The crystal structures of Y. pestis RipA mutants were determined to provide insights into the CoA transferase reaction pathway.
Keywords: Yersinia pestis, CoA transferase, mutational analysis, rip operon
Abstract
Yersinia pestis, the causative agent of bubonic plague, is able to survive in both extracellular and intracellular environments within the human host, although its intracellular survival within macrophages is poorly understood. A novel Y. pestis three-gene rip (required for intracellular proliferation) operon, and in particular ripA, has been shown to be essential for survival and replication in interferon γ-induced macrophages. RipA was previously characterized as a putative butyryl-CoA transferase proposed to yield butyrate, a known anti-inflammatory shown to lower macrophage-produced NO levels. RipA belongs to the family I CoA transferases, which share structural homology, a conserved catalytic glutamate which forms a covalent CoA-thioester intermediate and a flexible loop adjacent to the active site known as the G(V/I)G loop. Here, functional and structural analyses of several RipA mutants are presented in an effort to dissect the CoA transferase mechanism of RipA. In particular, E61V, M31G and F60M RipA mutants show increased butyryl-CoA transferase activities when compared with wild-type RipA. Furthermore, the X-ray crystal structures of E61V, M31G and F60M RipA mutants, when compared with the wild-type RipA structure, reveal important conformational changes orchestrated by a conserved acyl-group binding-pocket phenylalanine, Phe85, and the G(V/I)G loop. Binary structures of M31G RipA and F60M RipA with two distinct CoA substrate conformations are also presented. Taken together, these data provide CoA transferase reaction snapshots of an open apo RipA, a closed glutamyl-anhydride intermediate and an open CoA-thioester intermediate. Furthermore, biochemical analyses support essential roles for both the catalytic glutamate and the flexible G(V/I)G loop along the reaction pathway, although further research is required to fully understand the function of the acyl-group binding pocket in substrate specificity.
1. Introduction
Bubonic plague is a pathogenic disease that has resulted in millions of deaths throughout history in several great pandemics (Perry & Fetherston, 1997 ▶). In the modern day, antibiotics and much-improved health standards have decreased its prevalence; however, endemic threats remain in rural parts of North and South America, Africa and Asia (Perry & Fetherston, 1997 ▶). The enterobacterium Yersinia pestis, the causative agent of plague, has both intracellular and extracellular phases of infection; in the early stages of infection, phagocytosed Y. pestis can survive and replicate in macrophages (Cavanaugh & Randall, 1959 ▶; Janssen & Surgalla, 1969 ▶; Straley & Harmon, 1984 ▶; Charnetzky & Shuford, 1985 ▶) before escaping into the extracellular environment (Williamson & Oyston, 2013 ▶; Bliska & Casadevall, 2009 ▶). Furthermore, various virulence factors allow Y. pestis to disseminate in the host and evade its innate immune defense mechanisms (Perry & Fetherston, 1997 ▶; Williamson & Oyston, 2013 ▶). The pigmentation (pgm) locus, which is critical for early-phase infection (Perry et al., 1999 ▶) and replication in postactivated (interferon γ-induced) macrophages (Pujol et al., 2005 ▶), contains many virulence factors. Specifically, Y. pestis replication in postactivated macrophages has been correlated with reduced host nitric oxide (NO) levels in pgm + Y. pestis despite the upregulation of macrophage-inducible NO synthase (iNOS) expression. Interestingly, a Y. pestis Δpgm mutant replicates in postactivated iNOS−/− macrophages, whereas its growth is severely attenuated in wild-type (WT) macrophages, suggesting that killing of the Δpgm mutant is NO-dependent. Within the pgm locus, studies demonstrate that the ripA gene is necessary for Y. pestis replication in postactivated macrophages and for a decrease in host NO levels. Recently, RipA was characterized as a putative butyryl-CoA transferase (butyryl-CoAT; Torres et al., 2011 ▶), thus alluding to a role for Y. pestis butyrate production in observed host NO reduction, as butyrate is a known anti-inflammatory that has been shown to reduce NO levels in interferon γ-activated macrophages (Park et al., 2007 ▶; Stempelj et al., 2007 ▶).
RipA belongs to a class of CoA transferase (CoAT) enzymes that transfer the CoAS− anion from a CoA-thioester donor to a free acid acceptor. Additionally, RipA is a member of the family I CoATs (Heider, 2001 ▶), all members of which have similar two-domain αβ folds and a conserved catalytic glutamate (Fig. 1 ▶ a) important in facilitating several covalent intermediates (Solomon & Jencks, 1969 ▶). The multistep reaction (Fig. 2 ▶) follows a series of nucleophilic attacks on acceptor carbonyl C atoms starting with attack of the catalytic glutamate on the CoA-donor carbonyl C atom, forming the first glutamyl-anhydride intermediate. The covalent glutamyl CoA-thioester intermediate complex is then formed by attack of CoA-thiolate on the carbonyl C atom of glutamate in conjunction with the release of the donor carboxylic acid. A new acyl-CoA is then formed through the reverse reaction via a second anhydride intermediate.
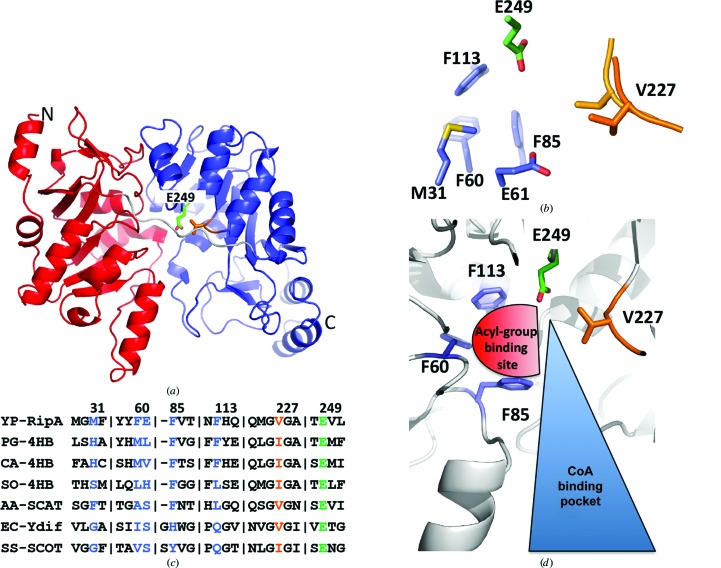
Figure 1.
Structure of apo WT RipA (PDB entry 3qli). (a) Monomeric structure with the N-terminal and C-terminal domains colored blue and red, respectively. The catalytic glutamate Glu249 and Val227 of the G(V/I)G loop are in stick representation with C atoms colored green and orange, respectively, and O and N atoms red and blue, respectively. (b) The active site of WT RipA with the hydrophobic pocket residues forming the acyl-group binding pocket shown as blue sticks, the catalytic glutamate shown as green sticks and the two conformations of the flexible G(V/I)G loop shown in orange as sticks and ribbon. (c) A truncated structure-based sequence alignment with the corresponding acyl-group binding-pocket residues, the catalytic glutamate and the conserved G(V/I)G loop shown with the residues colored as in (a) and (b). YP-RipA, Y. pestis RipA; PG-4HB, Porphyromonas gingivalis 4-HB-CoAT; CA-4HB, C. aminobutyricum 4-HB-CoAT; SO-4HB, S. oneidensis 4-HB-CoAT; AA-SCAT, A. aceti SCACT; EC-Ydif, E. coli Ydif CoAT; SS-SCOT, S. scrofa SCOT. (d) The active site from a previous MD simulation of RipA with the open acyl-group binding site shown as a red half circle and the CoA binding pocket shown as a blue triangle. The structure is shown as a cartoon in white with the hydrophobic pocket residues, the catalytic glutamate and flexible G(V/I)G loop shown as sticks and colored as above.
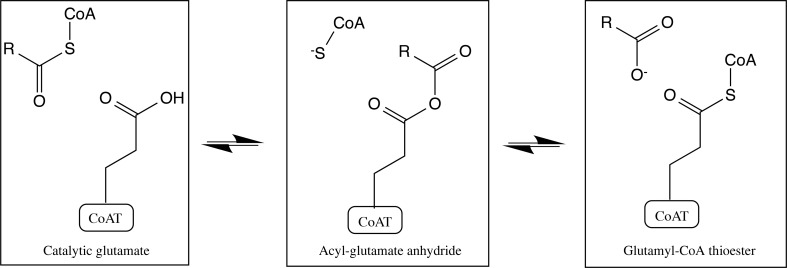
Figure 2.
Family I CoA transferase intermediates for general half-reaction. From left to right the initial acyl-CoA in the active site passes through two intermediates, the acyl-glutamyl anhydride and the glutamyl-CoA thioester, with the catalytic glutamate before exchanging the acyl group. The reverse reaction forms the same intermediates from right to left to form the new acyl-CoA.
To date, several crystal structures of family I CoATs have been solved (Macieira et al., 2009 ▶; Rangarajan et al., 2005 ▶; Bateman et al., 2002 ▶), including two with CoA bound in their trapped glutamyl CoA-thioester conformations (Rangarajan et al., 2005 ▶; Fraser et al., 2010 ▶). Additionally, structures of Clostridium aminobutyricum 4-hydroxybutyrate-CoA transferase (4-HB-CoAT; Macieira et al., 2012 ▶) and Acetobacter aceti succinyl-CoA:acetate CoA transferase (SCACT; also known as AarC; Mullins & Kappock, 2012 ▶) exhibit apo (open) and CoA-bound (closed) conformations facilitated by the movement of the conserved flexible G(V/I)G loop. The previously solved apo RipA structure contains two monomers in the asymmetric unit, where each monomer has a different G(V/I)G loop conformation (Fig. 1 ▶ b), suggesting open and closed configurations; this loop flexibility was also observed in apo RipA molecular-dynamics (MD) simulations (Torres et al., 2011 ▶). Furthermore, the conformational flexibility of the G(V/I)G loop has been proposed to be important for the protection of reaction intermediates from hydrolysis in its closed state (Macieira et al., 2012 ▶; Torres et al., 2011 ▶) and suggests that varying degrees of open to closed conformations may occur as the CoAT transitions through different intermediates along its reaction pathway.
Here, we present three different structural conformations of RipA, a putative family I butyryl-CoAT, which represent the open apo form, the closed glutamyl-anhydride intermediate and the open flexible CoA-thioester intermediate. Notably, the two binary structures reveal CoA moieties in two distinct configurations. We also observe conformational diversity of a conserved phenylalanine (Phe85) which gates access to the acyl-group binding pocket, and of the G(V/I)G loop, which facilitates access to the catalytic active site. Taken together, this structural analysis illustrates the distinct conformational changes required for RipA and other CoATs to cycle through their reaction mechanism.
2. Materials and methods
2.1. Site-directed mutagenesis
Site-directed mutagenesis was performed using the QuikChange II Site-Directed Mutagenesis Kit (Stratagene). Briefly, PCR was performed with 125 ng of each mutant primer pair, 10 ng of WT RipA plasmid DNA as the template, dNTP mix, dH2O, reaction buffer and PfuUltra II polymerase (Stratagene). Following the PCR, the product was digested with DpnI and then transformed into Top10 Escherichia coli cells. The resulting positive clones were then sequenced and transformed into E. coli BL21(DE3) Gold cells (Stratagene) for expression.
2.2. Overexpression and purification of RipA mutants
The RipA mutants were overexpressed as described previously for WT RipA (Torres et al., 2011 ▶). In brief, the constructs in a pET28 plasmid were grown in E. coli BL21(DE3) Gold cells (Stratagene) at 310 K in LB medium containing 30 µg ml−1 kanamycin. Protein expression was induced by adding 1 mM IPTG at an OD600nm of ∼1.0 and the cells were grown at 291 K overnight before harvesting. The cells were pelleted at 5000 rev min−1 for 10 min and then resuspended in wash buffer (50 mM Tris pH 7.4, 350 mM NaCl, 10 mM imidazole, 10% glycerol) containing phenylmethylsulfonyl fluoride and hen egg-white lysozyme. The cells were lysed by sonication and centrifuged at 13 000 rev min−1 for 40 min followed by filtration (0.22 µm) of the cell lysate to remove cell debris before purification. The cell lysate was loaded onto an Ni2+-charged HiTrap column (5 ml) and washed with wash buffer before the protein was eluted with an 10–500 mM linear gradient of imidazole (100 ml), in which the purified protein eluted between 200 and 300 mM imidazole. The fractions were collected and concentrated to between 5 and 7 mg ml−1 in a Centricon (15 ml).
2.3. CoA transferase activity assay
RipA enzymatic activity assays were carried out as described previously (Torres et al., 2011 ▶). The transfer of CoA from an acyl-CoA (Sigma) to acetate was determined by the coupled release of CoASH from the condensation of oxaloacetate by citrate synthase (Sigma) and was detected at 412 nm using 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB). Each 100 µl of reaction mixture contained 50 µM acyl-CoA, 100 mM sodium acetate, 100 mM Tris pH 7.0, 1 mM oxaloacetate, 0.5 U citrate synthase and 1 mM DTNB. Reactions were initiated by the addition of 10 µM RipA and were monitored for 30 min at 412 nm at room temperature, detecting the formation of the nitrothiobenzoate dianion. The negative control was the same reaction mixture without citrate synthase. Kinetic experiments were carried out for RipA (5 µM), in which the concentrations of butyryl-CoA and propionyl-CoA were varied. All experiments were performed in at least three independent assays. All kinetic data were determined using the GraphPad Prism software (v.5.0 for Mac).
2.4. Crystallization of RipA mutants
M31G RipA crystals were grown over a period of one week at room temperature by hanging-drop vapor diffusion with a reservoir consisting of 0.2 M ammonium acetate, 24% PEG 3350, 1 mM succinyl-CoA and crystallization drops consisting of a 1 µl:1 µl ratio of protein solution to reservoir solution. E61V RipA mutant crystals were similarly grown in 0.3 M sodium malonate pH 5, 22% PEG 3350, 1 mM propionyl-CoA. F60M RipA crystals were grown similarly in 25% PEG 3350, 0.4 M ammonium phosphate monobasic, 1 mM butyryl-CoA. E249A RipA crystals were grown similarly in 0.1 M bis(2-hydroxyethyl)amino-tris(hydroxymethyl)methane (bis-tris) pH 5.5, 21% PEG 3350, 0.2 M ammonium sulfate, 1 mM succinyl-CoA. E249D RipA crystals were grown similarly in 0.1 M bis-tris pH 5.5, 27% PEG monomethyl ether 2000, 1 mM propionyl-CoA. Crystals were mounted and collected under cryoconditions with the addition of 25% glycerol to the reservoir condition as a cryoprotectant.
2.5. Data collection, structure determination and refinement
Data sets for E61V RipA, M31G RipA, E249A RipA, E249D RipA and F60M RipA were collected at the synchrotron at a wavelength of 1 Å. Data reduction was carried out with the HKL-2000 suite (Otwinowski & Minor, 1997 ▶), resulting in a greater than 98% complete data set to 2.4, 2.0, 2.0, 2.0 and 2.8 Å resolution for E61V, M31G, F60M, E249A and E249D RipA, respectively. The structures were solved by molecular replacement using WT RipA as a search model in phenix.automr (Adams et al., 2010 ▶) followed by automatic building in phenix.autobuild and iterative cycles of phenix.refine (Adams et al., 2010 ▶) for refinement and manual building in Coot (Emsley et al., 2010 ▶). The final data-collection and refinement statistics are summarized in Table 1 ▶. The final stereochemistry and geometry for each model was validated with MolProbity (Chen et al., 2010 ▶). All figures were produced using the PyMOL molecular-graphics program (DeLano, 2002 ▶), with ligand alignments performed with the LigAlign plugin (Heifets & Lilien, 2010 ▶).
Table 1. Data-collection and refinement statistics.
Values in parentheses are for the outer shell.
| Mutant | E61V | M31G | F60M | E249A | E249D |
|---|---|---|---|---|---|
| Ligand | None | CoA, acetate | CoA | None | None |
| Data collection | |||||
| Wavelength () | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| Resolution range () | 502.4 (2.492.40) | 502.01 (2.082.01) | 502.0 (2.072.00) | 502.0 (2.032.00) | 502.8 (2.852.80) |
| Space group | C2 | C2 | P31 | C2 | P3121 |
| Unit-cell parameters | |||||
| a () | 118.9 | 117.4 | 197.3 | 118.2 | 197.2 |
| b () | 109.0 | 107.2 | 197.3 | 108.2 | 197.2 |
| c () | 85.0 | 87.8 | 59.9 | 84.9 | 59.2 |
| () | 90 | 90 | 90 | 90 | 90 |
| () | 120 | 120 | 90 | 120 | 90 |
| () | 90 | 90 | 120 | 90 | 120 |
| Total reflections | 136447 | 224554 | 860262 | 233367 | 362778 |
| Unique reflections | 36693 | 61943 | 173491 | 62152 | 32103 |
| Multiplicity | 3.7 (3.7) | 3.6 (2.7) | 5.0 (3.5) | 3.8 (3.5) | 11.3 (11.2) |
| Completeness (%) | 99.73 (98.31) | 99.55 (95.96) | 97.9 (79.5) | 99.16 (96.25) | 99.87 (98.74) |
| Mean I/(I) | 20.74 (10.58) | 37.22 (8.35) | 16.39 (3.21) | 14.65 (2.39) | 18.02 (3.72) |
| Wilson B factor (2) | 26.24 | 21.00 | 27.60 | 26.69 | 66.59 |
| R merge † | 0.061 (0.159) | 0.089 (0.293) | 0.082 (0.396) | 0.071 (0.442) | 0.147 (0.802) |
| Refinement | |||||
| R factor‡ | 0.1515 (0.1660) | 0.1351 (0.1886) | 0.1727 (0.2345) | 0.1477 (0.2015) | 0.1820 (0.2585) |
| R free | 0.1916 (0.2273) | 0.1799 (0.2438) | 0.1980 (0.2681) | 0.1759 (0.2721) | 0.2192 (0.2705) |
| No. of atoms | 6796 | 14445 | 15160 | 6792 | 6662 |
| No. of waters | 320 | 599 | 167 | 569 | 31 |
| No. of protein residues | 880 | 880 | 1774 | 880 | 862 |
| R.m.s.d., bonds () | 0.010 | 0.016 | 0.017 | 0.012 | 0.005 |
| R.m.s.d., angles () | 1.04 | 1.40 | 1.44 | 1.19 | 0.98 |
| Ramachandran favored (%) | 98 | 97 | 97 | 97 | 96 |
| Ramachandran outliers (%) | 0 | 0.34 | 0.23 | 0 | 0 |
| B factors (2) | |||||
| Average | 33.5 | 30.30 | 34.6 | 35.2 | 79.60 |
| Macromolecules | 33.5 | 29.70 | 33.8 | 34.7 | 79.70 |
| Solvent | 33.1 | 35.60 | 40.5 | 40.8 | 58.70 |
| PDB code | 4n8h | 4n8i | 4n8j | 4n8k | 4n8l |
R
merge = 
 .
.
R
work = 
 . R
free was computed identically except that all reflections belonged to a test set consisting of a 5% random selection of the data.
. R
free was computed identically except that all reflections belonged to a test set consisting of a 5% random selection of the data.
3. Results
3.1. Mutations within the proposed active site of RipA alter reactivity and kinetics
The role of the conserved catalytic glutamate residue in CoA transfer for family I CoATs has been well established (Solomon & Jencks, 1969 ▶); however, additional vital residues that contribute to CoAT activity are not well documented. Previously, we demonstrated the preference of RipA for short and unbranched acyl-CoA substrates (butyryl-CoA and propionyl-CoA) and proposed that an acyl-group binding pocket adjacent to the catalytic glutamate (Glu249) may confer substrate specificity in RipA (Torres et al., 2011 ▶; Figs. 1 ▶ b and 1 ▶ d). This pocket consists of three phenylalanine residues (Phe60, Phe85 and Phe113) and two additional residues (Met31 and Glu61) that are proposed to form contacts with the acyl group. Furthermore, it was suggested that the conserved flexible G(V/I)G loop governs access to the active site (Torres et al., 2011 ▶; Figs. 1 ▶ b and 1 ▶ d). To examine the effects of these residues on RipA CoAT activity, a comprehensive series of select single-point mutants were constructed. Moreover, amino-acid substitutions within the acyl-group binding pocket were constructed based on aligned residue equivalents in other family I CoAT members (Fig. 1 ▶ c), as the initial Ala mutations resulted in insoluble protein (data not shown). Finally, mutations of the catalytic glutamate (Glu249) and Val227 in the G(V/I)G loop were chosen to inactivate RipA.
The transferase activity of each RipA mutant was determined utilizing the coupled DTNB assay in the presence of the potential substrates butyryl-CoA or propionyl-CoA as described previously (Torres et al., 2011 ▶). No activity was observed for the proposed acyl-group binding-pocket mutations of RipA: M31H, F60V, F85H, F85Y, F113L and F113Q (Figs. 1 ▶ b and 1 ▶ c; data not shown). Similarly, catalytic glutamate E249A and E249D and flexible G(V/I)G loop V227G and V227W RipA mutants were rendered inactive (data not shown). In contrast, E61V, M31G and F60M RipA mutants resulted in increased activities compared with WT RipA (Table 2 ▶); these mutants were further characterized by determining V max and K m values for both CoA substrates (Table 2 ▶). Distinct from WT RipA, the K m values for M31G and F60M RipA are 2–2.5-fold higher for butyryl-CoA and ∼1.5-fold lower for propionyl-CoA. E61V RipA, on the other hand, revealed similar binding abilities for both butyryl-CoA and propionyl-CoA. Furthermore, all three mutants exhibited significant 2–10-fold increases in their enzyme efficiencies (k cat/K m).
Table 2. Kinetic data for WT RipA and RipA mutants.
| WT | E61V | F60M | M31G | |
|---|---|---|---|---|
| Butyryl-CoA† | ||||
| k cat (min1) | 3.50 0.22 | 21.4 3.4 | 13.7 1.0 | 40.5 19.1 |
| K m (M) | 149 32 | 145 9 | 302 12 | 375 157 |
| k cat/K m (M 1s1) | 4.0 102 1.1 102 | 2.4 103 2.4 102 | 7.6 102 8.6 101 | 1.8 103 1.1 102 |
| Propionyl-CoA† | ||||
| k cat (min1) | 3.5 0.18 | 20.4 1.36 | 9.24 0.72 | 22.1 5.50 |
| K m (M) | 272 19 | 132 23 | 203 22 | 249 77 |
| k cat/K m (M 1s1) | 2.1 102 3.5 101 | 2.6 103 2.5 102 | 8.0 102 2.9 101 | 1.8 103 1.8 102 |
The CoA donor for the transfer reaction is acetate.
3.2. RipA mutant structures reveal mechanistic insights
Previously, WT RipA was determined to be tetrameric in solution and the X-ray crystal structure of apo WT RipA (Figs. 1 ▶ a and 3 ▶ a) also demonstrated a tetramer; however, there were two molecules in the asymmetric unit, with each monomer manifesting a different conformation of the conserved flexible G(V/I)G loop (Torres et al., 2011 ▶; Fig. 1 ▶ b). Furthermore, the apo structure was a result of crystallizing WT RipA in the presence of succinyl-CoA (1 mM), although there was no observable electron density for the substrate within the model (Torres et al., 2011 ▶). This present study builds upon previous work by demonstrating RipA conformational flexibility utilizing E61V, F60M and M31G RipA mutant structures, where each structure described below represents an intermediate along the reaction pathway (Table 1 ▶, Fig. 3 ▶).
Figure 3.
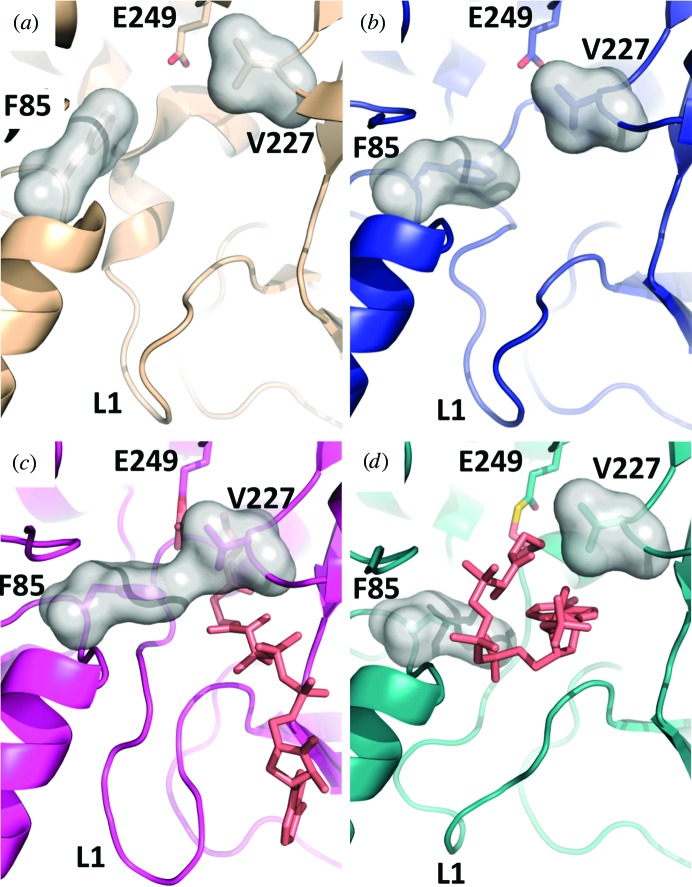
Comparison of the monomeric structures of WT RipA with RipA mutants. The overall structures are shown in cartoon representation, in which WT RipA (PDB entry 3qli) is colored wheat (a), E61V RipA is colored dark blue (b), M31G RipA is colored magenta (c) and F60M RipA is colored teal (d). The catalytic glutamate in each structure is labeled and shown in stick representation, with C, O and N atoms colored yellow, red and blue, respectively. C atoms of the anhydride and CoA in the M31G RipA structure and the CoA-thioester in the F60M RipA structure are colored white, whereas O and N atoms are colored red and blue, respectively. The flexible G(V/I)G loop is circled in black and the flexible loop 1 (L1) is circled in red.
E61V RipA was crystallized in the presence of propionyl-CoA in space group C2, and the structure was solved to 2.4 Å resolution with an r.m.s.d. of 0.16 Å compared with WT RipA over all Cα atoms. Similar to WT RipA, each E61V RipA monomer within the asymmetric unit exhibits a distinct G(V/I)G loop conformation and no observable electron density for propionyl-CoA (Fig. 3 ▶ b). The only observable structural difference is the rotation of the conserved phenylalanine side chain, Phe85, away from the catalytic glutamate by ∼60° in the E61V RipA structure (Fig. 4 ▶ a), resulting in a volume increase of the hydrophobic acyl-group binding pocket. This phenomenon was previously observed in the MD simulation for WT RipA (Torres et al., 2011 ▶) and is proposed to prime the pocket for accepting the acyl group from an incoming acyl-CoA. Thus, we hypothesize that the fully open conformation comprises the opening of the G(V/I)G loop, as observed in one monomer of both WT RipA (Fig. 5 ▶ a) and E61V RipA. Additionally, we propose that the alternate G(V/I)G loop orientation of E61V RipA represents a partially closed conformation with an accessible acyl-binding pocket (Fig. 5 ▶ b).
Figure 4.
An enlarged view comparing the active sites of the WT RipA structure in stick representation colored in wheat with an overlay of (a) the E61V RipA mutant colored in dark blue, (b) the M31G RipA mutant colored in magenta and (c) the F60M RipA mutant colored in teal, with O, N and S atoms colored red, blue and yellow, respectively. The CoA from the F60M RipA structure and the CoA and anhydride from the M31G RipA structure are shown in a white stick representation. The mutant residues in each structure are labeled in blue, with arrows indicating the changes in the residue positions between the WT RipA and mutant structures.
Figure 5.
Different conformations of the flexible G(V/I)G loop. (a) The WT RipA structure with the open G(V/I)G loop in wheat, (b) the E61V RipA structure with the partially closed G(V/I)G loop in dark blue, (c) the M31G RipA structure with the closed G(V/I)G loop in magenta and (d) the F60M RipA structure with the open G(V/I)G loop in teal are shown as cartoons. Val227, Phe85 and Glu249 are shown as sticks, while the surface representations of Phe85 and Val227 are colored gray. The CoA from the F60M RipA structure and the CoA and anhydride from the M31G RipA structure are shown in salmon as sticks. The flexible loop 1 is labelled L1.
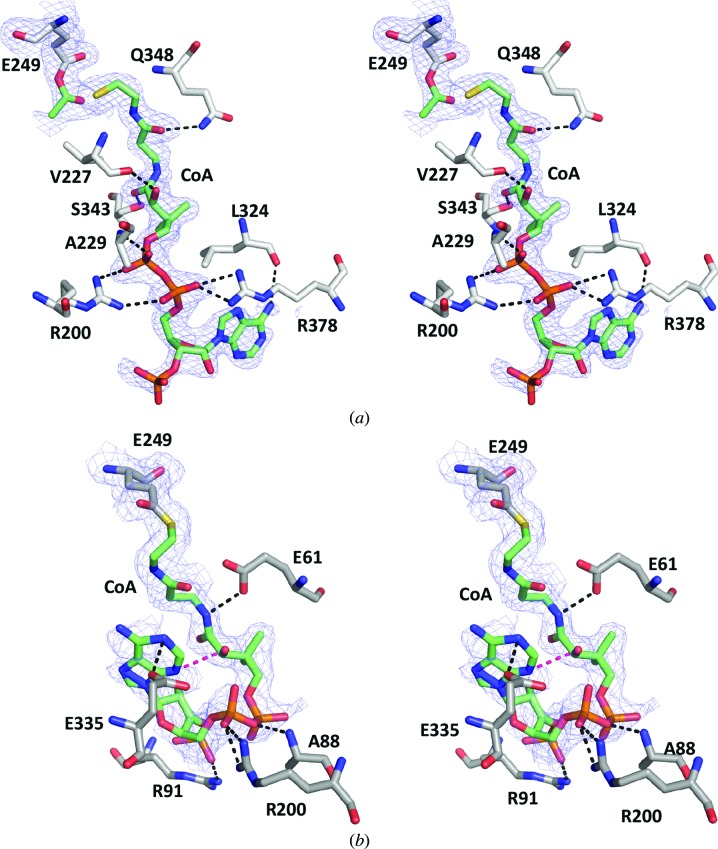
M31G RipA was crystallized in the presence of succinyl-CoA in space group C2, and the structure was solved to 2.0 Å resolution with an r.m.s.d. of 0.59 Å compared with WT RipA over all Cα atoms (Fig. 3 ▶ c). Within the active site, there is observable electron density for the acyl-anhydride intermediate between an acetate molecule and the catalytic glutamate (Glu249), as well as the resulting CoA-thiolate. The CoA molecule is modeled in an extended conformation such that the OE2 O atom of Glu249 is approximately 23 Å from the C2A C atom of the CoA adenosine moiety, which is located near the binding-pocket entrance (Fig. 6 ▶ a). Additionally, several residues (Arg200, Val227, Ala229, Leu324, Ser343, Gln348 and Arg378) bind CoA through electrostatic interactions (Table 3 ▶, Fig. 6 ▶ a). The M31G RipA mutant structure represents a fully closed conformation, which is defined by two separate events. Firstly, the CG2 C atom of Val227 from the flexible G(V/I)G loop in the M31G RipA structure moves ∼3.5 Å into the pocket (Figs. 4 ▶ b and 5 ▶ c), similar to the partially closed conformation discussed for the E61V mutant in Fig. 5 ▶(b). Secondly, the Phe85 side chain flips 60° out of the acyl-group binding pocket with a concomitant 2.4 Å shift of the Cα backbone towards Val227 (Figs. 4 ▶ b and 5 ▶ c). Furthermore, the M31G mutation appears to affect additional residues within the acyl-group binding pocket, whereby the Phe60 side chain is flipped 120° to compensate for the mutated Met31 and the side chain of Phe113 is rotated 50° into the binding pocket, reducing its volume (Fig. 4 ▶ b).
Figure 6.
2mF o − ΔF c composite OMIT electron-density map for bound ligands calculated with Glu249 and ligands omitted. (a) The catalytic glutamate and interacting residues from the M31G RipA structure are shown as white sticks, while the acetate molecule from the glutamyl-anhydride and the CoA moiety are shown as green sticks with the O, N, P and S atoms colored red, blue, orange and yellow, respectively. The electron density colored as a blue mesh is contoured at 1.0σ. (b) The F60M RipA structure is shown as gray sticks, while the CoA moiety is shown as green sticks with the O, N, P and S atoms colored red, blue, orange and yellow, respectively. The electron density colored as a blue mesh is contoured at 0.9σ. Electrostatic interactions between the CoA and the protein residues are shown as black dashed lines, while an intramolecular CoA hydrogen bond is shown as a magenta dashed line.
Table 3. Coordinating ligands of RipA M31G and F60M mutants in complex with CoA.
| RipA atom | CoA atom | Distance () |
|---|---|---|
| M31G mutant | ||
| Gln348NE2 | O5P | 2.8 |
| Ser343OG | O9P | 2.8 |
| Ala229N | O5A | 3.0 |
| Arg200NH | O4A | 2.8 |
| Arg200NH2 | O1A | 2.9 |
| Arg378NH1 | O2A | 2.9 |
| Arg378NH2 | O2A | 3.0 |
| Leu324O | N6A | 2.8 |
| Val227O | OAP | 2.8 |
| F60M mutant | ||
| Glu61OE2 | N8P | 3.1 |
| Glu335OE2 | N7A | 2.8 |
| Arg91NH1 | O8A | 2.9 |
| Arg91NH2 | O8A | 2.7 |
| Ala88N | O3A | 3.0 |
| Arg200NH1 | O2A | 2.9 |
| Arg200NH2 | O2A | 3.1 |
Compared with WT RipA, these M31G RipA conformational changes result in a very distinct constriction in the vicinity of the G(V/I)G loop, preventing additional solvent access to the active site (Fig. 5 ▶ c). This constriction is evident when comparing the shortest Phe85 to Val227 distances between the various structures, which range from 11.6 Å in the fully open apo WT RipA conformation (Fig. 5 ▶ a) to 6.6 Å in the partially closed E61V RipA mutant (Fig. 5 ▶ b) and 3.8 Å in the fully closed M31G RipA mutant (Fig. 5 ▶ c). Moreover, multiple smaller rearrangements along the CoA-binding pocket are observed in M31G RipA, including within flexible loop 1 (L1), that allow CoA to bind in its extended orientation (Fig. 5 ▶ c).
F60M RipA was crystallized in the presence of butyryl-CoA in space group P31, and the structure was solved to 2.0 Å resolution with an r.m.s.d. of 0.43 Å compared with WT RipA over all Cα atoms (Fig. 3 ▶ d). As for M31G RipA, there is observable density for CoA in the F60M RipA structure; however, the modeled CoA is trapped as an intermediate covalently bound to the catalytic glutamate, Glu249, via a thioester bond. Furthermore, unlike the extended CoA conformation observed in M31G RipA (Figs. 5 ▶ c and 6 ▶ a), F60M RipA features a J-like CoA conformation, in which the bound CoA is bent back on itself at the pyrophosphate group such that an intramolecular hydrogen bond (3.2 Å) forms between the N7A N atom of the adenosine group and the O3A O atom of the carboxyl of the extended pantetheine arm, which points back towards the catalytic site (Figs. 5 ▶ d and 6 ▶ b). The bound CoA forms electrostatic interactions with residues Glu61, Ala88, Arg91, Arg200 and Glu335 along the CoA binding pocket (Table 3 ▶), which remains in the fully open conformation as assessed by the orientation of Phe85 as well as the 11 Å distance between Phe85 and Val227 (Fig. 5 ▶ d). Of note, there is weak electron density for the adenine group of CoA, presumably owing to its flexibility within the open CoA binding pocket (Fig. 6 ▶ b). Furthermore, L1 has a different conformation compared with the WT RipA and M31G RipA structures (Fig. 5 ▶), resulting in part from the collapsed CoA conformation that induces a 2.2 Å movement and rotamer rotation of Glu335 that forms an electrostatic interaction with N1A N atom of the CoA adenosine group (Fig. 6 ▶ b). Additionally, the Glu61 side chain rotates 90° to interact with the O5P O atom of the pantetheine group of CoA.
3.3. Essentiality of the catalytic glutamate
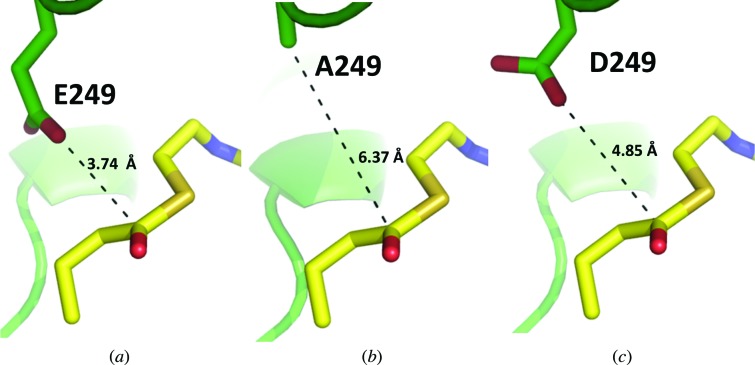
Functional assays reveal that both the E249A and E249D RipA mutants are rendered inactive, and the structures reinforce the importance of the key catalytic glutamate residue. E249A RipA was crystallized in the presence of succinyl-CoA in space group C2, and the structure was solved to 2.0 Å resolution with an r.m.s.d. of 0.148 Å compared with WT RipA over all Cα atoms. E249D RipA was also crystallized in the presence of succinyl-CoA in space group P31, and the structure was solved to 2.8 Å resolution with an r.m.s.d. of 0.468 Å compared with WT RipA over all Cα atoms. Both structures are nearly identical to WT RipA except for the mutated Glu249 residue: the substituted catalytic residue is located too distantly (6.37 and 4.85 Å for E249A and E249D, respectively) from the docked butyryl-CoA previously described for WT RipA (Torres et al., 2011 ▶) to attack the carboxylate of the acyl-CoA (Fig. 7 ▶).
Figure 7.
Modeled butyryl-CoA in the WT RipA (Torres et al., 2011 ▶) and E249A/D RipA mutant structures. Stick representations of the (a) WT RipA, (b) E249A RipA and (c) E249D RipA catalytic glutamate or mutant residue and modeled butyryl-CoA are shown with the side chain to carbonyl electrophile distance shown in black. RipA and CoA C atoms are colored green and yellow, respectively, while all O, N and S atoms are colored red, blue and tan, respectively.
4. Discussion
4.1. Comparison of RipA mutant structures with other CoA transferases
WT RipA and all three mutant structures exist as tetramers in solution (Torres et al., 2011 ▶), in contrast to the dimers formed by other characterized family I CoATs (Mullins & Kappock, 2012 ▶; Macieira et al., 2009 ▶). Apart from this difference, the new apo and CoA-bound RipA mutant structures described here share many traits previously observed in other family I CoAT members. In particular, the E61V RipA structure is almost identical to that of WT RipA and, as such, is comparable to other CoATs, especially homologous 4-HB-CoATs (Fig. 1 ▶ c), as previously described for WT RipA (Torres et al., 2011 ▶). Moreover, unlike in WT RipA, the E61V RipA acyl-group binding-pocket residue Phe85 is in a similar orientation as the conserved phenylalanines from both the C. aminobutyricum (Macieira et al., 2009 ▶) and Shewanella oneidensis 4-HB-CoATs (PDB entry 2oas; Northeast Structural Genomics Consortium, unpublished work).
The M31G RipA structure is very similar to that described for the closed CoA-bound complexes of 4-HB-CoAT from C. aminobutyricum (Macieira et al., 2012 ▶) and SCACT from A. aceti (Mullins & Kappock, 2012 ▶). The CoA substrates in all three structures superimpose in the same extended orientation along the closed CoA pocket from the C-terminal side of the catalytic glutamate (Fig. 8 ▶ a). While all three structures feature a similar closed G(V/I)G loop, M31G RipA Phe85 requires an additional movement to assume a fully closed position, while the corresponding phenylalanines in both 4-HB-CoAT and SCACT have no alternate conformations. For the fully closed RipA conformation, the distance from the CZ C atom of Phe85 to the CG2 C atom of Val227 is 3.8 Å, reminiscent of the distances of 2.9 and 4.7 Å in C. aminobutyricum 4-HB-CoAT (Macieira et al., 2012 ▶) and A. aceti SCACT (Mullins & Kappock, 2012 ▶), respectively, suggesting that the compensatory Phe85 movement observed in the M31G RipA structure is necessary for RipA function. Additionally, the M31G RipA structure has electron density for the glutamyl-anhydride as observed for A. aceti SCACT, while C. aminobutyricum 4-HB-CoAT has water molecules in place of the anhydride. The M31G RipA structure also has distinct CoA contacts, with interactions from residues Arg200 and Arg378 pinching the CoA phosphodianion (Table 3 ▶), which is not observed in the RipA homologs.
Figure 8.
Superimposition of two distinct protein-bound CoA conformations. (a) Stick representation of M31G RipA with the catalytic glutamate shown in magenta and the CoA shown in white with the superimposed A. aceti SCACT (PDB entry 4eu6; Mullins & Kappock, 2012 ▶) and C. aminobutyricum 4-HBCoAT (PDB entry 3qdq; Macieira et al., 2012 ▶) CoAs shown in gray and wheat, respectively. (b) Stick representation of F60M RipA with the catalytic glutamate shown in teal and the CoA shown in white with the superimposed M. tuberculosis malate synthase G (PDB entry 2gq3; Anstrom & Remington, 2006 ▶) and M. tuberculosis FadB (PDB entry 4b3i; Venkatesan & Wierenga, 2013 ▶) CoAs shown in gray and wheat, respectively. O, N, S and P atoms are colored red, blue, yellow and orange, respectively.
The F60M RipA structure with the covalently bound CoA-thioester intermediate shares the same open conformation as the S. oneidensis 4-HBCoAT (PDB entry 2oas) and A. aceti SCACT (Mullins & Kappock, 2012 ▶) structures, albeit possessing a unique J-like CoA conformation among CoATs. However, a similar J-like CoA conformation has been reported in Mycobacterium tuberculosis malate synthase G (PDB entry 2gq3; Anstrom & Remington, 2006 ▶) and M. tuberculosis FadB (PDB entry 4b3i; Venkatesan & Wierenga, 2013 ▶) (Fig. 8 ▶ b), where equivalent intramolecular hydrogen bonds are present as observed in the F60M RipA-bound CoA (Fig. 6 ▶ b). This variability of CoA conformations within RipA is most likely to be a consequence of the flexibility of CoA itself as well as the open-state conformation within CoATs.
The proposed family I CoAT acyl-binding site is partially conserved. RipA Phe85 is invariant in the other homologs, while RipA Phe60 and Phe113 are substituted with hydrophobic residues in C. aminobutyricum 4-HB-CoAT (Macieira et al., 2009 ▶; Fig. 1 ▶ c). However, RipA Met31 is replaced with His31 in C. aminobutyricum 4-HB-CoAT, which may account for the difference in substrate preference (Fig. 9 ▶). Additionally, a possible co-substrate pocket has been proposed in C. aminobutyricum 4-HB-CoAT which consists of a conserved glutamine (Gln224) that forms an electrostatic interaction with an adjacent asparagine (Asn321) (Macieira et al., 2009 ▶). Notably, the Q224S RipA mutant did not show any activity (data not shown), although a structural role for this conserved glutamine cannot be ruled out.
Figure 9.
Overlay of the putative acyl-binding pockets from WT RipA modeled with butyryl-CoA and C. aminobutyricum 4-HB-CoAT. The corresponding WT RipA and 4-HB-CoAT residues are represented as wheat and dark green sticks, respectively. The molecular surface of WT-RipA is shown in gray with the three conserved phenylalanines (Phe60, Phe85 and Phe113) from the proposed acyl-group binding site and the possible co-substrate pocket highlighted by a red dashed line.
4.2. CoA transferase mechanism
Since the different observed conformations in the WT RipA and mutant structures correspond to different intermediates in the general reaction mechanism described for family I CoATs (Buckel et al., 1981 ▶; Fig. 2 ▶), mechanistic insights can be obtained. To start, the presence of an open state observed in the CoA-thioester intermediate and a closed state observed for the anhydride intermediate suggests that the CoA binding pocket must open and close at different steps of the reaction. These structural dynamics were previously proposed in early enzymatic work on the family I CoAT Sus scrofa succinyl-CoA:3-ketoacid transferase (SCOT), in which the conformational change was suggested to be the result of the utilization of noncovalent binding energy of CoA to accelerate the rate-limiting acyl transfer by compressing the substrate thioester tightly against the catalytic glutamate (White & Jencks, 1976 ▶; White et al., 1976 ▶), with a large portion of this energy attributed to the interaction of the noncatalytic pantetheine and nucleotide portions of CoA (Fierke & Jencks, 1986 ▶). This conformational change has subsequently also been observed in two recent CoAT structures, suggesting that the mechanism is conserved in family I CoATs (Macieira et al., 2012 ▶; Mullins & Kappock, 2012 ▶). The conserved G(V/I)G loop therefore plays an important part in this mechanism as it appears to be responsible for this observed conformational change and serves two functional purposes: (i) to form the rate-enhancing electrostatic and hydrophobic interactions with the non-reactive pantetheine moiety of CoA and (ii) to close the catalytic site, extruding it from solvent to allow stabilization of the intermediates. The use of a flexible loop in the active site to control enzyme activity seems to be a conserved mechanism for all family I CoATs (Mullins & Kappock, 2012 ▶; Macieira et al., 2012 ▶) and other phosphodianion moiety-containing enzymes such as orotidine 5′-monophosphate decarboxylase, triosephosphate isomerase and α-glycerol phosphate dehydrogenase (Malabanan et al., 2010 ▶; Amyes & Richard, 2013 ▶).
The CoA transferase mechanism for RipA starts in the open state (WT RipA structure; Figs. 3 ▶ a and 5 ▶ a) ready to accept the CoA substrate. Residue Phe85, however, is positioned such that the acyl-group binding-site access is blocked, as suggested by previous butyryl-CoA docking (Torres et al., 2011 ▶). In the subsequent step, rotation of Phe85 increases the volume of the acyl-binding hydrophobic pocket (E61V RipA structure; Figs. 4 ▶ a and 5 ▶ b), a phenomenon also observed in MD simulations of WT RipA (Torres et al., 2011 ▶), and thus primes the active site for acyl-CoA binding. Interestingly, although this phenylalanine is a conserved aromatic residue in several CoATs, the flipped-in conformation is only observed in WT RipA; however, a similar pocket-activating/inactivating phenylalanine conformational switch is observed in the protein kinase DGF motif (Jura et al., 2011 ▶; Kornev et al., 2006 ▶), suggesting that a similar role is possible for RipA. Following the opening of the acyl-group binding pocket, the positive electric isopotential surface positioned above the active site of RipA attracts the negatively charged acyl-CoA into the binding cleft (Torres et al., 2011 ▶) and both the conserved G(V/I)G loop and Phe85 move to close the active site, positioning the acyl-CoA carbonyl for attack by the catalytic glutamate, forming the first anhydride intermediate (M31G RipA structure; Figs. 4 ▶ b and 5 ▶ c). The CoA-thiolate then attacks the carbonyl of the anhydride, forming the CoA-thioester intermediate, and the enzyme returns to the open state for the concurrent release/acceptance of the donor/acceptor acyl-acid in the open state (F60M RipA structure; Figs. 4 ▶ c and 5 ▶ d). The new acyl-acid follows the reverse reaction, forming the second anhydride intermediate again in the closed state (M31G RipA structure) to yield a newly formed acyl-CoA, which is eventually released in the open state.
4.3. Mutational effects on transferase activity
The RipA mutational studies revealed expected as well as unexpected transferase activity results. As predicted, activity was lost when Glu249, a conserved catalytic residue in all family I CoATs, was mutated to an alanine or aspartate, suggesting that the position and charge of the glutamate carboxylate is important for activity and is consistent with mutational studies of other family I CoATs (Mullins & Kappock, 2012 ▶; Macieira et al., 2009 ▶). Structural studies with modeled CoA (Torres et al., 2011 ▶) also suggest that CoA is positioned such that only a glutamate can attack the carboxylate of the acyl-CoA (Fig. 7 ▶). Furthermore, loss of function for mutants of Val227 (V227G and V227W), a residue within the conserved G(V/I)G loop required for gating, suggests that loop flexibility and residue size are essential for the catalytic cycle of RipA. The bulkier V227W RipA mutant is most likely to block access of CoA to the catalytic glutamate, preventing turnover, while the smaller V227G mutation together with the acyl-group binding pocket Phe85 probably loses the ability to form a closed conformation to protect intermediates in the active site during catalysis. Interestingly, transferase activity for acyl-binding site mutants was either abolished or increased in comparison to WT RipA. The lack of activity for several mutants (i.e. M31H, F60V, F85H, F85Y, F113L and F113Q) suggests that these residues provide a favorable electrostatic environment for acyl substrate binding or aid in the formation of a closed active site. In contrast, the increased activity of E61V, M31G and F60M RipA suggests these residues play a role in the acyl-transfer reaction, although the exact mechanism is not understood.
5. Conclusion
Although further research is needed to understand the effects of mutations at the CoA acyl-group binding site, this study has shown that there are indeed distinct conformational changes during the progression of the CoAT mechanism. In addition, it has been shown that cooperative movements between the conserved residues Val227 and Phe85 are essential for function and that the acyl-group binding-pocket residues play a role in RipA activity. As more information is unraveled on how these CoATs function and bind specific CoA substrates, more specific inhibitors for CoATs can be designed. This is especially noteworthy as RipA has been suggested as a possible drug target for Y. pestis.
Supplementary Material
PDB reference: Y. pestis RipA, E61V mutant, 4n8h
PDB reference: M31G mutant, 4n8i
PDB reference: F60M mutant, 4n8j
PDB reference: E249A mutant, 4n8k
PDB reference: E249D mutant, 4n8l
Acknowledgments
This study was funded by the National Institutes of Health (AI-65359, PI Alan Barbour, subaward to CWG). We thank the Advanced Light Source (ALS) at Berkeley National Laboratories for invaluable help in data collection. The Advanced Light Source is supported by the Director, Office of Basic Energy Sciences, of the US Department of Energy under Contract No. DE-AC02-05CH11231. We would also like to thank all of the staff at Stanford Synchrotron Radiation Lightsource (SSRL) for their invaluable help in data collection. Portions of this research were carried out at the SSRL, a Directorate of SLAC National Accelerator Laboratory and an Office of Science User Facility operated for the US Department of Energy Office of Science by Stanford University. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS or NIH.
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Amyes, T. L. & Richard, J. P. (2013). Biochemistry, 52, 2021–2023 [DOI] [PMC free article] [PubMed]
- Anstrom, D. M. & Remington, S. J. (2006). Protein Sci. 15, 2002–2007. [DOI] [PMC free article] [PubMed]
- Bateman, K. S., Brownie, E. R., Wolodko, W. T. & Fraser, M. E. (2002). Biochemistry, 41, 14455–14462. [DOI] [PubMed]
- Bliska, J. B. & Casadevall, A. (2009). Nature Rev. Microbiol. 7, 165–171. [DOI] [PubMed]
- Buckel, W., Dorn, U. & Semmler, R. (1981). Eur. J. Biochem. 118, 315–321. [DOI] [PubMed]
- Cavanaugh, D. C. & Randall, R. (1959). J. Immunol. 83, 348–363. [PubMed]
- Charnetzky, W. T. & Shuford, W. W. (1985). Infect. Immun. 47, 234–241. [DOI] [PMC free article] [PubMed]
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- DeLano, W. L. (2002). PyMOL. http://www.pymol.org.
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Fierke, C. A. & Jencks, W. P. (1986). J. Biol. Chem. 261, 7603–7606. [PubMed]
- Fraser, M. E., Hayakawa, K. & Brown, W. D. (2010). Biochemistry, 49, 10319–10328. [DOI] [PubMed]
- Heider, J. (2001). FEBS Lett. 509, 345–349. [DOI] [PubMed]
- Heifets, A. & Lilien, R. H. (2010). J. Mol. Graph. Model. 29, 93–101. [DOI] [PubMed]
- Janssen, W. A. & Surgalla, M. J. (1969). Science, 163, 950–952. [DOI] [PubMed]
- Jura, N., Zhang, X., Endres, N. F., Seeliger, M. A., Schindler, T. & Kuriyan, J. (2011). Mol. Cell, 42, 9–22. [DOI] [PMC free article] [PubMed]
- Kornev, A. P., Haste, N. M., Taylor, S. S. & Eyck, L. F. (2006). Proc. Natl Acad. Sci. USA, 103, 17783–17788. [DOI] [PMC free article] [PubMed]
- Macieira, S., Zhang, J., Buckel, W. & Messerschmidt, A. (2012). Arch. Microbiol. 194, 157–166. [DOI] [PubMed]
- Macieira, S., Zhang, J., Velarde, M., Buckel, W. & Messerschmidt, A. (2009). Biol. Chem. 390, 1251–1263. [DOI] [PubMed]
- Malabanan, M. M., Amyes, T. L. & Richard, J. P. (2010). Curr. Opin. Struct. Biol. 20, 702–710. [DOI] [PMC free article] [PubMed]
- Mullins, E. A. & Kappock, T. J. (2012). Biochemistry, 51, 8422–8434. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Park, J.-S., Lee, E.-J., Lee, J.-C., Kim, W.-K. & Kim, H.-S. (2007). Int. Immunopharmacol. 7, 70–77. [DOI] [PubMed]
- Perry, R. D., Balbo, P. B., Jones, H. A., Fetherston, J. D. & DeMoll, E. (1999). Microbiology, 145, 1181–1190. [DOI] [PubMed]
- Perry, R. D. & Fetherston, J. D. (1997). Clin. Microbiol. Rev. 10, 35–66. [DOI] [PMC free article] [PubMed]
- Pujol, C., Grabenstein, J. P., Perry, R. D. & Bliska, J. B. (2005). Proc. Natl Acad. Sci. USA, 102, 12909–12914. [DOI] [PMC free article] [PubMed]
- Rangarajan, E. S., Li, Y., Ajamian, E., Iannuzzi, P., Kernaghan, S. D., Fraser, M. E., Cygler, M. & Matte, A. (2005). J. Biol. Chem. 280, 42919–42928. [DOI] [PubMed]
- Solomon, F. & Jencks, W. P. (1969). J. Biol. Chem. 244, 1079–1081. [PubMed]
- Stempelj, M., Kedinger, M., Augenlicht, L. & Klampfer, L. (2007). J. Biol. Chem. 282, 9797–9804. [DOI] [PubMed]
- Straley, S. C. & Harmon, P. A. (1984). Infect. Immun. 45, 649–654. [DOI] [PMC free article] [PubMed]
- Torres, R., Swift, R. V., Chim, N., Wheatley, N., Lan, B., Atwood, B. R., Pujol, C., Sankaran, B., Bliska, J. B., Amaro, R. E. & Goulding, C. W. (2011). PLoS One, 6, e25084. [DOI] [PMC free article] [PubMed]
- Venkatesan, R. & Wierenga, R. K. (2013). ACS Chem Biol. 8, 1063–1073. [DOI] [PubMed]
- White, H. & Jencks, W. P. (1976). J. Biol. Chem. 251, 1688–1699. [PubMed]
- White, H., Solomon, F. & Jencks, W. P. (1976). J. Biol. Chem. 251, 1700–1707. [PubMed]
- Williamson, E. D. & Oyston, P. C. (2013). Clin. Exp. Immunol. 172, 1–8. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: Y. pestis RipA, E61V mutant, 4n8h
PDB reference: M31G mutant, 4n8i
PDB reference: F60M mutant, 4n8j
PDB reference: E249A mutant, 4n8k
PDB reference: E249D mutant, 4n8l