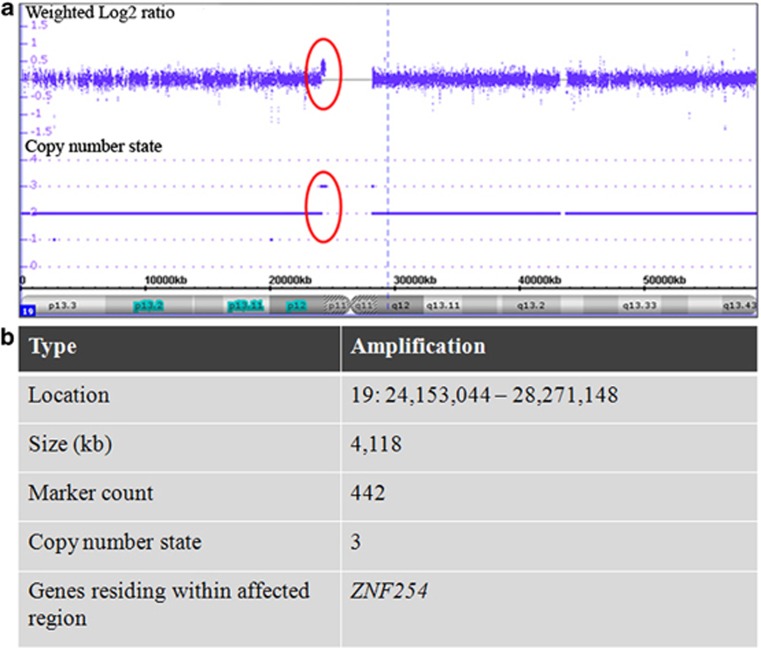
With a single exception, all eight oligoclones with DLC1 integrations did not show significant alterations in genome copy number determined by hybridization on high-density single-nucleotide polymorphism arrays (average probe spacing 1148 bp; Affymetrix CytoScan HD array). A single amplification (copy number state of 3) in the peri-centromeric region of chromosome 19 (spanning 4118 kb) was present in oligoclone 10 (Figure 4). Only a single gene, ZNF254, resided within this amplified region but its expression was not changed by more than two-fold.
Figure 4.
Copy number change in chromosome 19 in CLEC oligoclone 10. (a) Copy number profile of chromosome 19 of CLEC oligoclone 10 with transgene integration at chromosome 8p22 generated on Affymetrix Cytoscan HD Array Set. Weighted log2 signal intensity ratios and copy number state across chromosome 19 are depicted on the vertical axis, and cytobands and distance from the start of the chromosome on the horizontal axis. A single locus with significant amplification (identified by at least 50 consecutive probes showing concordant change) is circled. (b) Characteristics of the significantly amplified genomic region. The genomic location, physical size, number of probes and copy number state across the affected region in chromosome 19 as well as the single gene residing within this region are tabulated.
Tumourigenicity and FVIII expression in vivo
FVIII-expressing DLC1-integrated oligoclonal CLECs were implanted in the subcutaneous nuchal region of NOD-SCID IL2Rγ null (NSG) mice. Survival and engraftment of implanted cells was monitored by measuring levels of transgenic hybrid FVIII secreted by implanted CLECs into murine plasma using a FVIII-antigen capture assay specific for human FVIII that did not cross-react with murine FVIII. Untreated mice and mice implanted with CLECs that were electroporated without plasmid DNA served as negative controls for tumour formation and plasma hybrid FVIII measurements, respectively. NSG mice implanted with Hs746T, a human cancer cell line, served as positive tumourigenic controls.
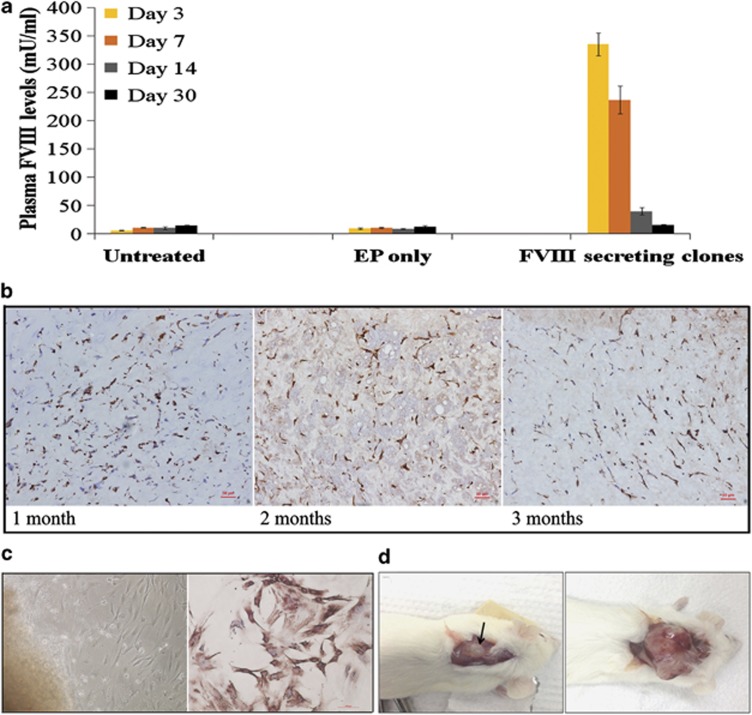
Hybrid FVIII was readily detected at high levels in murine plasma 3 days post-implantation (335.65±19.98 mU ml–1; Figure 5a). Although plasma levels of transgenic FVIII showed a declining trend (that is, 236.9±24.67 mU ml–1 on day 7 and 40.17±6.29 mU ml–1 on day 14), day 14 levels remained significantly higher than untreated mice (10.5±2.45 mU ml–1; P=0.0094) and mice implanted with unmodified CLECs (8.67±1.06 mU ml–1; P=0.0063). Plasma FVIII levels were not different from control mice by day 30 (P>0.05) although CLECs were demonstrable at implantation sites for at least 3 months after implantation (Figure 5b). Loss of transgenic FVIII levels in plasma may not have been caused by silencing of FVIII transgene expression in implanted CLECs. We recovered and re-established secondary cultures from implanted cells, and documented that they retained the capacity to secrete FVIII in vitro (1304±34.05 mU FVIII/106 cells per 24 h). Recovered CLECs were positive for human vimentin by immunohistochemical staining (Figure 5c). Collectively, these data indicate that a proportion of the initially implanted cells did survive and established long-term engraftment but were not sufficiently numerous to secrete detectable levels of transgenic FVIII in murine plasma. Although no tumours were observed for up to 3 months after implantation of 3 × 106 unmodified (n=4) or FVIII-expressing CLECs (n=16), large tumours were readily observed as early as 3 weeks in mice implanted with the same number of Hs746T cells (n=4; Figure 5d). These data showed that oligoclonal DLC1-integrated CLECs, which had been cultured in vitro for at least 2 months were unlikely to be tumourigenic.
Figure 5.
Assessment of in vivo FVIII secretion and tumourigenicity following implantation of CLECs stably secreting transgenic hybrid FVIII in NSG mice. (a) Plasma FVIII levels of NSG mice that were unimplanted (n=4) or implanted subcutaneously with 3 × 106 cells per animal of (i) Matrigel-encapsulated CLECs that received electroporation only (EP only, n=4); or (ii) FVIII-secreting oligoclonal CLECs (n=16) with pattBhybrid FVIII integrated at 8p22 were measured using an assay specific for human FVIII (Visulize FVIII antigen ELISA kit) on days 3, 7, 14 and 30 post-implantation. Plasma FVIII levels of mice implanted with FVIII-secreting cells were significantly higher (P<0.05, Student's unpaired t-test) on days 3 (yellow bar), 7 (orange bar) and 14 (grey bar) compared with control mice that were either unimplanted or implanted with cells electroporated without any plasmid DNA (EP only). Data are mean±s.e.m. (b) Immunohistochemical staining of Matrigel implants retrieved by excision from implanted mice at the indicated time points post-implantation show evidence (brown staining) for the presence of engrafted CLECs expressing human vimentin. Scale bar=50 μm. (c) (Left) Brightfield view of outgrowth cells from a culture of Matrigel implants retrieved 2 months post-implantation; and (right) immunohistochemical staining of explant cultured cells for human vimentin expression. Scale bar=100 μm. (d) No tumours were observed in mice implanted with Matrigel-encapsulated 3 × 106 genome-modified CLECs at day 90 post-implantation (left) while nodular tumours developed in mice implanted with equal numbers of Matrigel-encapsulated Hs746T, a known tumourigenic cell line, 1-month post-implantation (right). Arrow points to Matrigel implant.
DLC1 integration in primary adult cells
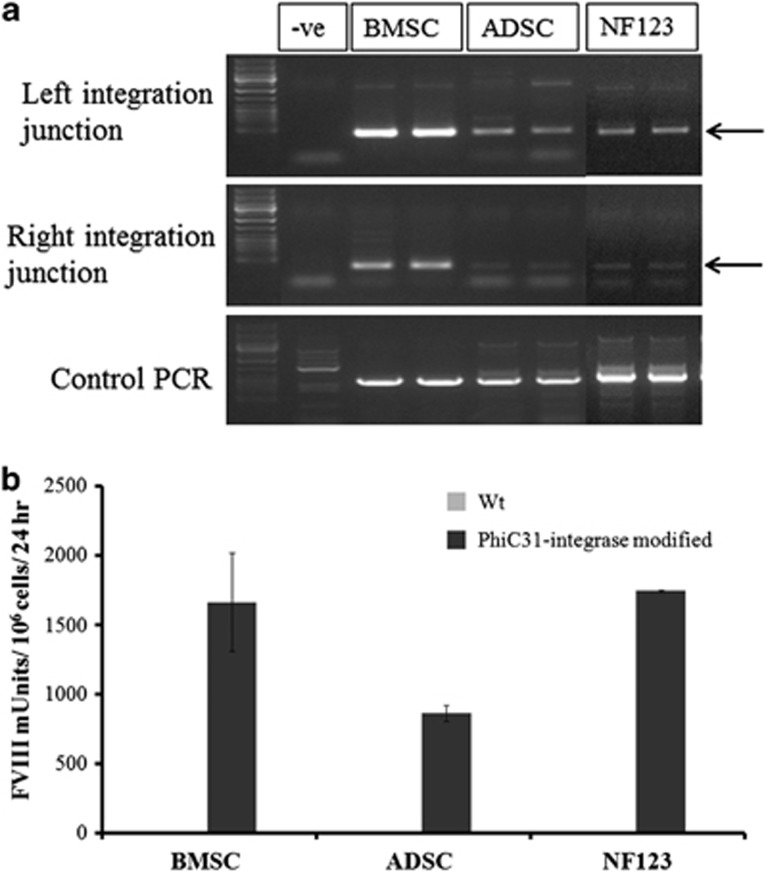
Primary cultures of bone marrow-derived stromal cells, adipose-derived stromal cells and dermal fibroblasts were co-electroporated with pattBhybrid FVIII and phiC31 integrase plasmids, then selected with G418 to derive stable cell cultures. Integration junction PCR was positive for site-specific transgene integration in intron 7 of DLC1 in all three primary cell types (Figure 6a), showing that this site was permissive for transgene integration in a range of adult primary human cell types. Moreover, these stably modified cells secreted FVIII expression for at least 1 month (Figure 6b) indicating that the human ferritin light chain promoter was not silenced and could stably drive hybrid FVIII expression in several cell types.
Figure 6.
Transgene integration at 8p22 and durable FVIII secretion in phiC31 integrase-modified primary adult human cells. (a) G418-resistant stable cells derived from primary cultures of bone marrow-derived stromal cells (BMSC), adipose-derived stromal cells (ADSC) and normal dermal fibroblasts (NF123) co-electroporated with pattBhybrid FVIII and phiC31 integrase were examined for evidence of transgene integration into intron 7 of DLC1 by left and right integration junction PCR. ‘–ve' refers to minus template amplification. Left and right integration junctions were amplified with vector-specific and genomic DNA-specific primers while control genomic PCR was performed with a pair of genome specific-primers amplifying a 900-bp region within chromosome 19. Amplified products were electrophoresed on 1% agarose gels and imaged using BioRad Gel Doc 2000 transilluminator and QuantityOne software. A composite image of the original gel is shown. Black arrows indicate the predicted integration junction PCR amplified bands. (b) FVIII activity in overnight conditioned media of untreated (Wt) and G418-resistant (phiC31 integrase-modified) BMSC, ADSC and NF123, determined 1 month after co-electroporation with pattBhybrid FVIII and phiC31 integrase. Data are mean±s.e.m.; n=3 per group.
Discussion
Oncogenic complications that have marred several clinical cell therapy trials have renewed concerns about the risks of randomly integrating gene delivery vectors. Developing non-viral methods capable of modifying genomes efficiently and with precision will be necessary to advance the biosafety of gene-based cell therapies.
Although phiC31 integrase is effective in mediating stable transgene integrations and expression in a variety of mammalian cells, its activity in integrating donor DNA into multiple genomic sites could be perceived to be a barrier to clinical applications. Along with others,12, 13, 20 we17 have noted that intron 7 of DLC1 in 8p22 is a frequent site for phiC31 integrase-mediated transgene insertion. Its permissiveness to genome modifications is possibly due to wide expression of DLC1 in normal human tissues.26, 27 Here we show that the use of phiC31 integrase for stable transgene integration could still be useful and rendered safer by screening, identifying and evaluating clonal cells that have integrated the transgene into a specific site, that is, DLC1. Our data suggest that, given the high frequency of integration in DLC1, such cells could justifiably be evaluated for insertional oncogenesis and efficacy of transgene expression ex vivo by multiple techniques to determine their suitability for cell therapy. DLC1 is not an obvious safe genomic harbour site, being a tumour suppressor. Moreover, it was not known if this site is capable of sustaining durable transgene expression. It was therefore of interest to assess the genotype and phenotype of primary human cells that had integrated a hybrid FVIII transgene within DLC1. We have previously shown that transgene integrations into 8p22 were either mono- or biallelic.17 Hence, the foregoing genomic and transcriptome analyses in this study reflected genome modification at this single site, unconfounded by the effects of simultaneous transgene integrations into heterogeneous sites. We used integration junction PCR as a screening method to identify cells with transgene integration at 8p22, followed by FISH with transgene-specific probes to confirm integration in chromosome 8. Although these techniques were facile as screening methods for single site-specific integration, Southern blotting could have detected the occurrence of transgene integrations at multiple genomic sites.
Some oligoclones (#55, 20, 21, 24 and 30) appeared to be positive only for the left integration junction and not the right integration junction. In principle, deletion of vector and/or genomic DNA sequences where the integration junction primers were designed to anneal would cause the integration junction PCR to fail. Deletions of vector sequences as well as genomic DNA following phiC31 integrase-mediated integration have been noted in our current study (Supplementary Figure S1c) and in previous studies.17, 29 The vector-specific and genome-specific primer sequences for the right integration junction were 37 and 264 bp, respectively, from the integration (cross-over) region. Deletions to the vector of >37 bp or to the genomic DNA of >264 bp would thus have deleted the primer binding sites, resulting in negative amplification of the right integration junction.
We showed by RT-PCR and transcriptome analyses that DLC1 expression in nine oligoclonal transgene-integrated CLEC cultures was 0.6–1.6-fold relative to naive control CLECs. Insertional oncogenesis events that complicated recent clinical studies have been ascribed to transactivation of neighbouring oncogenes such as LMO2, CCND2, BMI1 and the MDS-EVI1 complex.7, 8, 9 Our transcriptome data show that intronic integration had little effect on DLC1 expression but also did not dysregulate transcription of neighbouring genes within 1 Mb centred on the integration site. Moreover, transcriptome profiles of oligoclonal CLECs with intragenic DLC1 transgene integration did not bear the hallmarks of transformed cell lines as a large majority of cell cycle genes were downregulated, suggesting that these genome-modified cells were likely to be less proliferative than transformed cells. Genome-wide copy number profiling of oligoclonal CLECs on a high-resolution single-nucleotide polymorphism array revealed no significant alteration in seven of eight oligoclonal CLECs. Only a minor amplification of a 4118-kb peri-centromeric region in chromosome 19 was observed in a single oligoclone that did not alter transcription of the only gene in close proximity, ZNF254. Given the high-density coverage of probes used in the array, the absence of significant copy number changes suggests that intragenic DLC1 integration did not induce structural rearrangements of chromosomes such as translocations as these are usually unbalanced in nature. Although structurally abnormal chromosomes have been reported with other phiC31 integrase studies,28, 29 the karyotypes reported were not selective for cells with 8p22 transgene integration.
The benign safety profile of phiC31 integrase-modified oligoclonal CLECs suggested by in vitro analyses was confirmed by absence of in vivo tumourigenicity in NSG mice. Although no tumours were observed for at least 4 months post-implantation of 3 million transgene-modified oligoclonal CLECs, parallel implantation of 3 million of a tumourigenic carcinoma cell line (Hs746T) resulted in tumour formation within 3 weeks. Given that tumours formed in this strain of mice implanted with as few as 100–1000 cancer cells,30, 31 even small numbers of genome-modified CLECs would have generated tumours if they had been oncogenically transformed by transgene integration in DLC1. Taken together, our biosafety assessments indicated that intragenic DLC1 transgene integration did not predict insertional oncogenicity, and genome-modified CLECs were non-tumourigenic in vivo. The eventual decline of plasma FVIII levels to basal levels by day 30 may have resulted from loss of a substantial proportion of implanted cells because of sub-optimal engraftment and vascularization in a xenogeneic host. Transgene silencing in vivo was another possible cause. We noted that although secondary cultures established from explanted tissues still secreted FVIII in vitro (1304±34.05 mU FVIII/106 cells per 24 h) following engraftment in mice for at least 3 months, these levels were lower than pre-implantation levels (2610–5724 mU FVIII/106 cells per 24 h), suggesting some level of transgene silencing following implantation. Collectively, these data indicate that a proportion of the initially implanted cells did survive and established long-term engraftment but were not sufficiently numerous to secrete detectable levels of transgenic FVIII in murine plasma. A comprehensive evaluation of durable FVIII expression in vivo may require long-term engraftment of genome-modified cells in an autologous model.
Unlike semi-random transgene integrations mediated by viral vectors, several non-viral methods using meganucleases,32 zinc-finger nucleases,33 nickases,34 transcription activator-like effector nucleases35 and the CRISPR/Cas9 system36 have been developed recently to enable site-directed integration into a pre-determined genomic locus. The choice of genomic sites that are predictably safe from insertional oncogenesis and that will not silence the integrated transgene remains challenging. Thus far, AAVS1 (19q13.42) and CCR5 (3p21.31) are the two most common genomic loci selected for site-directed integrations.10 Of these, integration into the AAVS1 locus is thought to be benign although there is as yet no comprehensive determination of its safety. This locus, which falls within the first intron of PPP1R12C, supports robust transgene expression in several cell types. However, as the function of the PPP1R12C protein is largely unknown, cellular and organismal consequences of its altered expression remain to be ascertained. Integration in AAVS1, CCR5 and DLC1 sites support transgene expression probably because they are intragenic and transcriptionally active in many cell types, unlike intergenic sites that may be heterochromatic or otherwise unpermissive for transgene expression.37 Intragenic transgenes are also less likely to perturb intergenic regions whose functions have yet to be identified and understood. Validation of candidate safe genomic harbours is urgently needed for gene and cell therapy to progress to clinical practice with acceptable risk–benefit calculus. Our characterization of transgene integration into DLC1 has several features that could be expected of a genomic safe harbour. Moreover, we show that DLC1 integration supports durable transgene expression comparable to other putative genomic safe harbours (AAVS1 and CCR5). It resulted in minimal changes to the transcriptome and genome copy number, and CLECs with DLC1-integrated transgene were not tumourigenic in vivo. This was not surprising because although DLC1 is a tumour suppressor, its expression was not extinguished by the integrated transgene. However, in the event that DLC1 expression could decrease significantly in other situations, concurrent expression of the closely related DLC2 (13q12), a RhoGAP with similar specificity for RhoA and Cdc42,38 may functionally compensate for reduced activity of DLC1.
Increasing the number of genomic sites into which transgenes can be integrated without insertional oncogenesis will be helpful for the potentially wide range of gene and cell therapy applications. The same site may well be selectively oncogenic in some, but not all, cell types because gene products often have cell type-specific functions. Intronic integration that does not change the expression of the host gene, as in our DLC1 data, is doubly advantageous as it is more likely to be functionally benign yet permissive of transgene integration and durable expression.
Our approach suggests that integrating a therapeutic transgene in primary human cells ex vivo with precision at a preselected single genomic site should mitigate risks of direct in vivo gene delivery by enabling comprehensive characterization of the transcriptome, genome copy number, karyotype and cellular behaviour. This strategy is feasible in primary cell types that are simple to procure, expandable in vitro to clinically meaningful numbers and genomically stable. Clonal cultures of genome-modified cells could also be more readily derived from primary cells having these characteristics. However, oligoclonal CLECs cultured for about 2 months had downregulated transcription of cell cycle genes and reduced proliferation. This development can be anticipated also for other primary human cells such as fibroblasts, tissue-specific progenitor and stromal cells after long-term culture. Nonetheless, finite expandability of CLECs does not necessarily preclude their application for haemophilia therapy. Estimates based on FVIII expression from transgene-modified CLECs (2131 mU FVIII/106 cells per 24 h) suggest that approximately 100–400 million cells are required to achieve a steady-state plasma FVIII level of 10% in a 20–40 kg child. Given that approximately 20 million transgene-modified CLECs can be derived from each oligoclonal CLEC identified by our screening procedure, this can be achieved by identifying 5–20 oligoclonal CLECs to generate the cell numbers required for clinically effective cell therapy. Based on our current study, identification of 5–20 oliogoclonal CLECs by screening for transgene integration at 8p22 is technically feasible. These clonal cells can be readily expanded to 100–400 million cells in total before growth begins to slow down. Alternatively, induced pluripotent stem cells that have unlimited proliferation capacity may be better suited in practice for transgene integration and clonal selection of cells with site-specific integration at 8p22, provided concerns pertaining to tumourigenicity are addressed.
We chose haemophilia A as a model disease caused by deficiency of a single protein to demonstrate the feasibility of autologous cell therapy using genome-modified CLECs. Secretion of BDD-hybrid FVIII was at least fivefold higher than BDD-human FVIII. Given the inherently poor expression of recombinant human FVIII,39, 40, 41 the more efficiently secreted BDD-hybrid FVIII42 could be the preferred transgene in future preclinical studies as fewer cells would be required to achieve therapeutically significant plasma FVIII levels.
The convergence of new methods for precision genome modification, high-throughput techniques for characterizing whole genomes and transcriptomes and improved design of donor transgenes could advance gene-based cell therapy of inherited and acquired disorders among paediatric and adult patients.
Materials and methods
Hybrid FVIII plasmid assembly
The hybrid porcine–human FVIII construct consisted of porcine A1 and A3 domains, human signal peptide, A2, and partial B domains (retaining the C-terminal 266 amino acids with eight glycosylation sites), C1 and C2 domains. Human FVIII components were derived by PCR amplification from the full-length sequence in pSP64-F8 (American Type Culture Collection, Manassas, VA, USA). Porcine FVIII domains were derived by RT-PCR of porcine liver RNA. An overlap PCR strategy was used to assemble components of the hybrid FVIII cDNA. The construct, expressed from the human ferritin light chain promoter, was eventually cloned in a vector containing the 350-bp attB fragment22 and neomycin resistance gene. The resulting construct was named pattBhybrid FVIII.
Cell culture
Primary human dermal fibroblasts (KF1 and NF123), human adipose-derived stromal cells, human bone marrow-derived stromal cells and CLECs were provided by CellResearch Corporation, Singapore. Dermal fibroblasts, adipose-derived stromal cells and bone marrow-derived stromal cells were cultured in Dulbecco's modified Eagle's medium and 25 mM glucose supplemented with 10% foetal bovine serum. The isolation and culture of CLECs has been described previously.17, 43, 44
Gene transfer and stable cell generation
All gene transfer studies were performed using a 4D-Nucleofector device (Lonza). CLECs were electroporated with Nucleofector solution P1 and setting CM113, bone marrow-derived stromal cells and adipose-derived stromal cells with Nucleofector solution P2 and setting EW104 while dermal fibroblasts (NF123) with Nucleofector solution P2 and setting FF113. Transfection efficiency was gauged by fluorescence-activated cell sorting analysis for green fluorescent protein-positive cells 24 h post-electroporation with 5 μg of pmaxGFP vector (Lonza). Cells stably modified by phiC31-integrase were generated by co-electroporation with 12 μg of pattBhybrid FVIII and 2.5 μg pCMV-Int plasmid22 followed by selection with 1 mg ml–1 of G418 for 5 days starting from day 6 post-electroporation.
Integration junction PCR and sequencing
Integration junction PCR was performed on 200 ng of genomic DNA (DNA extraction kit; Favorgen Biotech Corp., Ping-Tung, Taiwan) using DyNAzyme EXT DNA polymerase (Thermo Scientific, Rockford, IL, USA). Left integration junction PCR (454-bp amplicon) was performed using primer pairs specific to a locus on chromosome 8p22 (forward: 5′-GGGCTCTGGAGTAAAGGTGAAA-3′) and donor vector (reverse: 5′-GTTCGCCGGGATCAACTACC-3′). Right integration junction PCR (333-bp amplicon) was performed using primer pairs specific to the vector sequence (forward: 5′-TCGACGATGTAGGTCACGG-3′) and chromosome 8p22 genomic DNA (reverse: 5′-GCATGGCCTCATTTCCGTCT-3′). Transgene-specific PCR was performed with intron-spanning primers specific to the C1-domain of the integrated hybrid FVIII cDNA (forward: 5′-AGCTTTCAGAAGAGAACCCGACAC-3′ reverse: 5′-TCCCAGGGGAGTCTGACACTTCTTGCTGTACACCAGGAAAGT3′). Control genomic PCR (900-bp amplicon) was performed with genomic primers (forward: 5′-AAGAAGCGCACCACCTCCAGGTTCT-3′ reverse: 5′-ATGACCTCATGCTCTTGGCCCTCGTA-3′). Amplified products were electrophoresed on 1% agarose gels and imaged using BioRad Gel Doc 2000 transilluminator (Bio-Rad Laboratories, Hercules, CA, USA).
Screening of oligoclonal CLECs with transgene integration at 8p22
To screen oligoclonal CLECs for transgene integration at chromosome 8p22 locus,45 G418-resistant stable population of CLECs (pattBhybrid FVIII+pCMV-Int, G418 selected) were flow sorted (four cells per well) into individual wells of 96-well plates and allowed to expand in culture. A replica plate for further analysis of clones was established by detaching and re-seeding cells into a second 96-well plate. Cells for 8p22 integration screening (in 96-well plates) were lysed in situ with 60 μl of lysis buffer and screened by direct PCR for the presence of integration junctions and the integrated transgene using Phusion Human Specimen Direct PCR kit (Thermo Scientific) and the same primers above.
Fluorescence in situ hybridization
FISH to detect integration of pattBhybrid FVIII was performed on interphase nuclei of genome-modified CLECs with fluorescein-12-dUTP (PerkinElmer, Waltham, MA, USA) random prime labelled probe generated by PCR amplification of neomycin cDNA from pattB hybrid FVIII (817-bp PCR product: forward primer 5′-TTGCACGCAGGTTCTCCGGC-3′ reverse primer 5′-GGCGTCGCTTGGTCGGTCAT-3′). Texas-Red-5-dUTP-labelled human chromosome 8 centromeric probe (Children's Hospital Oakland Research Institute, Oakland, CA, USA) was used to determine colocalization with chromosome 8. Hybridized slides were counterstained with 4,6-diamino-2-phenylindole and visually enumerated for probe signals with an Olympus BX61 epifluorescence microscope (Olympus, Tokyo, Japan).
FVIII assay
Factor VIII activity in conditioned media was measured using a Chromogenix Coamatic Factor VIII kit whose detection limit was 1% FVIII activity (Chromogenix, Molndal, Sweden). Human-specific FVIII antigen levels in murine plasma was quantified using Visulize FVIII antigen ELISA kit having a detection limit of 0.8% Factor VIII antigen (Affinity Biologicals, Ancaster, ON, Canada). Both assays were performed according to the manufacturers' protocols.
Transcriptome analysis
Total RNA isolated from control CLECs (electroporated without plasmid DNA and having the same number of population doublings) and genome-modified oligoclonal CLECs with integrations at 8p22 served as starting material for transcript profiling on GeneChip PrimeView Human Gene Expression array (Affymetrix) following the recommended protocol. Transcription expression data were analyzed using GeneChip Operating Software (Affymetrix). Transcripts whose expression levels differed by >2.5-fold in genome-modified oligoclonal CLECs compared with naive CLECs were considered significantly altered and were further analyzed. DAVID (Database for Annotation, Visualization and Integrated Discovery) 2.1 Functional Annotation Tool (http://david.abcc.ncifcrf.gov) was used to ascribe functions and other annotations for significantly altered transcripts and for pathway mapping. Altered transcripts were also referenced to a compilation of known proto-oncogenes and tumour-suppressor genes (http://www.bushmanlab.org/links/genelists).
Genome copy number change analysis
Genome-modified oligoclonal CLECs with 8p22 integrations and control CLECs (electroporated without plasmid DNA and having the same number of population doublings) were evaluated using Cytoscan HD array and Chromosome Analysis Suite for data analysis (Affymetrix). Regions of copy number gain or loss were defined as segments of ⩾50 consecutive probes concordant for significant copy number abnormalities. Log2 signal intensity ratios >0.3 and ⩽–0.3 were criteria for significant copy number gain and loss, respectively.
RT-PCR analysis
RT-quantitative PCR was used to determine changes in DLC1 transcript levels in clonal CLECs with transgene integration in 8p22. CLECs electroporated without plasmid DNA and having the same number of population doublings were controls. In brief, total RNA was extracted using TriPure isolation reagent (Roche Applied Science, Indianapolis, IN, USA), treated with DNase I (Qiagen, Hilden, Germany) to remove contaminating genomic DNA and purified using RNeasy Mini kit (Qiagen). Reverse transcription was performed on approximately 1 μg of DNAse-treated RNA using iScript Advanced cDNA synthesis kit (Bio-Rad Laboratories), according to the manufacturer's instructions. Quantitative-PCR was performed using 2 μl of first strand cDNA in a 20 μl reaction volume using GoTaq qPCR Master Mix (Promega Corp., Madison, WI, USA) and 45 cycles at an annealing and extension temperature of 62 °C using CFX96 Real-Time PCR detection system (Bio-Rad Laboratories). Intron-spanning exonic primers were used to amplify DLC1 exons 1–2 (forward: 5′-TCCTGCCCCAATGGAATGTC-3′ reverse: 5′-GTTGGTGTGCCTGATGGAGA-3′), exons 8–9 (forward: 5′-GAAGGGGATGCAGCGGATAG-3′ reverse: 5′-AGCAGGGCCGTTAGCTTTAG-3′) and a housekeeping gene, GAPDH exon 6–7 (forward: 5′-GCCTCCTGCACCACCAACT-3′ reverse: 5′-CGCCTGCTTCACCACCTTC-3′). DLC1 transcript levels were normalized to GAPDH expression levels and the fold-change in DLC1 transcript levels in clonal CLECs with 8p22 integration was reported relative to DLC1 transcript levels in control CLECs, using the ‘delta-delta Ct method'.46
Cell proliferation assay
Oligoclonal CLECs that had been in culture for at least 2 months and control CLECs that were electroporated only and cultured for only 1 month were seeded at 1000 cells per well and allowed to proliferate for 3 days, after which MTS assay (CellTiter 96 Aqueous One Solution Cell Proliferation Assay, Promega Corp.) was performed to quantify live cells by absorbance readings at 490 nm.
Mice and implantation studies
All animal studies were conducted in accordance with ethical and international animal use guidelines following approval by SingHealth Institutional Animal Care and Use Committee (IACUC). The tumourigenic potential of hybrid FVIII-expressing clonal CLECs with transgene integration at 8p22 locus was investigated by implanting 3 × 106 Matrigel-encapsulated (BD Biosciences, San Jose, CA, USA) transgenic cells into the subcutaneous nuchal region of NOD-SCID IL2Rγ null (NSG) mice (Jackson Laboratories, Bar Harbor, ME, USA; n=16). Untreated mice (n=4) and mice implanted with CLECs that received electroporation only (n=4) served as negative controls for tumour formation and plasma hybrid FVIII measurements, respectively. NSG mice implanted with a tumourigenic cell line (Hs746T, American Type Tissue Culture, Manassas, VA, USA) served as positive tumourigenic controls. Survival and engraftment of implanted cells was monitored by measuring levels of transgenic hybrid FVIII (secreted by implanted CLECs) in murine plasma using a FVIII-antigen capture ELISA assay (Affinity Biologicals). Mice were monitored weekly for visible tumours at implantation sites. Matrigel implants were excised under sterile conditions to re-establish secondary cultures and for immunohistochemistry. Secondary explants cultures were established by dicing excised Matrigel implants into 0.5 mm cubes, seeding them onto 10-cm tissue culture dishes and culturing them exactly as for primary CLECs.
Immunohistochemistry
Immunohistochemical staining to identify implanted human cells was performed on formalin-fixed paraffin-embedded tissues excised from implantation sites. Five-micron tissue sections blocked in 10% goat serum for 30 min were incubated with 1:3 diluted mouse anti-human vimentin antibody (Clone V9, Zymed, San Francisco, CA, USA) for 1 h at 37 °C followed by 30-min incubation with 1:2 diluted DAKO REAL EnVision rabbit/mouse horseradish peroxidase-conjugated secondary antibody (Dako, Glostrup, Denmark). Tissue sections were washed three times with phosphate-buffered saline between each incubation step. Slides were stained with DAB chromogen (1 part DAB chromogen+3 parts DAKO REAL substrate buffer) for 3 min, counterstained with Mayer's haematoxylin and mounted with DPX mount solution. Tissue sections were visualized and imaged with an inverted microscope (Axiovert 25CFC, Carl Zeiss, Oberkochen, Germany) under × 20 magnification using the KS400 software (Carl Zeiss).
Statistical analyses
Results are expressed as mean±s.e.m. or s.d. where appropriate, with ‘n' indicating the total number of samples or measurements. Student's unpaired t-test with two-tailed P-values and 95% confidence interval was used for comparison between two groups. P<0.05 was considered statistically significant.
Acknowledgments
This work was supported by a research grant (CIRG 1326/2012) from the National Medical Research Council, Singapore to OLK. We thank Michele P Calos (Stanford University) for gifts of pTA-attb and pCMV-Int, Mickey Koh and Madelaine Niam (Health Sciences Authority, Singapore) for cell sorting and Jeya Kumar (CellResearch Corporation) for preparation of primary cells and culture media.
TTP who is the inventor of cord-lining epithelial cells technology, is a co-founder and shareholder of CellResearch Corporation.
Footnotes
Supplementary Information accompanies this paper on Gene Therapy website (http://www.nature.com/gt)
Supplementary Material
References
- Cavazzana-Calvo M, Hacein-Bey S, de Saint Basile G, Gross F, Yvon E, Nusbaum P, et al. Gene therapy of human severe combined immunodeficiency (SCID)-X1 disease. Science. 2000;288:669–672. doi: 10.1126/science.288.5466.669. [DOI] [PubMed] [Google Scholar]
- Gaspar H, Parsley K, Howe S, King D, Gilmour K, Sinclair J, et al. Gene therapy of X-linked severe combined immunodeficiency by use of a pseudotyped gammaretroviral vector. The Lancet. 2004;364:2181–2187. doi: 10.1016/S0140-6736(04)17590-9. [DOI] [PubMed] [Google Scholar]
- Aiuti A, Cattaneo F, Galimberti S, Benninghoff U, Cassani B, Callegaro L, et al. Gene therapy for immunodeficiency due to adenosine deaminase deficiency. N Engl J Med. 2009;360:447–458. doi: 10.1056/NEJMoa0805817. [DOI] [PubMed] [Google Scholar]
- Ott MG, Schmidt M, Schwarzwaelder K, Stein S, Siler U, Koehl U, et al. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat Med. 2006;12:401–409. doi: 10.1038/nm1393. [DOI] [PubMed] [Google Scholar]
- Boztug K, Schmidt M, Schwarzer A, Banerjee PP, Díez IA, Dewey RA, et al. Stem-cell gene therapy for the Wiskott-Aldrich syndrome. N Engl J Med. 2010;363:1918–1927. doi: 10.1056/NEJMoa1003548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartier N, Hacein-Bey-Abina S, Bartholomae CC, Veres G, Schmidt M, Kutschera I, et al. Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science. 2009;326:818–823. doi: 10.1126/science.1171242. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Garrigue A, Wang GP, Soulier J, Lim A, Morillon E, et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J Clin Invest. 2008;118:3132–3142. doi: 10.1172/JCI35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe SJ, Mansour MR, Schwarzwaelder K, Bartholomae C, Hubank M, Kempski H, et al. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J Clin Invest. 2008;118:3143–3150. doi: 10.1172/JCI35798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein S, Ott MG, Schultze-Strasser S, Jauch A, Burwinkel B, Kinner A, et al. Genomic instability and myelodysplasia with monosomy 7 consequent to EVI1 activation after gene therapy for chronic granulomatous disease. Nat Med. 2010;16:198–204. doi: 10.1038/nm.2088. [DOI] [PubMed] [Google Scholar]
- Sadelain M, Papapetrou EP, Bushman FD. Safe harbours for the integration of new DNA in the human genome. Nat Rev Cancer. 2011;12:51–58. doi: 10.1038/nrc3179. [DOI] [PubMed] [Google Scholar]
- Chalberg TW, Portlock JL, Olivares EC, Thyagarajan B, Kirby PJ, Hillman RT, et al. Integration specificity of phage phiC31 integrase in the human genome. J Mol Biol. 2006;357:28–48. doi: 10.1016/j.jmb.2005.11.098. [DOI] [PubMed] [Google Scholar]
- Ortiz-Urda S, Thyagarajan B, Keene DR, Lin Q, Calos MP, Khavari PA, et al. PhiC31 integrase-mediated nonviral genetic correction of junctional epidermolysis bullosa. Hum Gene Ther. 2003;14:923–928. doi: 10.1089/104303403765701204. [DOI] [PubMed] [Google Scholar]
- Ortiz-Urda S, Thyagarajan B, Keene DR, Lin Q, Fang M, Calos MP, et al. Stable nonviral genetic correction of inherited human skin disease. Nat Med. 2002;8:1166–1170. doi: 10.1038/nm766. [DOI] [PubMed] [Google Scholar]
- Ishikawa Y, Tanaka N, Murakami K, Uchiyama T, Kumaki S, Tsuchiya S, et al. Phage phiC31 integrase-mediated genomic integration of the common cytokine receptor gamma chain in human T-cell lines. J Gene Med. 2006;8:646–653. doi: 10.1002/jgm.891. [DOI] [PubMed] [Google Scholar]
- Held PK, Olivares EC, Aguilar CP, Finegold M, Calos MP, Grompe M, et al. In vivo correction of murine hereditary tyrosinemia type I by phiC31 integrase-mediated gene delivery. Mol Ther. 2005;11:399–408. doi: 10.1016/j.ymthe.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Keravala A, Chavez CL, Hu G, Woodard LE, Monahan PE, Calos MP, et al. Long-term phenotypic correction in factor IX knockout mice by using ΦC31 integrase-mediated gene therapy. Gene Therapy. 2011;18:842–848. doi: 10.1038/gt.2011.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivalingam J, Krishnan S, Ng WH, Lee SS, Phan TT, Kon OL, et al. Biosafety assessment of site-directed transgene integration in human umbilical cord-lining cells. Mol Ther. 2010;18:1346–1356. doi: 10.1038/mt.2010.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez CL, Keravala A, Chu JN, Farruggio AP, Cuéllar VE, Voorberg J, et al. Long-term expression of human coagulation factor VIII in a tolerant mouse model using the ϕC31 integrase system. Hum Gene Ther. 2012;23:390–398. doi: 10.1089/hum.2011.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertoni C, Jarrahian S, Wheeler TM, Li Y, Olivares EC, Calos MP, et al. Enhancement of plasmid-mediated gene therapy for muscular dystrophy by directed plasmid integration. Proc Natl Acad Sci USA. 2006;103:419–424. doi: 10.1073/pnas.0504505102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thyagarajan B, Olivares EC, Hollis RP, Ginsburg DS, Calos MP. Site-specific genomic integration in mammalian cells mediated by phage phiC31 integrase. Mol Cell Biol. 2001;21:3926–3934. doi: 10.1128/MCB.21.12.3926-3934.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doering CB, Denning G, Dooriss K, Gangadharan B, Johnston JM, Kerstann KW, et al. Directed engineering of a high-expression chimeric transgene as a strategy for gene therapy of hemophilia A. Mol Ther. 2009;17:1145–1154. doi: 10.1038/mt.2009.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth AC, Olivares EC, Thyagarajan B, Calos MP. A phage integrase directs efficient site-specific integration in human cells. Proc Natl Acad Sci USA. 2000;97:5995–6000. doi: 10.1073/pnas.090527097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao YC, Lo SH. Deleted in liver cancer-1 (DLC-1): a tumor suppressor not just for liver. Int J Biochem Cell Biol. 2008;40:843–847. doi: 10.1016/j.biocel.2007.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z, Huang Y, Sharfstein ST. Regulation of recombinant monoclonal antibody production in chinese hamster ovary cells: a comparative study of gene copy number, mRNA level, and protein expression. Biotechnol Prog. 2006;22:313–318. doi: 10.1021/bp0501524. [DOI] [PubMed] [Google Scholar]
- Wang GP, Garrigue A, Ciuffi A, Ronen K, Leipzig J, Berry C, et al. DNA bar coding and pyrosequencing to analyze adverse events in therapeutic gene transfer. Nucleic Acids Res. 2008;36:e49. doi: 10.1093/nar/gkn125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan BZ, Miller MJ, Keck CL, Zimonjic DB, Thorgeirsson SS, Popescu NC, et al. Cloning, characterization, and chromosomal localization of a gene frequently deleted in human liver cancer (DLC-1) homologous to rat RhoGAP. Cancer Res. 1998;58:2196–2199. [PubMed] [Google Scholar]
- Low JSW, Tao Q, Ng KM, Goh HK, Shu XS, Woo WL, et al. A novel isoform of the 8p22 tumor suppressor gene DLC1 suppresses tumor growth and is frequently silenced in multiple common tumors. Oncogene. 2011;30:1923–1935. doi: 10.1038/onc.2010.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Jeppesen I, Nielsen K, Jensen TG. Phi c31 integrase induces chromosomal aberrations in primary human fibroblasts. Gene Therapy. 2006;13:1188–1190. doi: 10.1038/sj.gt.3302789. [DOI] [PubMed] [Google Scholar]
- Ehrhardt A, Engler JA, Xu H, Cherry AM, Kay MA. Molecular analysis of chromosomal rearrangements in mammalian cells after phiC31-mediated integration. Hum Gene Ther. 2006;17:1077–1094. doi: 10.1089/hum.2006.17.1077. [DOI] [PubMed] [Google Scholar]
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, et al. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–1037. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- Hafez M, Hausner G. Homing endonucleases: DNA scissors on a mission. Genome. 2012;55:553–569. doi: 10.1139/g2012-049. [DOI] [PubMed] [Google Scholar]
- Palpant NJ, Dudzinski D. Zinc finger nucleases: looking toward translation. Gene Therapy. 2013;20:121–127. doi: 10.1038/gt.2012.2. [DOI] [PubMed] [Google Scholar]
- Kim E, Kim S, Kim DH, Choi BS, Choi IY, Kim JS, et al. Precision genome engineering with programmable DNA-nicking enzymes. Genome Res. 2012;22:1327–1333. doi: 10.1101/gr.138792.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung JK, Sander JD. TALENs: a widely applicable technology for targeted genome editing. Nat Rev Mol Cell Biol. 2013;14:49–55. doi: 10.1038/nrm3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo A, Cesana D, Genovese P, Di Stefano B, Provasi E, Colombo DF, et al. Site-specific integration and tailoring of cassette design for sustainable gene transfer. Nat Methods. 2011;8:861–869. doi: 10.1038/nmeth.1674. [DOI] [PubMed] [Google Scholar]
- Leung THY, Ching YP, Yam JWP, Wong CM, Yau TO, Jin DY, et al. Deleted in liver cancer 2 (DLC2) suppresses cell transformation by means of inhibition of RhoA activity. Proc Natl Acad Sci USA. 2005;102:15207–15212. doi: 10.1073/pnas.0504501102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch CM, Israel DI, Kaufman RJ, Miller AD. Sequences in the coding region of clotting factor VIII act as dominant inhibitors of RNA accumulation and protein production. Hum Gene Ther. 1993;4:259–272. doi: 10.1089/hum.1993.4.3-259. [DOI] [PubMed] [Google Scholar]
- Hoeben RC, Fallaux FJ, Cramer SJ, van den Wollenberg DJ, van Ormondt H, Briët E, et al. Expression of the blood-clotting factor-VIII cDNA is repressed by a transcriptional silencer located in its coding region. Blood. 1995;85:2447–2454. [PubMed] [Google Scholar]
- Tagliavacca L, Wang Q, Kaufman RJ. ATP-dependent dissociation of non-disulfide-linked aggregates of coagulation factor VIII is a rate-limiting step for secretion. Biochemistry. 2000;39:1973–1981. doi: 10.1021/bi991896r. [DOI] [PubMed] [Google Scholar]
- Doering CB, Healey JF, Parker ET, Barrow RT, Lollar P. Identification of porcine coagulation factor VIII domains responsible for high level expression via enhanced secretion. J Biol Chem. 2004;279:6546–6552. doi: 10.1074/jbc.M312451200. [DOI] [PubMed] [Google Scholar]
- Reza HM, Ng BY, Gimeno FL, Phan TT, Ang LPK. Umbilical cord lining stem cells as a novel and promising source for ocular surface regeneration. Stem Cell Rev. 2011;7:935–947. doi: 10.1007/s12015-011-9245-7. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Gan SU, Lin G, Lim YT, Masilamani J, Mustafa FB, et al. Characterization of human umbilical cord lining derived epithelial cells and transplantation potential. Cell Transplant. 2011;20:1827–1841. doi: 10.3727/096368910X564085. [DOI] [PubMed] [Google Scholar]
- Sclimenti CR, Thyagarajan B, Calos MP. Directed evolution of a recombinase for improved genomic integration at a native human sequence. Nucleic Acids Res. 2001;29:5044–5051. doi: 10.1093/nar/29.24.5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.