Abstract
The nucleus is a complex organelle containing numerous highly dynamic, structurally stable domains and bodies, harboring functions that have only begun to be defined. However, the molecular mechanisms for their formation are still poorly understood. Recently it has been shown that a nuclear body can form de novo by self-organization. But little is known regarding what triggers the formation of a nuclear body and how subsequent assembly steps are orchestrated. Nuclear bodies are frequently associated with specific active gene loci that directly contribute to their formation. Both coding and noncoding RNAs can initiate the assembly of nuclear bodies with which they are physiologically associated. Thus, the formation of nuclear bodies occurs via recruitment and consequent accumulation of resident proteins in the nuclear bodies by nucleating RNA acting as a seeder. In this chapter I describe how to set up an experimental cell system to probe de novo biogenesis of a nuclear body by nucleating RNA and nuclear body components tethered on chromatin.
Keywords: Nuclear body, Cajal body, RNA transcription, Gene expression, Nuclear organization
1 Introduction
The dynamic spatial organization of the cell nucleus plays a primary role in genome function and maintenance [1, 2]. Within the nuclear environment, which is characterized by a lack of defining membranes, chromosomes occupy specific nonrandom territories. These chromosome territories harbor a variety of distinct nuclear bodies (NBs) involved in various aspects of genome activity, regulation and maintenance form in their closest association [3, 4]. NBs are highly dynamic structures, of which their components show rapid turnover with the surrounding nucleoplasm. Their structural integrity is mediated by transient low affinity protein–protein and protein–RNA interactions [5, 6]. Importantly, NBs do not assemble as preformed structural entities but rather emerge as a direct reflection of specific activities associated with gene expression and genome maintenance [7]. The molecular mechanisms of how NBs form and maintain their structural integrity are still poorly understood. More specifically, little is known regarding how an NB formation is initiated and how following building steps in an NB assembly is orchestrated.
Recently two opposite models of NB formation have been proposed [7]: (a) ordered assembly model in which an NB may form by a tightly controlled series of sequential building steps with very limited number of molecules which can act as an initiator of the assembly and, alternatively, (b) a stochastic self-organization assembly model in which an NB is built by the random interactions of individual components without a strict hierarchical order of assembly. In this model many components of an NB can initiate the formation. These two models were experimentally validated on the formation of the Cajal body (CB) [8], which is one of the major NB involved in biogenesis and recycling of many small nuclear RNAs [5, 9, 10]. By tethering individual CB components fused to the Escherichia coli Lac repressor (LacI) to a Lac operator (LacO)-repeat array gene locus in living cells, it was demonstrated that any CB component can initiate the formation of the entire NB. This finding provided conclusive evidence that an NB is formed by self-organization, but it does not address which initiation event triggers the physiological formation of a CB [8].
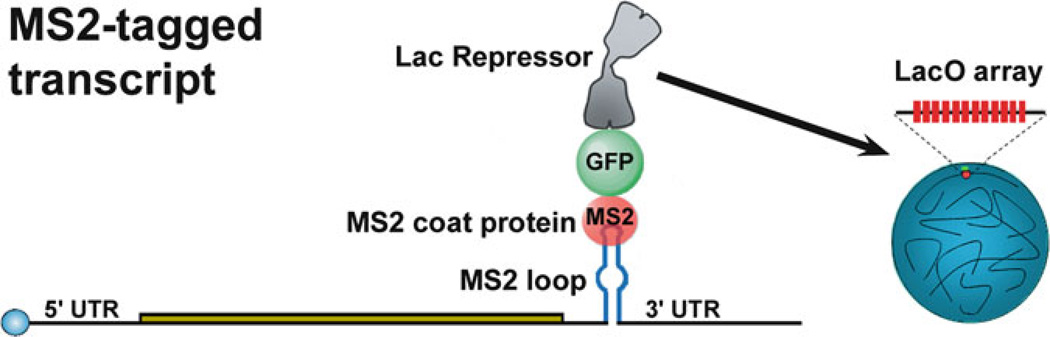
A critical step in NB formation is an initial nucleation event which serves to immobilize critical components to provide a seeding platform to trigger recruitment and retention of additional building blocks. Importantly, many NBs are formed at sites of active transcription at which their activities contribute to their formation. To investigate whether specific functionally related RNAs are sufficient to form major NBs, we developed an experimental cell system in which a specific RNA is tagged with a bacteriophage MS2 stem–loop sequence. When the MS2-tagged transcript is co-expressed with the LacI-NLS-GFP-MS2 coat protein, which selectively binds the MS2 loop, it is targeted to and accumulates on the 256 repeats of the LacO-binding sequence within the LacO array (Figs. 1 and 2a).
Fig. 1.
Schematic representation of RNA-tethering system used to probe the contribution of specific RNAs in the formation of nuclear bodies. The hypothetical transcript is tagged with one bacteriophage MS2 stem loop in the 3′ UTR. The monomeric LacI-NLS-GFP-MS2 coat protein selectively binds the MS2 loop and targets the MS2-tagged transcripts to the LacO array where they accumulate
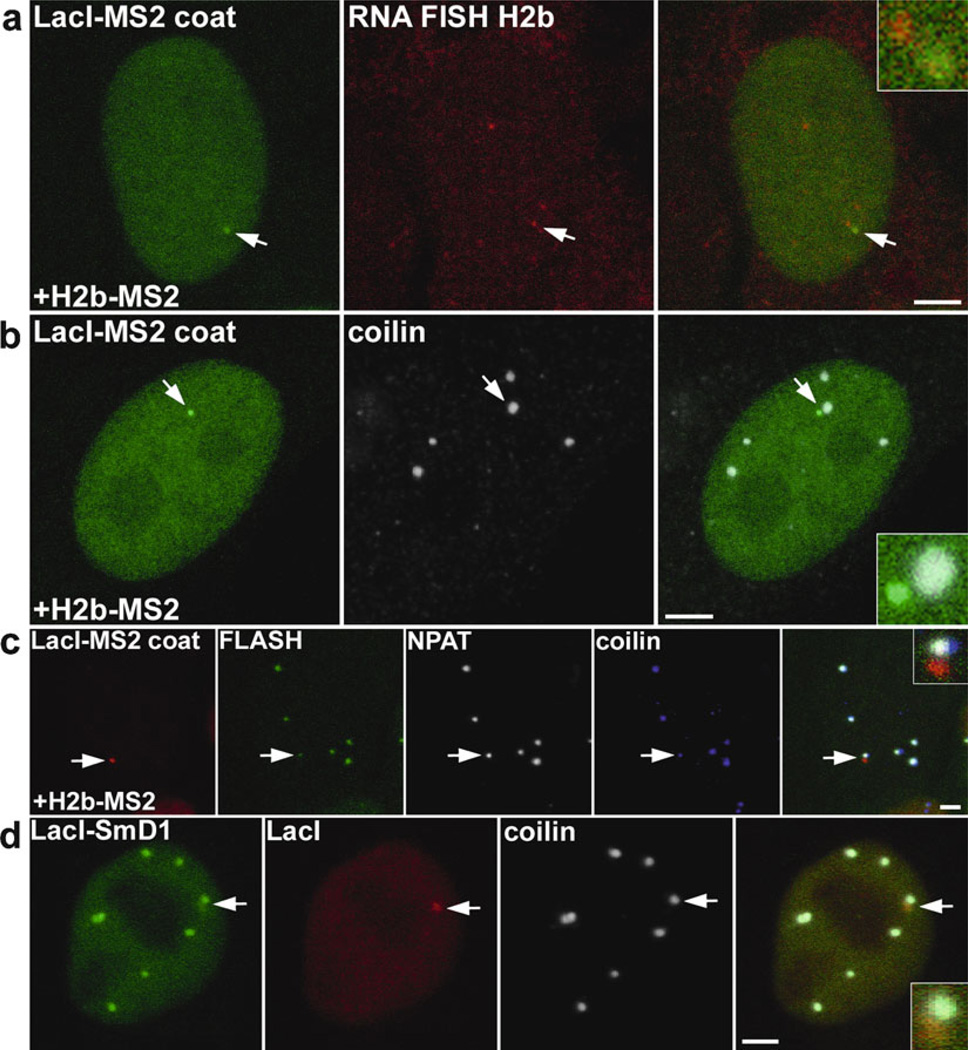
Fig. 2.
Immobilization of specific RNAs to chromatin leads to the formation of nuclear bodies. (a) Histone H2b transcripts tagged with the MS2 loop were transiently co-expressed with the monomeric LacI-NLS-GFP-MS2 coat protein in HeLa cell containing a stably integrated LacO array. Tethering of H2b-MS2 transcripts on the LacO array was detected by RNA FISH using a specific DNA FISH probe against H2b pre-mRNA (arrow indicates higher magnification shown in inset). (b) Tethering of H2b-MS2 pre-mRNA to the LacO array in HeLa cells leads to de novo formation of a Cajal body (CB) detected by an antibody against a CB marker protein coilin (arrow; shown in inset). (c) Immobilization of H2b-MS2 pre-mRNA on the LacO array in HeLa cells leads to de novo formation of two nuclear bodies, a CB with a physically associated histone locus body (HLB). HLB is detected by GFP-FLASH (green) and with an antibody against NPAT (white), two HLB marker proteins, and histone pre- mRNA processing factors. CB is detected with anti-coilin antibody (blue) (arrow, inset). (d) Tethering of a single component of a CB, a core protein of the Sm ring of spliceosomal U snRNPs, GFP-LacI-SmD1, on the LacO array (detected by mCherry-LacI) in HeLa cell leads to de novo CB formation. The CB is detected with an anti- coilin antibody (arrow, inset). Scale bar = 2 µm
This system has been used to tether coding histone H2b or β-globin pre-mRNAs, or the noncoding NEAT1 or SatIII RNAs, to a LacO array in living cells, in order to generate a specific NB. This approach led to the de novo nucleation of a histone locus body with associated CBs (Fig. 2b, c), nuclear speckle, paraspeckle, and nuclear stress bodies, respectively, at these tethering sites [11]. Additional evidence was provided by the visualization of the de novo formation of paraspeckles by inducing transcription of noncoding RNA Men ε/β (NEAT1 in human) [12]. Overall, these data indicate that several types of coding and noncoding RNAs can initiate the formation of some NBs with which they are physiologically associated and function as structural elements and as a nucleator of NBs. Expanding the use of these approaches will undoubtedly uncover the total composition of individual NBs, which will lead to the understanding of an entire interactive network of NB components and physiological processes responsible for their formation and ultimately their functions.
In this chapter I explain the standard requirements and overall technical steps for developing an experimental cell system to probe the ability of specific RNAs and individual NB components to nucleate a NB. This chapter will describe (a) subcloning of coding and noncoding RNAs that specifically associate with NBs and tagging them with the MS2 sequence, (b) fusion of specific components of NBs with GFP-LacI, (c) tethering of MS2-tagged RNAs on a genomic LacO array and their detection by RNA FISH, (d) tethering of MS2-tagged RNAs and LacI-fused NB components to the array with the detection of de novo NB formation, and (e) live-cell imaging of de novo NB formation initiated by an RNA molecule.
2 Materials
2.1 Cloning of Coding and Noncoding RNAs Which Specifically Associate with NBs
Genomic DNA extraction kit.
pcDNA3.1 vector (Invitrogen) or other convenient PCR cloning vector for PCR subcloning.
Mammalian expression vector such as pcDNA3.3-TOPO (Invitrogen) driven by a modified enhanced CMV promoter.
Restriction enzymes.
T4 DNA ligase.
Competent strain(s) of Escherichia coli for DNA transformation.
GFP-LacI-NLS and mCherry-LacI-NLS sucloned in the vector derived from pEGFP-C1 (Clontech). These plasmids can be obtained from Addgene.
Site-directed mutagenesis kit.
2.2 Cell Culture
Dulbecco’s Modified Eagle’s Medium (DMEM) for cell culture.
Fetal bovine serum (FBS).
Trypsin for releasing attached cells from culture plates.
PBS (phosphate-buffered saline) for washing cells.
Adherent cell line of preference with a stably integrated LacO array with 256 repeats. It is highly recommended to use human-transformed cell lines which can be obtained commercially from various sources such as ATCC.
Lipofectamine 2000 (Invitrogen) and Opti-MEM Reduced Serum Medium (Invitrogen).
0.1 M IPTG stock solution in sterile H2O.
2.3 RNA FISH
PBS for washing cells.
4 % paraformaldehyde in PBS: aliquot and freeze at −20 °C for later use.
0.2 % Triton X-100 in PBS: 40 µl/20 ml PBS. Store at 4 °C for later use.
20× Saline-sodium citrate buffer (SSC), pH 7.0.
Formamide and deionized formamide.
Yeast tRNA (Sigma): 10 mg/ml stock solution and freeze at −20 °C.
50 % dextran sulfate in sterile H2O: aliquot and store at −20 °C for later use.
Ice-cold absolute ethanol.
3 M sodium acetate (pH 5.2).
Rubber cement.
Hybridization moisture chamber with paper towels soaked with 2× SSC in bottom.
Fluorescently labeled DNA FISH probe against the target gene sequence. The plasmid with the gene of interest should be labeled by nick translation with Cy3 (or other convenient fluorophores such as Alexa) to produce a FISH probe of size ~200 bp. Use 100 ng of probe per a 22 × 22 mm square coverslip.
Hybridization solution: 200 µl of 20× SSC, 200 µl of 50 % dextran sulfate, 200 µl of 0.1 M Tris–HCl (pH 7.2), and 700 µl of nuclease-free H2O. It is possible to aliquot and freeze (−20 °C) the hybridization solution for later use.
Mounting medium with anti-fading agents (ProLong Gold or Vectashield) either with or without DAPI for DNA staining.
2.4 Microscopy
Fluorescent microscope of choice with either 63× or 100× Plan oil objectives. It is recommended to use a confocal microscope and collect vertical z-stacks to visualize de novo NB formation because the depth of field along z-axis is frequently important for NB detection on the chromatin locus. Since the nucleation of an NB is a very light-sensitive event, it is suggested to use a high-speed microscope equipped with a high-sensitivity CCD camera to reduce exposure time during live-cell imaging.
Microscope sample incubation chamber for live-cell imaging to maintain cells at 37 °C and 5 % CO2.
Live-cell imaging chambers with thin glass bottom.
DMEM buffered with 15 mM HEPES without phenol red.
Image analysis software such as Metamorph (Molecular Devices), Image J (NIH), or Imaris (Bitplane).
3 Methods
3.1 Subcloning of Coding and Noncoding RNAs That Specifically Associate with NBs and Their Tagging with the MS2 Sequence
Isolate the gene of interest from genomic DNA derived from cells of interest using the genomic DNA extraction kit or use commercially available genomic libraries. Amplify the gene by PCR and subsequently subclone it into a convenient PCR vector (e.g., pcDNA3.1) using a PCR subcloning kit. The sequence needs to be verified by sequencing and compared with genome information available in genome databases.
Analyze for predicted RNA secondary structures in the transcript transcribed from the gene of interest by Mfold (http://mfold.rna.albany.edu/?q=mfold/RNA-Folding-Form). This computational approach will identify suitable predicted RNA stem–loop structure(s) for the introduction of one bacteriophage MS2 loop sequence into the gene of interest. However, it is always essential to analyze and critically evaluate positions of important functional domains present in the gene such as splice sites, cleavage sites, regulatory regions, and binding sites and to avoid using them for the MS2 loop insertion (see Note 1).
By site-directed mutagenesis, introduce one bacteriophage MS2 stem–loop sequence (CGTACACCATCAGGGTACG) as an extension of a predicted RNA stem–loop structure in the transcript as predicted by Mfold. The correct insertion of the MS2 loop must be verified by sequencing and the whole gene of interest should be re-sequenced.
To determine which sequence and structural elements of RNA of interest are necessary and sufficient for de novo nucleation of an NB, a series of processing and deletion mutants derived from the original MS2 loop-containing wild-type transcript should be created at this stage by site-directed mutagenesis using the same technical approach as in step 3. Particularly, more stable (e.g., non-cleavable and unspliced) mutants of the transcript of interest should be generated and their efficiency to nucleate a de novo NB should be quantitatively compared to the wild-type-MS2 transcript.
Insert the wild-type MS2-tagged transcript and its processing and/or deletion mutants into a suitable mammalian expression vector such as pcDNA3.3-TOPO or others. Initially, a strong constitutive viral promoter such as the viral CMV should be used to achieve high expression levels. High levels of ectopic expression of RNA-MS2 provide functional evidence that tethering on chromatin leads to de novo NB formation. MS2-tagged transcripts can be expressed at a moderate level using an endogenous promoter to mimic endogenous levels of the native RNA of interest (see Note 2).
3.2 Fusion of NB Components with LacI
An alternative way to nucleate a de novo NB is to tether NB components fused with the LacI on the LacO array. This mimics a natural seeding event and thus initiates the assembly process of NB formation (Fig. 2d).
Clone or obtain a cDNA of an NB component of interest from various commercial sources (e.g., OriGene or Mammalian Gene Collection).
Obtain the GFP-LacI-NLS subcloned as an XhoI-EcoRI fragment into the pEGFP-C1 designed for fusion with proteins of interest from Addgene.
Using PCR add convenient available restriction sites present at the multicloning site of the pEGFP-C1 downstream of EcoRI, to the 5′ and 3′ ends of the NB component cDNA, for fusion with GFP-LacI-NLS. The prepared cDNA needs to have a stop codon to terminate the translation of the fusion product.
Insert the cDNA of the NB component into the GFP-LacI-NLS using the added restriction sites.
Verify the inserted clone by sequencing.
3.3 Cell Culture and Transfection
For preparation of cells before transfection, plate the cells containing a stably integrated LacO array (see Note 3) on 22 mm glass square coverslips, in medium supplemented with 10 % serum and 1 % glutamine without antibiotics, in a 6-well plate at 37 °C. Cells should reach 30–50 % confluency before transfection. Transfection efficiency is low when cell density is too high (see Note 4).
To transfect the cells on the following day, adjust the volume of medium in each well to 2 ml. Co-transfect the cells with plasmids encoding the MS2-tagged transcript and monomeric LacI-NLS-GFP-MS2 coat protein using Lipofectamine 2000. Briefly, dilute the plasmids (~1 µg in total) in 50 µl of Opti-MEM Reduced Serum Medium without serum and mix gently. The plasmid ratio of MS2-tagged transcript to LacI-NLS-GFP-MS2 coat protein should be 1:2 (see Note 5). In the second test tube, dilute 2 µl of Lipofectamine 2000 in 50 µl of Opti-MEM Reduced Serum Medium without serum, mix gently, and incubate for 5 min at RT. Then combine the diluted DNA with the diluted Lipofectamine 2000. Mix gently and incubate for 20 min at RT. Slowly add 100 µl of transfection complexes to each well containing cells and medium. Mix gently by rocking the plate back and forth. Incubate cells at 37 °C for 18–24 h.
Change the medium the next morning.
Fix cells for immunolocalization or visualize them live under the microscope 18–24 h after transfection.
3.4 Detection of Tethered MS2-Tagged RNA on the LacO Array by RNA FISH
To test whether MS2-tagged transcripts co-expressed with the LacI-NLS-GFP-MS2 coat protein are efficiently targeted and accumulated on the LacO array, perform RNA fluorescence in situ hybridization (FISH) using a specific DNA FISH probe against the MS2-tagged RNA (Fig. 2a) (see Note 6).
Wash cells briefly with PBS and fix with 4 % paraformaldehyde in PBS for 15 min at RT.
Wash cells three times with PBS at RT.
Permeabilize cells with 0.2 % Triton X-100 in PBS on ice for 5 min.
Wash cells twice with PBS and finally once with 2× SSC for 5 min at RT.
Prepare the hybridization solution.
Mix 100 ng of the DNA FISH probe against the target RNA (labeled with Cy3 by nick translation) and 40 µg yeast tRNA per coverslip and precipitate: 2 µl of probe, 4 µl of tRNA (10 mg/ml stock), 39 µl of sterile H2O, 5 µl of 3 M sodium acetate (pH 5.2), 100 µl of ice-cold absolute ethanol. Put it in a −70 °C freezer for at least 20 min. Spin at maximum speed for 20 min at 4 °C. Remove the supernatant. The pellet may be clear so be very careful not to remove the pellet with the supernatant. Air dry the pellet and reconstitute in 6 µl of deionized formamide.
Denaturate the DNA FISH probe by incubating the tube for 10 min at 80 °C. Vortex several times during heating. Chill in water-ice slurry immediately for 5 min.
Add 18 µl of the hybridization solution from the step 5 at RT, mix it, and spin it for 15 s at RT.
Use a petri dish to set up a moistened chamber. Place paper towels moistened with 2× SSC in the bottom of a plate.
Place 24 µl of the hybridization mixture from the step 8 onto each coverslip, and seal coverslips to microscopic slides by rubber cement and hybridize in a moistened chamber overnight at 37 °C.
30 min before washing, pre-warm the 2× SSC–50 % formamide to 37 °C.
Open the hybridization chamber, remove the rubber cement, and transfer the coverslips back to a 6-well plate with pre-warmed 2× SSC–50 % formamide.
Rinse coverslips twice for 30 min with 2× SSC–50 % formamide at 37 °C.
Rinse coveslips for 5 min with PBS at RT.
At this point the RNA FISH protocol is combined with indirect immunofluorescence to specifically visualize an NB or nuclear compartment functionally associated with immobilized MS2-tagged RNA. Incubate cells on coverslips with a primary antibody against a marker protein of the NB of interest, for 1 h at RT.
Wash cells three times with PBS at RT.
Incubate cells with an appropriate secondary antibody conjugated with Cy5 (or different convenient fluorophores such as Alexa) for 1 h at RT.
Wash cells three times with PBS at RT.
Mount coverslips in mounting medium with anti-fading agents either with or without DAPI. Leave slides overnight at 4 °C covered with aluminum foil to allow the medium to solidify.
Observe cells under a fluorescent microscope using either 63× or 100× Plan oil objectives with high numerical apertures. Check the localization of the LacO array as a single bright fluorescent spot in the nucleus with associated RNA FISH signal. Additional RNA FISH signals visible as bright foci, observed at the endogenous transcription sites of the gene of interest in the nucleoplasm, should be located and used as a positive control. The intensity of the RNA FISH signal at typical endogenous transcription sites should be approximately the same as the signal on the LacO array (Fig. 2a).
3.5 De Novo Nucleation of an NB by RNA or by Tethered NB Components
To test whether MS2-tagged transcripts or NB components immobilized on the LacO array are sufficient to de novo nucleate an NB, perform indirect immunofluorescence using the antibodies against marker protein(s) of NB of interest. To distinguish the position of the LacO array from endogenous NBs, where NB proteins fused with GFP-LacI should also be located, the cells need to be cotransfected with mCherry-LacI-NLS which marks the array specifically (Fig. 2d).
Wash transfected cells briefly with PBS and fix them with 2 % paraformaldehyde in PBS for 10 min at RT.
Wash cells three times with PBS at RT.
Permeabilize cells with 0.2 % Triton X-100 in PBS on ice for 5 min.
Wash cells three times with PBS at RT.
Incubate cells on coverslips with a primary antibody against a marker protein of NB for 1 h at RT.
Wash cells three times with PBS at RT.
Incubate cells with an appropriate secondary antibody conjugated with Cy5 for 1 h at RT.
Wash three times cells with PBS at RT.
Mount coverslips in mounting medium with anti-fading agents either with or without DAPI. Leave slides overnight at 4 °C covered with aluminum foil to allow the medium to solidify.
Observe cells under a fluorescent microscope using either 63× or 100× Plan oil objectives. Check the position of the LacO array as a single bright fluorescent spot in the nucleus where MS2-tagged RNAs are tethered. To distinguish the position of the LacO array from endogenous NBs labeled with GFP-LacI-NB proteins, check the localization of mCherry-LacI-NLS as a single bright red spot. Tethered GFP-LacI-NB fusion proteins are also clearly detectable on the array (Fig. 2d). The newly formed NB detected by specific antibodies against NB-specific components should be visible in strong association with the LacO array (Fig. 2b–d) but it is frequently located in different focal planes. Therefore vertical z-sections around the array should be acquired and all images should be combined and projected as a maximum intensity projection (see Note 7).
3.6 Kinetics of De Novo NB Formation by RNA
3.6.1 Time-Course of the Dynamics of De Novo NB Formation
To establish the kinetics of de novo NB formation, treat cells with isopropyl-β-d-thiogalactopyranoside (IPTG) which selectively prevents binding of LacI-fusion proteins to the LacO array, which in turn leads to the disassembly of newly nucleated NB. When IPTG is washed out extensively from the cells, LacI-fusion binding is reestablished and de novo formation of a NB can be directly visualized and analyzed using time-lapse microscopy (see Note 8).
Co-transfect cells containing a stably integrated LacO array growing on glass coverslips with LacI-NLS-GFP-MS2 coat protein and MS2-tagged RNA with Lipofectamine 2000 for 18–24 h.
Treat cells with IPTG (5 mM) for 16 h to prevent binding of LacI-NLS-GFP-MS2 coat protein with bound MS2-tagged RNAs to the LacO array.
Washout IPTG very extensively with PBS. This is time zero in the time-course experiment.
Perform a time-course series by fixing cells at: 15 min, 30 min, 1 h, 1 h 30 min, 2 h, 3 h and 4 h intervals. These time intervals would cover the assembly dynamics of a typical NB.
Perform indirect immunofluorescence localization using antibodies which detect marker proteins of an NB or nuclear sub-domain of interest. Observe cells under a fluorescent microscope using either 63× or 100× Plan oil objectives. Check localization of the LacO array as a single bright green spot detected by the LacI-NLS-GFP-MS2 coat protein in the nucleus and analyze the assembly dynamics of an NB on the LacO array detected by a specific anti-NB antibody at each time point. LacI-NLS-GFP-MS2 coat protein with bound MS2-RNA accumulates on the LacO array in HeLa cells within ~30 min after IPTG withdrawal. A typical NB nucleated by MS2-tagged transcripts tethered on the LacO array likely forms within approximately 1–2 h [10].
3.6.2 Live-Cell 4D Imaging of De Novo Formation of a Nuclear Body Nucleated by MS2-Tagged RNAs on the LacO Array
Plate cells stably expressing the integrated LacO array in chambers for live-cell imaging.
Co-transfect cells with a GFP-tagged marker protein for an NB of interest with LacI-NLS-mCherry-MS2 coat protein and MS2-tagged RNA with Lipofectamine 2000 for 18–24 h. The plasmid ratio of MS2-tagged transcripts to LacI-NLS-mCherry-MS2 coat protein and to GFP-NB marker protein should be approximately 1:2:2.
Treat cells with IPTG (5 mM) for 16 h to prevent binding of LacI-NLS-mCherry-MS2 coat protein with tethered MS2-tagged RNA to the LacO array.
Wash out the IPTG extensively with PBS at RT.
Change medium to phenol red-free DMEM buffered with HEPES supplemented with 10 % serum.
Transfer the transfected cells in the live-cell imaging chamber to a high-speed fluorescent microscope equipped with a stage temperature-controlled incubation chamber to maintain cells at 37 °C and 5 % CO2. Search for transfected cells with a strong but not overexpressed GFP signal detecting NBs and a diffuse nucleoplasmic red signal of LacI-NLS-mCherry-MS2 coat protein. The LacO array is not visible as a single bright spot in the nucleus immediately after IPTG withdrawal. LacI-NLS-mCherry-MS2 coat protein with bound MS2-tagged transcripts accumulates on the LacO array in HeLa cells within ~30 min after IPTG withdrawal. Use a 63× Plan oil objective to simultaneously visualize several cells in the field. Adjust imaging conditions for high-speed image acquisition and low laser intensity to prevent photodamage of cells. During the search for optimal cells, minimize their exposure to reduce the bleaching of fluorescent signals.
Determine experimental conditions for time-lapse imaging including number of fields, number of vertical z-stacks, time intervals, and number of repeats. Recommended parameters are z = 300–400 nm and 5–10 min intervals for up to 5 h for a typical NB.
All collected images from the z-stack should be projected in three dimensions as combined maximum intensity projections for each time point using specific image analysis software.
Acknowledgements
The laboratory of Miroslav Dundr is supported by NIH R01GM090156 grant from NIGMS.
Footnotes
Position of MS2 loop insertion. The right choice of site for MS2 loop insertion is critical for its interaction with a monomeric MS2 coat protein fused with the LacI and should not interfere with the proper processing and overall functionality of the MS2-tagged RNAs. It is recommended to insert one MS2 loop as an extension of a predicted helix region, or an RNA secondary structure located preferably outside of the coding region at the 3′ UTR, or portions of the coding region that are not involved in pre-mRNA processing or cleavage.
Promoter. The selection of a promoter will significantly affect the ectopic expression levels of tethered components and MS2-tagged RNAs. Therefore, it is essential at first to express both components at higher levels to assure that they would be sufficiently immobilized on the LacO array. Initially express these components under the strong constitutive viral CMV promoter with the ratio between plasmids encoding tethering components and MS2-tagged RNAs to 2:1. Once the tethering system proves that an NB of interest can be formed de novo, an endogenous promoter which typically produces moderate levels of expression can be used. This approach would mimic the endogenous levels of RNAs and thus more likely the biological conditions of NB formation.
Stable cell lines expressing the LacO-repeat array. In order to have a reproducible cell system for quantitative probing abilities of NB formation by different RNAs, a cell line with a stably integrated LacO array must be used. When use of special knockout cell lines or primary cells is essential, these cells can be transiently transfected with the plasmid encoding 256 repeats of the LacO. However, it is important to check the number of LacO repeats in the plasmid when transformed in bacteria because they tend to recombine and shorten the number of repeats. To minimize recombination this plasmid should be propagated in Stbl2 competent cells specifically designed for unstable inserts and cultured at 30 °C.
Efficiency of transfection. It is important to initially achieve a high level of transfection when using a transient transfection system. Therefore, it is always beneficial to use cells which are easy to transfect. It is recommended to use transformed cells such as HeLa or U2OS cells which are efficiently transfected with Lipofectamine 2000. As an alternative, it is possible to use electroporation which provides high transfection efficiency but requires a large number of cells for preparation. When primary cells, which are difficult to transfect, must be used it is recommended to use the Amaxa nucleofection system (Lonza) which has cell-type-specific reagents and good transfection efficiency for primary cells. The quality of plasmid DNA usually plays a critical role in high transfection efficiency. It is recommended to use a MidiPrep purification system from Zymo, Qiagen, Promega, etc. for high-purity plasmid DNA preparation.
LacI-GFP-NLS-MS2 coat protein transfection. To achieve successful targeting and accumulation of RNA-MS2 on the array, an appropriate level of LacI-GFP-NLS-MS2 coat protein needs to be expressed in the cells. It is suggested to use MidiPrep plasmid quality for high transfection efficiency and to adjust the plasmid ratio of MS2-RNA to LacI-GFP-NLS-MS2 coat protein to 1:2.
Detection of MS2-RNA on the LacO array by RNA FISH. Initially, it is important to verify whether MS2-tagged RNAs efficiently accumulate on the LacO array. LacI-NLS-GFP-MS2 coat protein marks the position of array as a single bright green spot in the nucleus, and the RNA FISH signal detecting MS2-RNA should be clearly visible in closed association with the array. Presence of endogenous and ectopic transcription sites visible as clear bright spots in the nucleoplasm detected by the FISH probe should be used as a positive control, and their signal intensity should be roughly equal to the FISH signal present on the array. Since the bacteriophage MS2 stem–loop sequence is relatively short (19 nucleotides), it is not a recommended target for RNA FISH.
Detection of de novo formed NB of interest on the array. For proper detection of de novo NB nucleation on the LacO array, endogenous NBs in the nucleus must be clearly detectable and used as a positive control. Suitable antibodies against the marker proteins of the NB of interest should be used. De novo NBs nucleated on the array are frequently positioned in different focal planes than the array. Therefore, it is important to visualize cell nuclei vertically by z-stack sectioning and to project the 3D volume around the LacO array. It is necessary to evaluate whether ectopic expression of MS2-tagged RNAs and tethering components do not affect localization, number, size, shape, and topology of endogenous NBs and the behavior of their components. Furthermore, it is essential to check whether the de novo formed NBs are functional by several criteria such as size, shape, composition, and kinetic behavior of their components.
Live-cell imaging of a NB formation by RNA. The nucleation of an NB is a highly photosensitive event. Therefore, it is suggested to use a high-speed microscope such as a fast- spinning disc confocal microscope equipped with a high-sensitivity CCD camera to minimize cell exposure during live-cell visualization. The number of vertical z-sections depends on the size of NB of interest and cell shape. It is suggested to start with a number of z-sections which cover the entire cell nucleus from the top to the bottom and a step-width of 0.3–0.4 µm. The light intensity and exposure time should be extremely low to prevent photodamage of the cells and bleaching of the signals but still high enough for the signal detection over the background. An optimal signal-to-noise ratio should be adjusted by low laser intensity and fast image acquisition.
References
- 1.Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 2.Rajapakse I, Groudine M. On emerging nuclear order. J Cell Biol. 2011;192:711–721. doi: 10.1083/jcb.201010129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mao YS, Zhang B, Spector DL. Biogenesis and function of nuclear bodies. Trends Genet. 2011;27:295–306. doi: 10.1016/j.tig.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matera AG, Izaguire-Sierra M, Praveen K, Rajendra TK. Nuclear bodies: random aggregates of sticky proteins or crucibles of macromolecular assembly? Dev Cell. 2009;17:639–647. doi: 10.1016/j.devcel.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dundr M. Nuclear bodies: multifunctional companions of the genome. Curr Opin Cell Biol. 2012;24:415–422. doi: 10.1016/j.ceb.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caudron-Herger M, Rippe K. Nuclear architecture by RNA. Curr Opin Genet Dev. 2012;22:179–187. doi: 10.1016/j.gde.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 7.Dundr M, Misteli T. Biogenesis of nuclear bodies. Cold Spring Harb Perspect Biol. 2010;2:a000711. doi: 10.1101/cshperspect.a000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaiser TE, Intine RV, Dundr M. De novo formation of a subnuclear body. Science. 2008;322:1713–1717. doi: 10.1126/science.1165216. [DOI] [PubMed] [Google Scholar]
- 9.Nizami Z, Deryusheva S, Gall JG. The Cajal body and histone locus body. Cold Spring Harb Perspect Biol. 2010;2:a000653. doi: 10.1101/cshperspect.a000653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Machyna M, Heyn P, Neugebauer KM. Cajal bodies: where form meets function. WIREs RNA. 2012 doi: 10.1002/wrna.1139. [DOI] [PubMed] [Google Scholar]
- 11.Shevtsov SP, Dundr M. Nucleation of nuclear bodies by RNA. Nat Cell Biol. 2011;13:167–173. doi: 10.1038/ncb2157. [DOI] [PubMed] [Google Scholar]
- 12.Mao YS, Sunwoo H, Zhang B, Spector DL. Direct visualization of the cotranscriptional assembly of a nuclear body by noncoding RNAs. Nat Cell Biol. 2011;13:95–101. doi: 10.1038/ncb2140. [DOI] [PMC free article] [PubMed] [Google Scholar]