Abstract
The kidney is a complex organ with over 30 different cell types, and understanding the lineage relationships between these cells is challenging. During nephrogenesis, a central question is how the coordinated morphogenesis, growth, and differentiation of distinct cell types leads to development of a functional organ. In mature kidney, understanding cell division and fate during injury, regeneration and aging are critical topics for understanding disease. Genetic lineage tracing offers a powerful tool to decipher cellular hierarchies in both development and disease because it allows the progeny of a single cell, or group of cells, to be tracked unambiguously. Recent advances in this field include the use of inducible recombinases, multicolor reporters, and mosaic analysis. In this review, we discuss lineage-tracing methods focusing on the mouse model system and consider the impact of these methods on our understanding of kidney biology and prospects for future application.
Keywords: kidney development, kidney disease, lineage tracing, mouse genetics, recombination
In a lineage-tracing experiment, a cell is identified by expression of a reporter gene. If that cell divides, expression of the reporter is passed on to all progeny, and migration of these marked cells can be easily measured. Typical reporter genes include β-galactosidase or a fluorescent protein. Historically, lineage tracing was used most in the field of developmental biology. In its earliest application, lineage tracing was accomplished not by the expression of a fluorescent reporter, but through direct observation of embryos by light microscopy. This allowed the generation of ‘fate maps' for cells from the one-cell stage until generation of germ layers.1
Subsequent advances in the field included application of dyes to the surface of an embryo in order to track the movement of groups of cells.2 However, direct observation is limited to the study of small, transparent embryos, and as a result lipid-soluble dyes were injected into the embryo and later sectioned.3 Ultimately, the introduction of genetic markers superseded dye experiments because of their advantages: they do not leak into neighboring cells and they are typically inherited by progeny where they are expressed at levels equal to that of the parental cell. These genetic markers were originally introduced by viral infection, direct injection, or transfection. Over time, techniques for lineage tracing have evolved to the point that they now possess remarkable sensitivity and offer sufficient resolution to track single cells, in real time. The subject of this review is the most commonly used lineage-tracing approach—genetic recombination.
GENETIC LINEAGE TRACING
The concept of genetic lineage tracing involves the expression of a recombinase enzyme in a cell-specific manner in order to activate the expression of a reporter gene. Cre recombinase (causes recombination of the bacteriophage P1 genome) has become an indispensible tool in the mouse model system, and it has recently been adapted for use in zebrafish.4, 5 Flippase is an alternate recombinase that has been used mostly in Drosophila and will not be discussed here. Cre recognizes a 34-base-pair nucleotide sequence called loxP that is not present in the mouse genome. When two loxP sites are oriented in a head-to-tail manner, Cre recombinase will excise the intervening DNA sequence while rejoining the ends together. This is very useful in turning genes on for lineage tracing. Normally, a reporter gene is located downstream of a ubiquitous promoter, such as the Rosa26 locus.6 If a strong transcriptional stop sequence, such as a triple polyadenylation sequence, is placed between two loxP sequences and located upstream of this reporter gene, this reporter gene will always be turned OFF. However, expression of Cre recombinase will cause deletion of the polyadenylation sequences, turning the reporter ON in that cell and all of its descendants.
By choosing cell-specific promoters for expression of Cre and combining it with a reporter, one can mark any cell population when that promoter becomes activated. An ever-growing variety of Cre driver mouse lines are available, and they have been created by several approaches: for example, by random integration of short promoter sequences or by much larger bacterial artificial chromosome transgenes. Of the two, bacterial artificial chromosome transgenics are more likely to faithfully recapitulate the endogenous expression pattern of the gene of interest, as unknown regulatory sequences are more likely to be included on the bacterial artificial chromosome. Knock-in approaches are also useful, and these are also highly likely to recapitulate endogenous expression patterns, as all nearby DNA sequences are intact at the locus of interest.
In many cases, it is useful to be able to inducibly mark a cell, and this is accomplished using a Cre enzyme fused to a modified form of the estrogen receptor (CreERt2), which causes Cre to be sequestered in the cytoplasm until it binds to its ligand tamoxifen, triggering nuclear translocation.7, 8, 9 Both a codon-optimized version (iCreERt2) and a codon-optimized version fused to two ERt2 domains (iERCreER) at the N and C termini are now available and offer improved expression and recombination frequency.10, 11 Alternatively, a constitutively active Cre may be regulated by a drug-controlled promoter such as tetracycline. Tetracycline-regulated systems in particular are quite robust in mouse but are generally used for protein overexpression experiments.12 They are less commonly used for lineage analysis because they may require three separate alleles in this application rather than two, making for a cumbersome (and expensive) breeding strategy. However this approach has been used with success along mouse nephron epithelia, for example, in which a Pax8-rtTA allele, which drives expression of the reverse tetracycline transactivator (rtTA) is crossed to a mouse line with an allele consisting of a tetracycline-response element driving Cre expression as well as a reporter mouse.13 In this case, administration of doxycycline binds to rtTA present in Pax8-positive renal epithelial cells only, and the doxycycline-rtTA complex drives Cre expression and subsequently activation of the reporter.
Among the important issues to consider before choosing an appropriate reporter is the anticipated method of detection. The reporter chosen must possess sufficient signal-to-noise ratio to enable unambiguous detection (Figure 1). The Escherichia coli lacZ gene, β-galactosidase, has been used extensively and produces an intense blue color when incubated with the substrate analog X-gal. Yet β-galactosidase does not provide cellular resolution in thin cells, because the reaction product does not entirely fill long processes but is enriched in the cell body.14 In addition, there is a lack of specific antibodies that would allow routine indirect immunofluorescence detection in all systems. Therefore, LacZ is not the best reporter to detect interstitial cells. Fluorescent reporters are now the norm, and in this case detection by epifluorescence is far superior to antibody-enhanced methods, which are subject to nonspecific binding of primary and secondary antibodies. Therefore, an adequate expression level of the reporter is critical. Although the Rosa26 locus drives strong expression of reporters in developing kidney, the expression level in adult kidney is reduced and may be inadequate (Humphreys, unpublished observations). Thus, straight Rosa26 reporters may not be ideal for experiments in adults. In such cases, reporters that have an additional CAG promoter knocked into the locus provide much stronger expression levels.
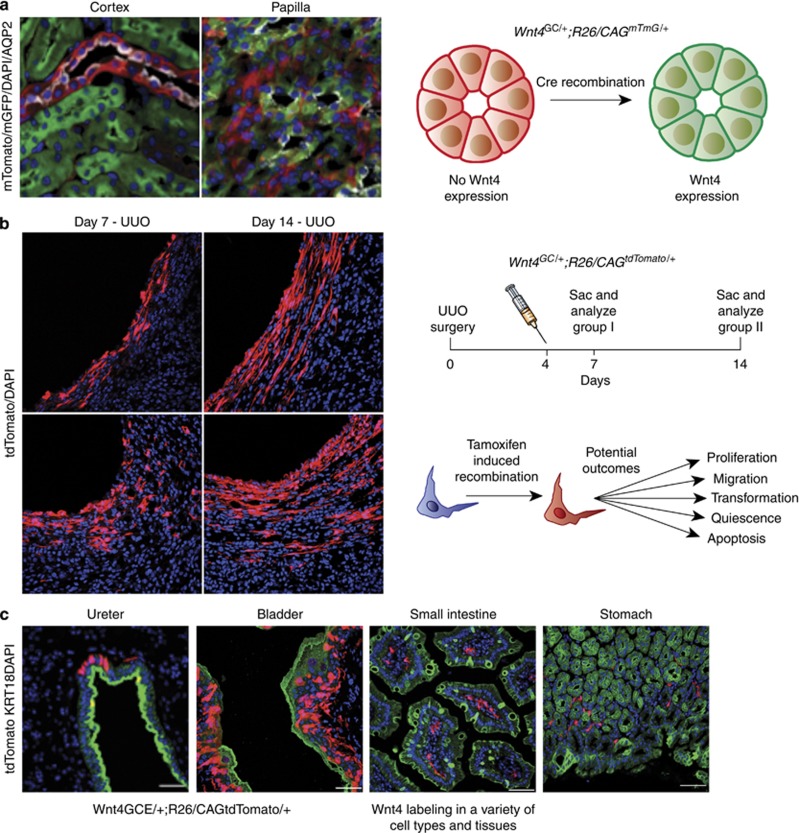
Figure 1.
Using conventional and tamoxifen-inducible Cre recombinase for cell-specific genetic labeling.(a) In the Wnt4GC/+; R26mTmG/+ mouse line, Cre expression is driven by Wnt4. Here, the reporter used was mT/mG (membrane-Tomato before Cre/membrane-GFP after Cre) expressed from the Rosa26 locus.16 In bigenic mice, cells that expressed Wnt4 at some point in development are green and cells that never expressed Wnt4 are red. As Wnt4 is expressed in the renal vesicle during development, all epithelial lineages are GFP-positive with the exception of the collecting duct, which is derived from ureteric bud, which remains red. In the kidney cortex, collecting ducts that are identified by positive staining for aquaporin 2 (seen in white) are all expressing red fluorescent protein. All surrounding tubules are green. However, in the papilla, the collecting ducts are now green, as Wnt4 is switched on in this region of the kidney. (b) Fate labeling is useful for quantitative analysis of specific cell populations. With a single dose of tamoxifen, administered at the same time point, cells in two experimental groups will be permanently labeled with the reporter gene, in this case tdTomato. Following the labeling event, analysis of the two groups at two distinct time points after the labeling may reveal differences in cell number, phenotype, and localization, among other cellular and molecular characteristics. (c) With systemic injections of tamoxifen in inducible Cre lines, recombination will occur in cell types throughout the organism. The Wnt4GCE/+;R26tdTomato/+mouse reveals tdTomato-positive Wnt4-expressing cells in the epithelial and stromal layers of the ureter and bladder and in the stromal compartments of the small intestine and stomach. DAPI, 4,6-diamidino-2-phenylindole; GFP, green fluorescent protein; tdTomato, tandem dimer of Tomato; UUO, unilateral ureteral obstruction.
Older reporters exist in which viral promoters such as CAG were generated as random integration transgenics, rather than knocked into the Rosa26 locus.15 These may suffer from mosaic expression as the integration locus may be inactive in certain cells, and this is an important limitation. The mT/mG reporter addresses this limitation because cells that have not undergone recombination express membrane-targeted tdTomato (mT), but after recombination they express membrane-targeted enhanced green fluorescent protein (eGFP) (mG, Figure 1a). In this way, mT expression verifies that cells of interest are capable of expressing the reporter allele.16
Fluorophores with strong epifluorescence also improve signal-to-noise ratio. Although eGFP and its variants continue to be quite useful, a new generation of fluorophores offer much brighter fluorescence. The most highly fluorescent reporter currently available is a tandem dimmer of Tomato (tdTomato), which is itself a mutagenized version of DsRed. tdTomato is nearly three times brighter than eGFP, is nontoxic to cells, and is photostable.17 A Rosa26 knock-in reporter mouse expressing tdTomato is available, in which a strong CAG promoter has been inserted to increase expression levels18 and a woodchuck hepatitis virus post-transcriptional regulatory element is present in order to further enhance mRNA stability (Figure 2).19 These mice can be maintained as homozygotes. The primary weakness of this line is that tdTomato fluorescence is so strong that it can easily bleed into the eGFP channel.
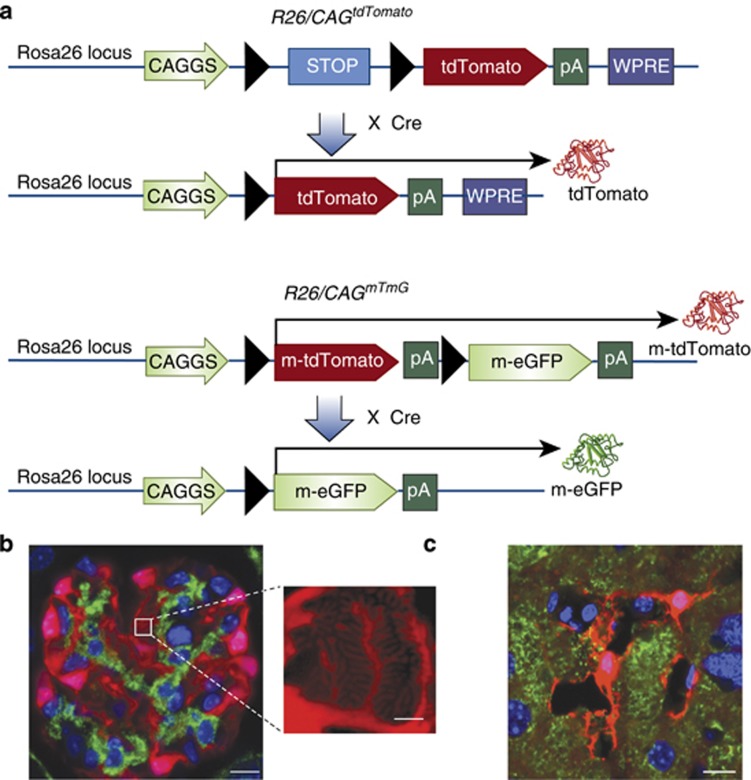
Figure 2.
Permanently labeling cells with the bright and photostable fluorescent protein tdTomato. (a) The tdTomato reporter cassette is knocked-in to the Rosa26 locus by homologous recombination. The CAGGS promoter, which contains the cytomegalovirus (CMV) early enhancer element and chicken beta-actin promoter, is included to enhance gene expression. A floxed STOP cassette is upstream of the tdTomato gene, thus preventing expression of the fluorophore in the absence of Cre recombinase. The Woodchuck hepatitis virus post-transcriptional regulatory element (WPRE) is included to enhance mRNA stability and protein expression. In the presence of Cre recombinase, the STOP sequence between the loxP sites is excised and the tdTomato protein is permanently and constitutively expressed in that cell and its descendents. In the R26mTmGconstruct, a membrane-targeted tdTomato reporter is expressed in the absence of Cre-mediated recombination. After recombination, the membrane tdTomato is excised and a membrane-targeted enhanced green fluorescent protein (eGFP) reporter protein will be expressed. (b) In Coll1α1GCE/+;R26tdTomato/+ mice, GFP-Cre-ERT2 is driven by the collagen1-α1 promoter and will result in the expression tdTomato protein after treatment with tamoxifen. Pictured are tdTomato+ podocytes adjacent to platelet derived growth factor beta positive (PDGFRβ+) mesangial cells labeled with a green antibody. The extremely bright and photostable tdTomato fluorophore enables high-resolution optical visualization of tertiary foot processes by recombining z-stacks obtained with conventional fluorescence microscopy.42 Scale bar=10 and 2 μmol/l in the inset image. (c) Using the Wnt4GCE/+;R26tdTomato/+mouse, Wnt4+ cells can be seen in liver sections. In this confocal micrograph, extensive processes are seen extending from the tdTomato+ cell body and are interacting with nearby structures. This likely hepatic stellate cell is located in between hepatocytes and bile duct epithelia. Cytokeratin 18 is highlighted in green. Scale bar=10 μmol/l.
CRITICAL STEPS IN LINEAGE TRACING
Nearly any kidney cell type can now be targeted through widely available Cre drivers and reporter strains, but it remains critical to characterize each system in order to interpret results accurately. For example, mosaic expression of Cre in target tissues can lead to incomplete recombination and false-negative results. This may either be due to silencing of the transgene in a subset of cells or due to low absolute expression levels. An even more serious problem is unfaithful expression outside of the cell type that should express Cre, causing false-positive results. Many Cre drivers are created with short transgenic promoters, and these may have unexpected expression patterns (‘promiscuous expression') owing to local effects of the integration site, which is random. In an extreme example of this phenomenon, 24 separate founders for one individual transgene consisting of regulatory elements for the thy1 gene had 24 different expression patterns.20 Fortunately, online databases that document Cre driver characterization are becoming available, and these can be of great assistance in planning an experiment (Table 1).21, 22
Table 1. Cre resourcesa 21, 22.
| Resource name | Number of Cre lines characterized | Web address |
|---|---|---|
| Mouse Genome Informatics CrePortal | 1920 | http://www.creportal.org |
| Gene Expression Nervous System Atlas Cre project | 258 | http://www.gensat.org/CrePipeline.jsp |
| Allen Brain Atlas | 215 | http://connectivity.brain-map.org |
| NIH Neuroscience Blueprint Cre Driver Network | 310 | http://www.credrivermice.org |
| International Mouse Strain Resource | 920 | http://www.findmice.org |
As of May 2013.
An important limitation of inducible Cre experiments is leakiness of the CreERt2. Some lines will have a basal rate of recombination even in the absence of tamoxifen. To control for this, tissues must be examined before and after the experimental manipulation from mice that have not been exposed to tamoxifen. An added complication is the potential toxicity of tamoxifen itself. For example, tamoxifen is toxic to the gastric epithelium and liver at high doses.23, 24 When administered during pregnancy, tamoxifen acts as a mixed estrogen agonist, and this can induce late-term abortions. One workaround is to administer half the dosage of progesterone along with tamoxifen in this setting.25
LINEAGE TRACING IN KIDNEY
Genetic lineage analysis has allowed fundamental discoveries in kidney development, homeostasis, and disease. With regard to kidney development, a fate-marking study by Mugford et al.26 used an OSR1-CreERt2 allele to show that intermediate mesoderm gives rise to the great majority of cell types in the metanephric kidney, including ureteric and metanephric epithelium, interstitial stromal cells, vasculature, and smooth muscle. Anatomic fate-mapping techniques in chick also demonstrate that kidney stromal lineage, which later becomes FoxD1+, is also specified before the formation of the metanephric kidney.27 In another experiment from the McMahon group, clonal analysis of Six2+ cap mesenchyme cells provided direct proof that these are multipotent, self-renewing kidney stem cells. Kobayashi et al.28 created a Six2-CreERt2 driver and administered submaximal tamoxifen in order to label single-cap mesenchyme cells. After kidney development was completed, labeled progeny from this single cell could be observed not only in cap mesenchyme but also in the podocyte, proximal, and distal tubule lineages. Similar conclusions were drawn using a Cited1-CreERt2-based lineage tracing strategy.29 More recently, Barker et al.30 used an inducible genetic strategy to show that Lgr5+ cells within the S-shaped body are an intratubular progenitor population that gives rise to the thick ascending limb and distal convoluted tubule.
Genetic recombination is particularly useful in defining cellular hierarchies in adult kidney after injury or stress, because the influx of inflammatory cells and transient state changes (i.e., dedifferentiation) complicate accurate cellular identification by antigen- or marker-based approaches in these settings. An early example of this was provided by Moeller et al.31, who genetically labeled podocytes and induced crescentic glomerulonephritis. The results indicated that cellular crescents are in part populated by podocytes themselves. More recently, lineage analysis showed that parietal cells also contribute to crescent formation, as well as to the formation of focal and segmental glomerulosclerosis lesions.32, 33 Finally, recent studies implicate glomerular parietal cells as a niche for podocyte progenitors. Genetically labeled parietal cells at postnatal day 5 were shown to migrate onto the glomerular tuft and become podocytes in adult mice, providing direct evidence for a new cellular source of podocytes.34
A growing body of work has documented epithelial hierarchies in acute kidney disease and chronic kidney fibrosis models using lineage analysis. The approach demonstrated that bone marrow cells do not meaningfully contribute to the epithelium after acute injury, for example, although low levels of cell fusion can be detected.35, 36 We have used fate mapping to establish that the cells responsible for repair of damaged nephron epithelial originate from within the tubule and not from any extratubular compartment.37 In kidney fibrosis, our fate-mapping approach showed that interstitial myofibroblasts did not originate from epithelial cells through epithelial to mesenchymal transition, but rather they originate from endogenous kidney pericytes and interstitial fibroblasts.38 These results have been substantiated using separate genetic fate-mapping approaches by other groups.39
Lineage tracing has also provided insight concerning the identification and plasticity of erythropoietin-producing cells in kidney interstitium and the origins of renal vasculature. Gomez used a renin-Cre line to show that renin-expressing cells give rise to multiple lineages during development, and those cells retain the capacity to synthesize renin during stress.40 More recently, Asada et al. provided lineage-tracing evidence that erythropoietin-producing cells originate from the neural crest, and upon injury differentiate into myofibroblasts while they lose the capacity for erythropoietin production. Notably, erythropoietin production could be rescued by the administration of dexamethasone or neuroprotective agents, suggesting a novel strategy to treat the anemia of chronic kidney disease.41 With sparse genetic recombination, visualization of cellular substructures is made possible. We recently showed that tertiary podocyte foot processes can be easily detected by light microscopy using such a strategy.42
LIVE-CELL IMAGING AND NEW LINEAGE-TRACING TECHNOLOGIES
The increasing sensitivity and fidelity of genetic reporters has made genetic recombination well suited for live-cell imaging. For example, Shan et al.43 used kidney organ culture from a Wnt4-GFPCre; Rosa26YFP reporter cross to show differentiation and expansion of Wnt4-positive renal vesicle and tubules. Similar strategies can be used to target most populations in developing kidney, and the ability of kidney to develop in culture makes it uniquely suited for these analyses.44 Live imaging of podocytes from isolated glomeruli, in which a podocin-Cre driver activated the expression of the mT/mG reporter, has been used to demonstrate rhythmic glomerular contractions ex vivo.45 The Barasch group coupled the expression of a gene that is induced by kidney injury (Ngal) to a luciferase reporter and generated a powerful mouse model in which kidney injury activates luciferase expression. This was not a Cre/lox approach; however, luciferase reporters exist and provide a powerful means to assess gene expression and cell localization noninvasively.46 In other tissues, the dynamic analysis of cell fate in living tissues has also been facilitated by multiphoton microscopy, which allows greater depth of penetration and reporter detection without phototoxicity.47 Peti-Peterdi and colleagues have reported the first such study in the kidney,48 and it is very likely that many more will follow.
The advent of multicolor reporter alleles has had a profound impact on the stem cell field, and they are sure to have a similar effect in the kidney. The concept is straightforward: by combining multiple different fluorescent reporters with alternating lox sites, mutually exclusive excision possibilities are created and Cre is forced to ‘choose' which reporters to delete. This strategy is made possible by the discovery that variant lox sequences will not recombine with the canonical loxP site, but will recombine with an identical copy of the variant. Cre recombinase will also invert a DNA segment between two inward-facing loxP sequences.49 This property can be exploited by linking two reporters in tandem with the opposite DNA orientation in between loxP sites that are facing each other. As long as Cre is present, this segment will invert the reporter DNA segment repeatedly. Once Cre activity is gone (i.e., after tamoxifen is degraded in a CreERt2 approach), the DNA segment will randomly stabilize in only one orientation. An important consideration here is that in the case of a straight Cre driver whose expression is maintained at the time of analysis, more than one reporter will be expressed because the inversion will continue as long as Cre is present (Figure 3). The original demonstration of this elegant approach was by Livet et al.47, who developed the Brainbow2.1 reporter and used it to color-code individual neurons and their axons with up to 90 different colors.
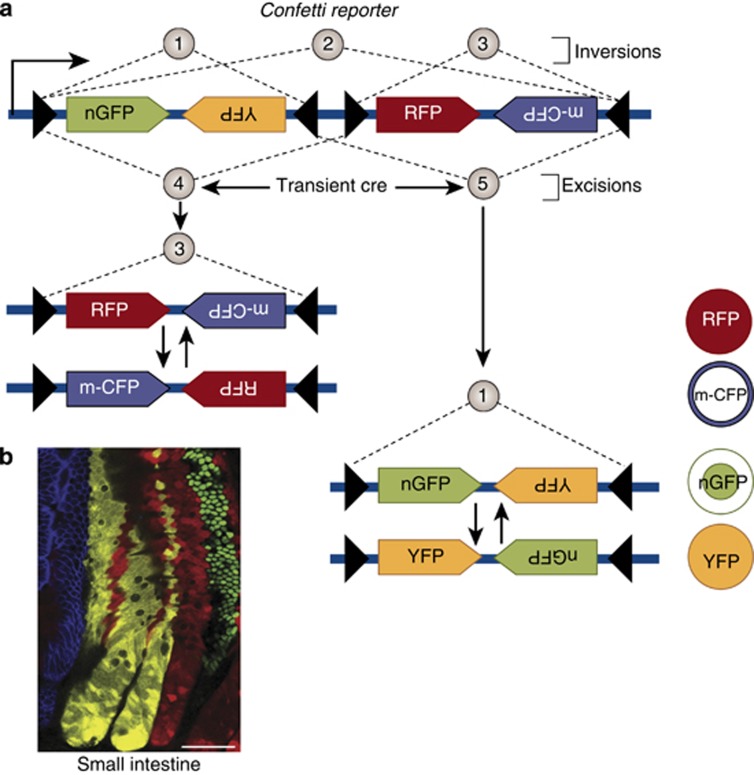
Figure 3.
Strategies for combinatorial expression of several fluorescent proteins. (a) When loxP sites are inward-facing, the intervening DNA inverts repeatedly as long as Cre activity is present, but stabilizes in only one orientation after it is removed. When loxP sites face the same direction, the intervening DNA is excised. The Brainbow2.1 construct in Confetti reporter mice takes advantage of these properties. Two cassettes, each containing two reporters placed in the opposite orientation, were separated by a series of loxP sites also in different orientations. This leads to different inversion possibilities (1, 2, and 3) and different excision possibilities (4, 5). An excision event leads to either outcome (3) or (1). Each of these in turn can undergo subsequent inversion events, but once Cre activity is removed it will stabilize in only one configuration, leading to stable expression of only one of the four reporters. Figure adapted from Lichtman et al.56 (b) An example of the Confetti reporter used to label individual crypt stem cells in the intestine. Each color represents progeny from a single labeled crypt stem cell30 (Reproduced with permission).
Multicolor reporters have allowed the assessment of contributions from multiple cell types, such as with mouse digit tip regeneration. Clevers and colleagues have modified the Brainbow2.1 by simplifying to only four possible colors and transferring the cassette to the Rosa26 locus downstream of the strong CAG promoter, the ‘Confetti' mouse. This approach enabled examination of the individual behavior of multiple stem cells in a single niche. In the case of Lgr5-positive intestinal crypt cells, the Confetti reporter revealed that crypt stem cells do not undergo asymmetrical division, but rather symmetrical division to create two identical daughter stem cells that subsequently compete for residency in the niche.50 This paradigm-shifting finding demonstrated that intestinal stem cell fate is not hierarchical (i.e., stem cell → transit amplifying cell → differentiated cell) but stochastic. Each daughter cell has equivalent potential to assume either fate, and the fate assumed is determined by neutral competition for residency within the niche. Several possible applications for multicolor reporters can be envisioned in the kidney. These include evaluation of the clonality of various proposed progenitor pools during nephrogenesis, such as Six2+ metanephric mesenchyme, FoxD1+ mesenchyme, and HoxB7+ ureteric bud lineages. In adult, several progenitor niches are known and one can imagine multicolor lineage tracing of parietal epithelium which may hold podocyte progenitors,34, 51 evaluation of proliferative potential of proximal tubule after injury,52 and interstitial mesenchymal cells some of which may be multipotent.53
A final Cre-based approach for lineage tracing involves interchromosomal recombination called mosaic analysis with double markers.54 This technique was pioneered in mice by Liquin Luo. Fluorescent reporters in this approach are artificially split by a loxP-containing artificial intron, and reciprocally chimeric genes that each encode the opposite half of the reporter are placed at identical positions in a pair of homologous chromosomes. Neither reporter is expressed until a Cre-mediated interchromosomal recombination event occurs, which reconstitutes the functional reporters. After the cell subsequently undergoes mitosis, each allele segregates to separate daughter cells and therefore express distinct colors. A powerful advantage of this approach is that many different mosaic analysis with double marker alleles on different chromosomes now exist, and they can be coupled to a mutation of interest such that homozygous mutant cells express GFP and daughter cells that are homozygous wild type express red fluorescent protein.55 Generating the appropriate crosses to execute mosaic analysis with double marker is time-consuming, and although no kidney study has yet been reported there is no other technique that allows single-cell labeling with mutant analysis.
CONCLUSIONS
Lineage analysis is a powerful tool that is now accessible to nearly all scientists. With a multitude of new Cre driver lines available, the time when any cell type in the kidney can be fate-mapped is on the immediate horizon. These techniques promise to help identify progenitor populations for regenerative medicine and to disambiguate complex cellular relationships in the kidney. Exciting new methods including multicolor reporters and single-cell mosaic analysis will answer mechanistic questions. However, a basic understanding of the limitations of these systems remains mandatory for the proper design and interpretation of a lineage-tracing experiment.
Acknowledgments
We apologize to those authors whose works we could not include owing to space constraints. BDH is supported by grants from the NIH (DK088923), the Harvard Stem Cell Institute, and an Established Investigator Award from the American Heart Association. DPD is supported by a research fellowship from the National Kidney Foundation (2011-D000691).
All the authors declared no competing interests.
References
- Conklin EG. The organization and cell lineage of the ascidian egg. J Acad Nat Sci Phil. 1905;12:1–119. [Google Scholar]
- Vogt W. Gestaltungsanalyse am Amphibienkeim mit ortlicher Vitalfarbung. II. Teil Gastrulation und Mesodermbildung bie Urodelen und Anuren. Wilhelm Roux Arch Entwicklungsmech Org. 1929;120:384–706. doi: 10.1007/BF02109667. [DOI] [PubMed] [Google Scholar]
- Axelrod D. Carbocyanine dye orientation in red cell membrane studied by microscopic fluorescence polarization. Biophys J. 1979;26:557–573. doi: 10.1016/S0006-3495(79)85271-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A. Cre recombinase: the universal reagent for genome tailoring. Genesis. 2000;26:99–109. [PubMed] [Google Scholar]
- Hans S, Freudenreich D, Geffarth M, et al. Generation of a non-leaky heat shock-inducible Cre line for conditional Cre/lox strategies in zebrafish. Dev Dyn. 2011;240:108–115. doi: 10.1002/dvdy.22497. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Littlewood TD, Hancock DC, Danielian PS, et al. A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res. 1995;23:1686–1690. doi: 10.1093/nar/23.10.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S, McMahon AP. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Dev Biol. 2002;244:305–318. doi: 10.1006/dbio.2002.0597. [DOI] [PubMed] [Google Scholar]
- Feil R, Wagner J, Metzger D, et al. Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochem Biophys Res Commun. 1997;237:752–757. doi: 10.1006/bbrc.1997.7124. [DOI] [PubMed] [Google Scholar]
- Shimshek DR, Kim J, Hubner MR, et al. Codon-improved Cre recombinase (iCre) expression in the mouse. Genesis. 2002;32:19–26. doi: 10.1002/gene.10023. [DOI] [PubMed] [Google Scholar]
- Casanova E, Fehsenfeld S, Lemberger T, et al. ER-based double iCre fusion protein allows partial recombination in forebrain. Genesis. 2002;34:208–214. doi: 10.1002/gene.10153. [DOI] [PubMed] [Google Scholar]
- Saunders TL. Inducible transgenic mouse models. Methods Mol Biol. 2011;693:103–115. doi: 10.1007/978-1-60761-974-1_7. [DOI] [PubMed] [Google Scholar]
- Traykova-Brauch M, Schonig K, Greiner O, et al. An efficient and versatile system for acute and chronic modulation of renal tubular function in transgenic mice. Nat Med. 2008;14:979–984. doi: 10.1038/nm.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder EY, Deitcher DL, Walsh C, et al. Multipotent neural cell lines can engraft and participate in development of mouse cerebellum. Cell. 1992;68:33–51. doi: 10.1016/0092-8674(92)90204-p. [DOI] [PubMed] [Google Scholar]
- Novak A, Guo C, Yang W, et al. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. Genesis. 2000;28:147–155. [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, et al. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- Shaner NC, Lin MZ, McKeown MR, et al. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat Methods. 2008;5:545–551. doi: 10.1038/nmeth.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng G, Mellor RH, Bernstein M, et al. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000;28:41–51. doi: 10.1016/s0896-6273(00)00084-2. [DOI] [PubMed] [Google Scholar]
- Murray SA, Eppig JT, Smedley D, et al. Beyond knockouts: cre resources for conditional mutagenesis. Mamm Genome. 2012;23:587–599. doi: 10.1007/s00335-012-9430-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffner CS, Herbert Pratt C, Babiuk RP, et al. Supporting conditional mouse mutagenesis with a comprehensive cre characterization resource. Nat Commun. 2012;3:1218. doi: 10.1038/ncomms2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh WJ, Khurana SS, Geahlen JH, et al. Tamoxifen induces rapid, reversible atrophy, and metaplasia in mouse stomach. Gastroenterology. 2012;142:e27. doi: 10.1053/j.gastro.2011.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MH, Kim JW, Kim JH, et al. Gene expression profiling of murine hepatic steatosis induced by tamoxifen. Toxicol Lett. 2010;199:416–424. doi: 10.1016/j.toxlet.2010.10.008. [DOI] [PubMed] [Google Scholar]
- Nakamura E, Nguyen MT, Mackem S. Kinetics of tamoxifen-regulated Cre activity in mice using a cartilage-specific CreER(T) to assay temporal activity windows along the proximodistal limb skeleton. Dev Dyn. 2006;235:2603–2612. doi: 10.1002/dvdy.20892. [DOI] [PubMed] [Google Scholar]
- Mugford JW, Sipila P, McMahon JA, et al. Osr1 expression demarcates a multi-potent population of intermediate mesoderm that undergoes progressive restriction to an Osr1-dependent nephron progenitor compartment within the mammalian kidney. Dev Biol. 2008;324:88–98. doi: 10.1016/j.ydbio.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillaume R, Bressan M, Herzlinger D. Paraxial mesoderm contributes stromal cells to the developing kidney. Dev Biol. 2009;329:169–175. doi: 10.1016/j.ydbio.2009.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi A, Valerius MT, Mugford JW, et al. Six2 defines and regulates a multipotent self-renewing nephron progenitor population throughout mammalian kidney development. Cell Stem Cell. 2008;3:169–181. doi: 10.1016/j.stem.2008.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle S, Misfeldt A, Chandler KJ, et al. Fate mapping using Cited1-CreERT2 mice demonstrates that the cap mesenchyme contains self-renewing progenitor cells and gives rise exclusively to nephronic epithelia. Dev Biol. 2008;313:234–245. doi: 10.1016/j.ydbio.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N, Rookmaaker MB, Kujala P, et al. Lgr5(+ve) stem/progenitor cells contribute to nephron formation during kidney development. Cell Rep. 2012;2:540–552. doi: 10.1016/j.celrep.2012.08.018. [DOI] [PubMed] [Google Scholar]
- Moeller MJ, Soofi A, Hartmann I, et al. Podocytes populate cellular crescents in a murine model of inflammatory glomerulonephritis. J Am Soc Nephrol. 2004;15:61–67. doi: 10.1097/01.asn.0000102468.37809.c6. [DOI] [PubMed] [Google Scholar]
- Smeets B, Uhlig S, Fuss A, et al. Tracing the origin of glomerular extracapillary lesions from parietal epithelial cells. J Am Soc Nephrol. 2009;20:2604–2615. doi: 10.1681/ASN.2009010122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeets B, Kuppe C, Sicking EM, et al. Parietal epithelial cells participate in the formation of sclerotic lesions in focal segmental glomerulosclerosis. J Am Soc Nephrol. 2011;22:1262–1274. doi: 10.1681/ASN.2010090970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appel D, Kershaw DB, Smeets B, et al. Recruitment of podocytes from glomerular parietal epithelial cells. J Am Soc Nephrol. 2009;20:333–343. doi: 10.1681/ASN.2008070795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F, Moran A, Igarashi P. Intrarenal cells, not bone marrow-derived cells, are the major source for regeneration in postischemic kidney. J Clin Invest. 2005;115:1756–1764. doi: 10.1172/JCI23015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffield JS, Park KM, Hsiao LL, et al. Restoration of tubular epithelial cells during repair of the postischemic kidney occurs independently of bone marrow-derived stem cells. J Clin Invest. 2005;115:1743–1755. doi: 10.1172/JCI22593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys BD, Valerius MT, Kobayashi A, et al. Intrinsic epithelial cells repair the kidney after injury. Cell Stem Cell. 2008;2:284–291. doi: 10.1016/j.stem.2008.01.014. [DOI] [PubMed] [Google Scholar]
- Humphreys BD, Lin SL, Kobayashi A, et al. Fate tracing reveals the pericyte and not epithelial origin of myofibroblasts in kidney fibrosis. Am J Pathol. 2010;176:85–97. doi: 10.2353/ajpath.2010.090517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koesters R, Kaissling B, Lehir M, et al. Tubular overexpression of transforming growth factor-beta1 induces autophagy and fibrosis but not mesenchymal transition of renal epithelial cells. Am J Pathol. 2010;177:632–643. doi: 10.2353/ajpath.2010.091012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequeira Lopez ML, Pentz ES, Nomasa T, et al. Renin cells are precursors for multiple cell types that switch to the renin phenotype when homeostasis is threatened. Dev Cell. 2004;6:719–728. doi: 10.1016/s1534-5807(04)00134-0. [DOI] [PubMed] [Google Scholar]
- Asada N, Takase M, Nakamura J, et al. Dysfunction of fibroblasts of extrarenal origin underlies renal fibrosis and renal anemia in mice. J Clin Invest. 2011;121:3981–3990. doi: 10.1172/JCI57301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grgic I, Brooks CR, Hofmeister AF, et al. Imaging of podocyte foot processes by fluorescence microscopy. J Am Soc Nephrol. 2012;23:785–791. doi: 10.1681/ASN.2011100988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan J, Jokela T, Skovorodkin I, et al. Mapping of the fate of cell lineages generated from cells that express the Wnt4 gene by time-lapse during kidney development. Differentiation. 2010;79:57–64. doi: 10.1016/j.diff.2009.08.006. [DOI] [PubMed] [Google Scholar]
- Costantini F, Watanabe T, Lu B, et al. Imaging kidney development. Cold Spring Harbor Protocols. 2011;2011:468–474. doi: 10.1101/pdb.top109. [DOI] [PubMed] [Google Scholar]
- Hohne M, Ising C, Hagmann H, et al. Light microscopic visualization of podocyte ultrastructure demonstrates oscillating glomerular contractions. Am J Pathol. 2013;182:332–338. doi: 10.1016/j.ajpath.2012.11.002. [DOI] [PubMed] [Google Scholar]
- Woolfenden S, Zhu H, Charest AA. Cre/LoxP conditional luciferase reporter transgenic mouse for bioluminescence monitoring of tumorigenesis. Genesis. 2009;47:659–666. doi: 10.1002/dvg.20545. [DOI] [PubMed] [Google Scholar]
- Livet J, Weissman TA, Kang H, et al. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450:56–62. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- Hackl MJ, Lam L, Burford J, et al. Cell fate tracking with serial multiphoton imaging of a new podocyte confetti mouse in vivo reveals podocyte proliferation and migration. J Am Soc Nephrol. 2012;23:SA-OR082. [Google Scholar]
- Hoess RH, Wierzbicki A, Abremski K. The role of the loxP spacer region in P1 site-specific recombination. Nucleic Acids Res. 1986;14:2287–2300. doi: 10.1093/nar/14.5.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snippert HJ, van der Flier LG, Sato T, et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143:134–144. doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- Ronconi E, Sagrinati C, Angelotti ML, et al. Regeneration of glomerular podocytes by human renal progenitors. J Am Soc Nephrol. 2009;20:322–332. doi: 10.1681/ASN.2008070709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys BD, Czerniak S, Dirocco DP, et al. Repair of injured proximal tubule does not involve specialized progenitors. Proc Natl Acad Sci USA. 2011;108:9226–9231. doi: 10.1073/pnas.1100629108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelekanos RA, Li J, Gongora M, et al. Comprehensive transcriptome and immunophenotype analysis of renal and cardiac MSC-like populations supports strong congruence with bone marrow MSC despite maintenance of distinct identities. Stem Cell Res. 2012;8:58–73. doi: 10.1016/j.scr.2011.08.003. [DOI] [PubMed] [Google Scholar]
- Zong H, Espinosa JS, Su HH, et al. Mosaic analysis with double markers in mice. Cell. 2005;121:479–492. doi: 10.1016/j.cell.2005.02.012. [DOI] [PubMed] [Google Scholar]
- Muzumdar MD, Luo L, Zong H. From the cover: modeling sporadic loss of heterozygosity in mice by using mosaic analysis with double markers (MADM) Proc Natl Acad Sci USA. 2007;104:4495–4500. doi: 10.1073/pnas.0606491104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtman JW, Livet J, Sanes JR. A technicolour approach to the connectome. Nat Rev Neurosci. 2008;9:417–422. doi: 10.1038/nrn2391. [DOI] [PMC free article] [PubMed] [Google Scholar]