SUMMARY
Unlike many viruses that suppress cellular protein synthesis, host mRNA translation and polyribosome formation are stimulated by human cytomegalovirus (HCMV). How HCMV impacts the translationally-regulated cellular mRNA repertoire and its contribution to virus biology remains unknown. We show using polysome profiling that HCMV presides over the cellular translational landscape, selectively accessing the host genome to extend its own coding capacity and regulate virus replication. Expression of the HCMV UL38 mTORC1-activator partially recapitulates these translational alterations in uninfected cells. The signature of cellular mRNAs translationally-stimulated by HCMV resembles pathophysiological states where translation initiation factor levels or activity increase such as cancer. In contrast, cellular mRNAs repressed by HCMV include those involved in differentiation and the immune response. Surprisingly, interfering with the virus-induced activation of cellular mRNA translation can either limit or enhance HCMV growth. The unanticipated extent to which HCMV specifically manipulates host mRNA translation may aid in understanding its association with complex inflammatory disorders and cancer.
INTRODUCTION
In addition to their absolute reliance on cellular ribosomes to produce viral polypeptides, viruses can profoundly impact host protein synthesis. To antagonize host defenses and promote their replication, viruses often impair host mRNA translation (Walsh & Mohr, 2011). Not only does this strategy foster viral mRNA translation, it restricts any potential contribution of host mRNA translation to virus biology. Conceptually, this has helped shape our understanding of how viruses manipulate host mRNA translation (Mohr & Sonenberg, 2012). Little is known, however, regarding how host mRNA translation might be perturbed by viruses that do not globally suppress ongoing cellular protein synthesis as part of their replicative program.
Unlike viruses that shutoff cellular protein synthesis, polyribosome formation is stimulated and host mRNA translation proceeds uninterrupted in HCMV-infected cells (Tanaka et al. 1975; Stinski, 1977). Furthermore, the abundance of the cellular translation initiation factor eIF4F, comprised of the cap-binding subunit eIF4E and the RNA helicase eIF4A bound to eIF4G, together with the polyadenylate binding protein PABP1 increase in response to HCMV infection (Kudchodkar et al., 2004; Walsh et al., 2005; Perez et al., 2011; McKinney et al., 2012). This HCMV-induced PABP increase stimulates eIF4F assembly, virus protein accumulation, and virus replication (McKinney et al., 2012, 2013). However, how HCMV infection impacts the global repertoire of translationally-regulated cellular mRNAs and their contribution, if any, to virus biology remains unknown. Here, we use polysome profiling to establish that viral functions exert an extensive, unforeseen level of specific control over which cellular mRNAs are recruited to or excluded from polyribosomes. The signature of cellular mRNAs translationally-activated by HCMV, which encode a select suite of proteins critical for DNA damage response, proliferation, ribosome biogenesis, chromatin organization, organelle function and vesicle transport, resembles pathophysiological states where translation initiation factor levels or activity increase, including cancer. Host mRNAs repressed by HCMV include those involved in differentiation and the acquired immune response. These alterations to host mRNA translation were partially recapitulated in uninfected cells by expressing the multifunctional HCMV UL38 protein. Significantly, we show that interfering with the virus-induced increase in cellular mRNA translation can either limit or, surprisingly, enhance productive HCMV growth. Thus, while viruses do not encode their own translation machinery, they can effectively manipulate which host mRNAs are recruited to or excluded from polysomes without globally suppressing cellular protein synthesis. Moreover, by presiding over the host translational landscape, HCMV selectively accesses the host genome, extending its own coding capacity to regulate virus replication.
RESULTS & DISCUSSION
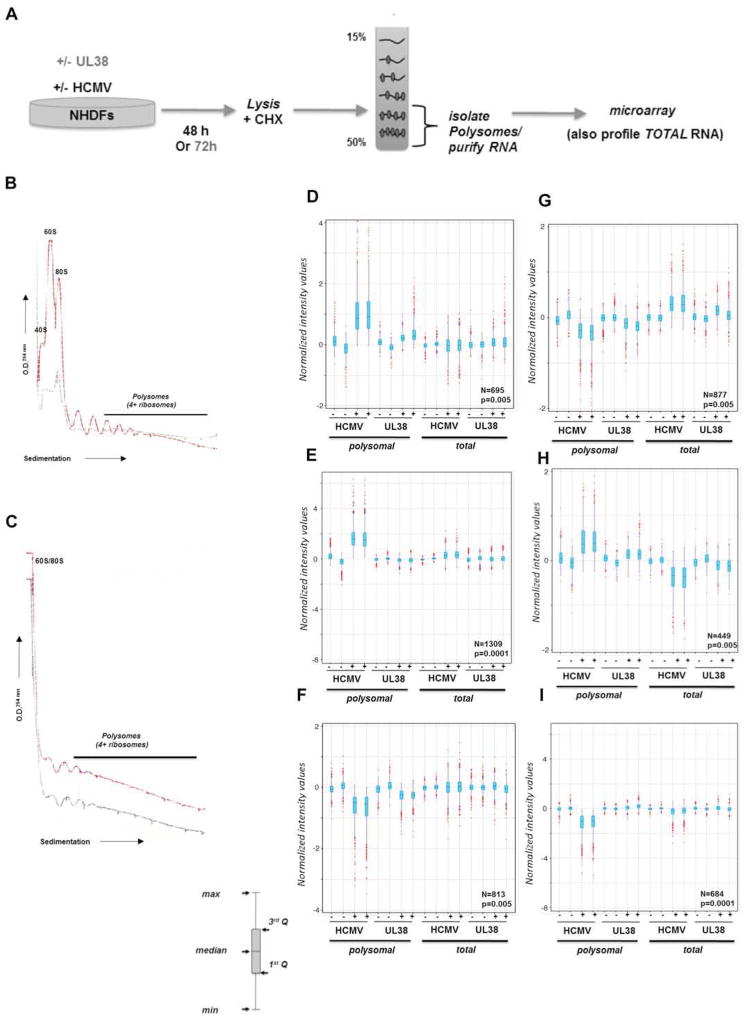
To determine if HCMV infection influenced host mRNAs selected for translation, cytosolic extracts prepared from primary, normal human fibroblasts (NHDFs) mock-infected or infected with HCMV at 48 h post-infection (hpi) were subject to sucrose gradient sedimentation (Fig. 1A). By 48 hpi, the HCMV-induced increase in PABP, eIF4F core subunit abundance, and eIF4F assembly was near maximal (Walsh et al., 2005; Perez et al., 2011; McKinney et al., 2012). Gradient fractionation while monitoring A254 revealed that the abundance of 40S and 60S ribosome subunits, 80S monoribosomes, and polyribosomes was enhanced in HCMV-infected cells (Fig. 1B). Thus, HCMV infection not only increased steady-state host translation factor levels, but substantially increased small and large ribosome subunit concentration, perhaps facilitating monosome and polysome assembly to stimulate mRNA translation. Interestingly, despite increasing 80S monosome formation, a substantial reservoir of free 40S and 60S subunits remained (Fig 1B).
Figure 1. Polysome profiling reveals extensive changes to ongoing host mRNA translation in response to HCMV infection.
A) Illustration depicting experimental procedure. NHDFs (+/− HCMV or transduced with a lentivirus that expresses the HCMV UL38 ORF from a dox-inducible promoter) were lysed in the presence of cycloheximide (48 hpi or 72 h post-induction of UL38). Extracts were sedimented through a 15–50% linear sucrose gradient that was subsequently fractionated and absorbance at 254 nm monitored. RNA isolated from fractions containing 4 or more ribosomes was used to prepare cRNA probes and hybridized to a DNA microarray. Total RNA isolated from parallel cultures was used to prepare cRNA probes and hybridized to a microarray to identify overall mRNA abundance changes in response to HCMV infection or UL38 induction. B) Absorbance tracing (254 nm) comparing mock-infected (gray line) vs HCMV-infected (red line) cultures. Migration of ribosome subunits (40S, 60S), monosomes (80S), and polyribosomes (4 or more ribosomes) are indicated. The top of the gradient is on the left. C) As in B but using uninfected cultures stably transduced with a doxycycline (dox)-inducible UL38-expression vector in the absence (gray line) or presence (red line) of dox. While polyribosome peaks were detected in uninfected cells, signals from 40S / 60S subunits and 80S monoribosomes were not resolved in the linear range of the tracing. D–I) Boxplot graphs based on normalized intensity values are shown with corresponding probe number (N) and Pavlidis Template Matching (PTM; Saeed et al., 2003) p-value indicated in the lower right corner. Outliers (in red), whiskers (indicating maximum and minimum), interquartile range (light blue shading) and median are shown. PTM algorithm (see methods) performed at p-values ranging from 5 × 10−3 to 10−5 identified specific profile patterns for groups of mRNAs in duplicate samples of polysomal and total RNA (−/+ HCMV; −/+ UL38 induction) that were elevated both in the polysomal fraction of HCMV-infected and dox+ (UL38-induced) cells (D); mRNAs that were elevated only in the polysomal RNA fraction from HCMV-infected cells (E); mRNAs reduced only in polysomal fraction from HCMV-infected and dox+ (UL38-induced) cells (F); mRNAs reduced in both polysomal RNA fraction from HCMV-infected and dox+ (UL38-induced) cells but elevated in the total RNA from the same conditions (G); elevated in both polysomal RNA fractions from HCMV-infected and dox+ cells but reduced in the total RNA from the same conditions (H); reduced only in polysomal RNA from HCMV-infected cells (I).
To interrogate if HCMV infection influenced the host polysome-associated mRNA population, RNA from fractions representing well-translated mRNAs associated with at least 4 or more ribosomes was used to generate a cRNA target set for hybridization to a human DNA microarray. In addition, microarrays were hybridized with cRNA prepared from total RNA isolated in parallel from mock or HCMV-infected cultures to measure overall mRNA abundance changes (Fig. 1A). Since attributing changes in gene expression to translation stimulation when mRNA levels increase and translational repression when mRNA abundance decreases is difficult, we restricted our analysis to four groups among cellular mRNAs that most likely represented translationally-controlled targets (Fig. 1D–I): mRNAs whose overall abundance i) remained relatively constant (no change as determined by Pavlidis template matching (PTM; Saeed et al., 2001) performed at p<0.005; see methods) that were recruited onto polysomes in response to infection (Fig. 1 D, E); ii) remained relatively constant (no change by PTM at p<0.005) whose presence on polysomes was restricted by infection (Fig 1F, I) ; iii) increased (PTM p<0,005, fold-change threshold >25%) yet whose recruitment onto polysomes diminished upon infection (Fig 1G); iv) decreased (PTM p<0,005, fold-change threshold >25%) but were selectively recruited onto polysomes upon infection (Fig 1H).
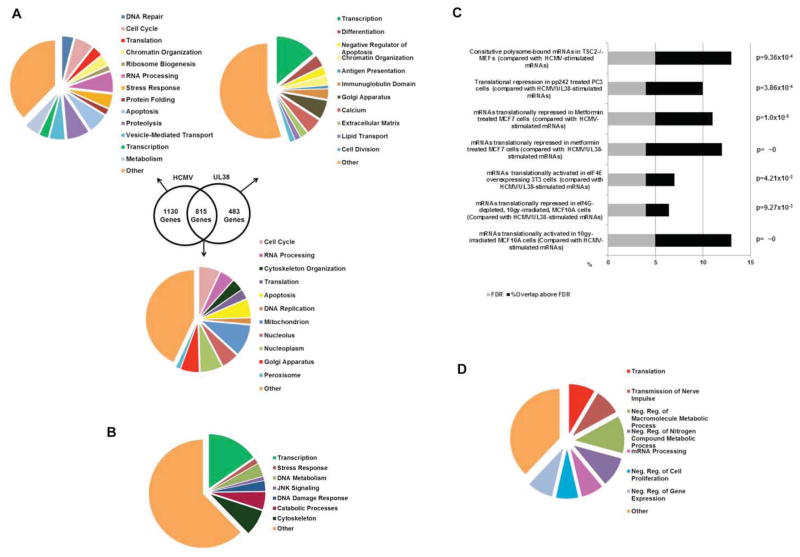
Remarkably, cellular mRNA translation was extensively reprogrammed by HCMV. Polysome profiling revealed approximately 2453 host mRNA targets whose association with polysomes was stimulated (≥ two fold enrichment) by HCMV infection (Fig. 1 D, E, H). In addition, polysome association of approximately 2374 cellular mRNA targets was reduced by HCMV (Fig. 1G, F, I). Significantly, most cellular mRNAs translationally-stimulated or repressed by HCMV clustered into functional groups that may promote viral replication, antagonize host defenses or conceivably represent host responses capable of limiting viral growth. Included amongst the translationally-activated mRNAs corresponding to 1945 host genes were those from DNA repair (p = 6.6 × 10−5; representing enrichment above random FDR; Huang et al., 2009), cell cycle control (p = 1.1 × 10−4), apoptosis (p = 2.4 × 10−4), stress response (p = 2.2 × 10−3), translation (p = 7.4 × 10−3), chromatin organization (p = 6.8 × 10−4), RNA processing (p = 8.3 × 10−13), vesicle mediated transport (p = 2.4 × 10−4), proteolysis (p = 8.7 × 10−3) and metabolism (p = 5.3 × 10−13) gene ontology (GO) categories (Fig. 2A; table S1, S2, S5). Notably, mRNAs involved in ribosome biogenesis were recruited to polysomes in HCMV-infected cells, likely accounting for the ribosome subunit increase observed (Fig 1B). By contrast, polysome-association of numerous host genes was restricted by HCMV infection. Among mRNAs translationally repressed by HCMV were those in GO categories involving calcium signaling (p = 2.2 × 10−2), cell division (p = 1.6 × 10−3), extracellular matrix (p = 4.5 × 10−2), lipid transport (p = 2.3 × 10−3), differentiation (p = 1.3 × 10−3), and antigen presentation (p = 3.7 × 10−2), the latter representing another potential strategy used by HCMV to evade host defenses (Fig 2B; Table S3). Finally, the gene set translationally-regulated in response to HCMV-infection was strikingly similar to those observed in different pathophysiological states where translation initiation factor levels or activity change (Silvera et al., 2010; Topisirovic & Sonenberg, 2011). Specifically, a statistically significant fraction of mRNAs whose translation was activated by HCMV was also stimulated in TSC-deficient cells (Bilanges et al., 2007), upon inducible eIF4E expression (Mamane et al., 2007), or following ionizing radiation (Badura et al., 2012; Fig 2C; table S9). Within this latter class, a subset of mRNAs whose translation responds to high eIF4G levels was stimulated (Badura et al., 2012). Finally, mRNAs in cancer cell lines whose translation was repressed by mTOR active-site inhibitors (Hsieh et al., 2012; Thoreen et al., 2012) or metformin (Larsson et al., 2012) were among those whose translation was activated by HCMV (Fig 2C). Indeed, many cellular mRNAs whose translation is stimulated in cancer cells or associated with cell proliferation were also activated by HCMV. HCMV is unusual among viruses in that it increases the abundance of ribosomes along with many translation factors, and this is required for its efficient replication. Thus similar signatures of translationally-controlled genes under different physiological conditions may reflect common underlying changes in the abundance and activity of ribosomes and translation factors.
Figure 2. Functional classification of cellular mRNAs translationally-regulated in response to HCMV or inducible UL38 expression.
(A) Gene symbol lists representing mRNAs translationally-stimulated by HCMV-infection or UL38 expression in uninfected cells (figs 1D, H), HCMV infection alone (fig 1E), or UL38 expression in uninfected cells (fig 1I) were functionally annotated to a curated list of biological processes using the Database for Annotation, Visualization and Integrated Discovery (DAVID) online bioinformatics resource. (B). As in A, but representing mRNAs translationally-repressed by HCMV-infection or UL38-expression in uninfected cells (figs 1F, G) (C) Gene symbol lists from (A) and (B) were analyzed for significant (χ2>3.84 for p<0.05) overlap, above a false rate of discovery (calculated in Experimental Procedures), with translationally regulated mRNAs in the indicated published studies. (D) TOP-containing mRNAs whose translation was stimulated by UL38 expression in uninfected cells and/or HCMV infection were identified using the UCSC Genome Browser and functionally classified as in (A).
PABP induction by HCMV is accompanied by translational activation of cellular mRNAs that contain a terminal oligopyrimidine (TOP) stretch at their 5′ terminus and encode translation factors, like PABP, and ribosomal proteins (McKinney et al., 2012). Both the increase in PABP and TOP mRNA translation depend upon mTORC1 activation by the HCMV multifunctional UL38 gene product, which also suppresses ER stress-induced cell death (Terhune et al., 2007; Moorman et al., 2008; Perez et al., 2011; Qian et al., 2011; McKinney et al., 2012, 2013). Furthermore, UL38 expression in uninfected cells confers TOP-like regulation on a reporter gene containing a functional 5′-TOP element (McKinney et al., 2012). Upon HCMV infection, 130 mRNAs containing a 5′ terminal TOP element, where the transcriptional start site is a C residue followed by four pyrimidines (Meyuhas, 2000), were mobilized onto polysomes (Table S7). These mRNAs are contained within GO categories for translation (p = 5.8 × 10−3), mRNA processing (p = 2.1 × 10−2), cell division (p = 3.3 × 10−2), and nerve impulse transmssion (p = 7.6 × 10−3) (Fig 2D; table S8). While TOP mRNA mobilization onto polysomes by HCMV was readily seen, statistically significant differences among mRNAs that contain a recently identified pyrimidine-rich translational element, PRTE, were not detected (Hsieh et al., 2012). Overall, cellular mRNAs whose access to polysomes was restricted in response to HCMV had similar GC content but an average 5′ UTR length between 10–22% longer than counterparts recruited onto polysomes or a randomly generated cellular mRNA list (p = 5.4 × 10−9 ; Table S6). Longer 5′-UTR’s among host mRNAs with diminished polysome association in HCMV-infected cells could confer more secondary structure, limiting ribosome scanning and subsequent translation initiation (Parsyan et al., 2011). Enrichment of cellular mRNAs with shorter, on average, 5′-UTRs onto polysomes in HCMV-infected cells, which have elevated eIF4F and PABP levels, is consistent with findings in yeast where mRNAs most dependent on eIF4G displayed an average 5′ UTR length at or below the mean for all yeast genes (Park et al., 2011).
To investigate if UL38 was responsible for the translational control of cellular mRNAs by HCMV, polysome profiling was performed using NHDFs transduced with a lentivirus expressing doxycyclin (dox)-inducible UL38. Analysis of A254 tracings revealed that inducible UL38 expression (+dox) was sufficient to stimulate polysome formation in uninfected cells (Fig 1C). Polysomal RNA from induced or uninduced (+/− dox) lentivirus-transduced NHDFs was used to generate probe sets for hybridization to DNA microarrays as described, and total RNA from parallel cultures used to normalize for mRNA abundance changes (Fig 1A). Significantly, of 1298 mRNAs whose translation was stimulated by UL38 expression in uninfected cells (Table S4), approximately 63% (815 genes) overlapped with those whose translation was stimulated by HCMV and were not restricted to only TOP mRNAs (Figs 1D, H; 2A). The remaining 37% (483 genes) of mRNAs translationally-activated upon UL38-induction were not stimulated in infected cells (Figs 1I;2A), indicating that their translational regulation in response to UL38 differs in the uninfected cell context. Notably, UL38 expression induced genes within the catabolic processes GO category not observed in infected cells (fig 2A; 24 genes; p=0.038). This implies that a different environment or mRNA population in HCMV-infected cells might curtail or sculpt the impact of UL38-mediated translational activation on host mRNAs. The host mRNAs translationally-repressed in response to UL38-expression in uninfected cells were similar to those repressed in infected cells (Fig. 2B, Table S3). Thus, expression of a single viral gene is sufficient to properly control a sizeable fraction of host mRNA translation in infected cells, and suggests that other translational control mechanisms, in addition to UL38-dependent regulation, may operate in HCMV-infected cells.
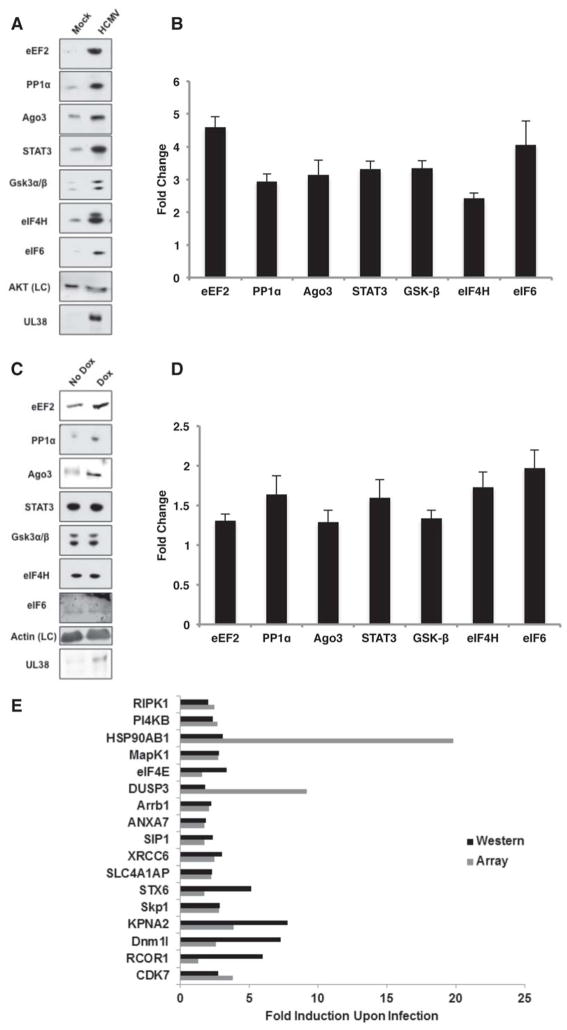
To validate that changes in host mRNA polysome association can alter steady-state protein levels, the abundance of select cellular proteins was evaluated by immunoblotting before and after HCMV infection (Fig 3A, B) or UL38 induction (Fig. 3C, D). Importantly, UL38 expressed from the lentivirus did not accumulate to the high level observed during HCMV infection (Fig 3C). Therefore, UL38 levels achieved through dox-induction were not supraphysiological, likely enabling identification of only highly UL38-responsive mRNAs and accounting for the lower magnitude of target gene induction compared to infected cells. Among mRNAs selected for validation, the abundance of eEF2, Protein Phosphatase 1α (PP1α) catalytic subunit (cs), Ago3, STAT3, Gsk3α/β, eIF4H and eIF6 polypeptides all increased in response to either HCMV-infection or UL38-induction in uninfected cells (Fig 3A–D). Real-time qPCR showed that mRNA abundance for each of these targets remained relatively constant following infection or UL38-induction, consistent with total RNA data from microarrays (Fig S2). While PP1α and eEF2 have been reported to increase in HCMV-infected cells, only eEF2 was shown to be translationally-controlled (Hakki & Geballe, 2008; McKinney et al., 2012). Finally, protein products of 17 additional mRNAs identified here as recruited onto polysomes in response to infection were shown by proteomic analysis to accumulate in HCMV-infected cells, while two proteins encoded by mRNAs whose polysome-association was reduced by infection (arginase 1, integrin β3) decreased, further validating our findings (Fig. 3E; Stanton et al., 2007).
Figure 3. HCMV-induced alterations in polysome profile of select mRNA targets results in altered steady-state protein levels in infected cells.
(A) NHDFs were mock-infected or infected with HCMV (MOI=3). At 48 hpi, total protein was collected, fractionated by SDS-PAGE and analyzed by immuno-blotting with the indicated antisera. Akt served as a loading control (LC). (B) Triplicate samples as in A were quantified by immunoblotting using the indicated primary antibodies and a secondary antibody covalently linked to an infrared fluorophore. The membrane was scanned and fold-change upon HCMV infection quantified using an Odyssey infrared imager. Each band was measured for raw intensity value and normalized to the loading control. (C) As in A except total protein was collected from NHDFs that express UL38 from a dox-inducible promoter 72 h post-treatment +/− dox. The LC for uninfected cells was actin. (D) Triplicate samples from C were quantified as described in B to determine the fold change upon UL38 induction in uninfected cells. (E) Average fold-induction of protein accumulation at 72 hpi of select upregulated cellular factors revealed by Power Blot analysis (Stanton et al., 2007) plotted alongside the average fold-recruitment of those representative mRNAs enriched onto polysomes at 48 hpi.
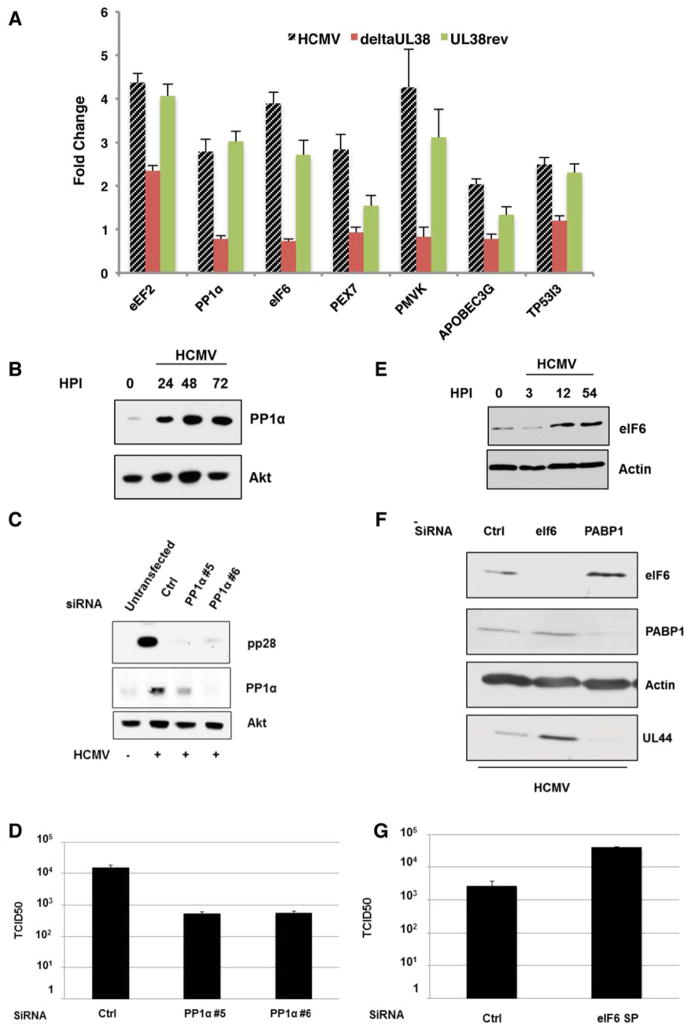
Having shown changes in host mRNAs associated with polysomes regulates cellular protein levels in response to HCMV infection or UL38 induction in uninfected cells, we investigated if any of these dynamic changes in polysome distribution i) were UL38-dependent in infected cells; and ii) contributed to virus infection biology or were instead biological noise that did not detectably contribute to productive viral growth. To determine if select host proteins induced by UL38 in uninfected cells (Figs 3C, D, S3) were expressed in a UL38-dependent manner in infected cells, their accumulation was measured in cells infected with WT HCMV, a UL38-deficient virus (ΔUL38), or a virus in which the UL38 deficiency was corrected. Indeed, the abundance of all seven cellular proteins in HCMV-infected cells was largely attenuated in cells infected with ΔUL38 and restored when the WT UL38 gene was present (Fig. 4A). Within this set, we first focused on PP1α (cs) which affects phosphorylation of many host and viral proteins by partnering with different regulatory subunits (Virshup & Shenolikar, 2009). Whereas PP1α protein accumulation was apparent by 24 hpi, PP1α mRNA levels increased less than two-fold in HCMV-infected cells (Fig 4B; S2). Importantly, PP1α-depletion in HCMV-infected cells using two different siRNAs reduced accumulation of an essential viral late protein, pp28, at 96 hpi and suppressed viral replication nearly 30-fold (Fig. 4C, D). As both measurements were made later than the 48 h point at which polysomal RNA was isolated, changes detected at 48 hpi have a lasting impact throughout the lengthy HCMV infection cycle. PP1α-induction by HCMV could explain why infected cells are more resistant to PP1α chemical inhibitors than uninfected cells (Hakki & Geballe, 2008). Importantly, PP1α-depletion did not detectably alter vaccinia virus protein accumulation in NHDFs, demonstrating that reducing PP1α abundance selectively impacted HCMV growth (Fig S4). Thus, preventing the virus-induced accumulation of PP1α restricts HCMV replication. This illustrates that HCMV infection promotes translation of a host mRNA whose protein product stimulates virus replication.
Figure 4. Translational regulation of PP1α cs and eIF6 in response to HCMV regulates productive viral growth.
(A) NHDFs were mock-infected or infected (MOI=3) with WT HCMV, a UL38-deficient mutant virus (deltaUL38), or a revertant virus where the UL38-deficiency was repaired by reintroducing a WT UL38 gene (UL38rev). At 48 hpi, total protein was isolated, fractionated by SDS-PAGE and analyzed by immunoblotting with the indicated antisera. Samples (n=3) were quantified using an Odyssey infrared imager. Each band was measured for raw intensity value, normalized to the loading control (Akt) and fold-change upon infection determined. (B) As in A except NHDFs were infected with WT HCMV and total protein harvested at the indicated HPI. (C) NHDFs transfected with a control, non-silencing siRNA (ctrl) or individual siRNAs targeting PP1α (#5, #6) were infected with HCMV (MOI=0.1). After 96 hpi samples were analyzed by immunoblotting as in A. D) Supernatants collected from three independent experiments detailed in C were assayed for viral particle production based upon their TCID50 (Kudchodkar et al., 2004). E) As in B. F) As in C but using siRNAs targeting PABP1 or eIF6. G) Supernatants collected from three independent experiments detailed in F were assayed for viral particle production as in D.
Because ribosome subunit concentration increased (Fig. 1B), we next examined if altering eIF6 levels influenced HCMV replication. In addition to its role in ribosome biogenesis, eIF6 is required for high level protein synthesis, is overexpressed in cancer cells and binds free 60S subunits to limit premature 80S ribosome formation (Loreni et al., 2013). While HCMV infection stimulated eIF6 protein accumulation in a UL38-dependent manner (Fig 4A, E), eIF6 mRNA levels remained relatively constant by qRT-PCR (Fig. S2). Unexpectedly, eIF6-depletion enhanced UL44 early-protein accumulation and stimulated virus replication approximately 20-fold relative to HCMV-infected cells treated with control siRNA (Fig 4F, G). In contrast, preventing the HCMV-induced PABP increase, which inhibits viral protein accumulation and replication (McKinney et al., 2012), severely reduced UL44 accumulation (Fig 4F). As 40S and 60S subunit concentration increased in response to HCMV (Fig. 1B), a commensurate eIF6 increase may be required to control 80S ribosome formation. The impact of eIF6-depletion was specific for HCMV, as it did not detectably augment protein synthesis or viral protein accumulation in HSV1-infected cells (Fig. S4). Thus, translational activation of host mRNAs in response to HCMV does not always promote viral replication, but can surprisingly restrict viral replication.
Recent delineation of hundreds of previously unidentified ORFs suggests that HCMV coding capacity is more complex than expected (Stern-Ginossaur et al., 2012). However, whereas α and γ herpesvirus subfamily members impair host proteins synthesis, cellular mRNA translation proceeds in HCMV-infected cells (Walsh & Mohr, 2011; Fig S1). Here we show that the potential liability of allowing host protein synthesis to proceed has been harnessed and exploited by the virus. After forcing the host to produce ribosomes and translation factors, viral functions selectively control which cellular transcripts access ribosomes without imposing a global shutoff of host protein synthesis. Thus, the translationally-regulated host mRNA landscape controls HCMV replication, giving a virus with a large DNA genome access to more than 1,000 host functions with previously undocumented roles in virus biology.
While it is intuitive to appreciate how virus-induced translational stimulation of host mRNA targets might stimulate viral replication, it was surprising that suppressing eIF6 accumulation enhanced viral replication. This implies that not all mRNAs translationally-activated by HCMV stimulate viral replication, but instead may antagonize viral growth. As eIF6-depletion enhanced viral replication, its induction by infection could represent a novel host defense to restrict virus growth. In this regard, proper translational regulation of host mRNAs can also be harnessed to suppress productive replication of the α-herpesvirus HSV1 and maintain viral latency in neurons (Kobayashi et al., 2012). Alternatively, virus-induced eIF6 accumulation could enable HCMV to prolong its lytic replication cycle and limit its virulence by moderating cellular protein synthesis capacity. By activating or restricting virus replication, translationally-controlled host mRNAs may influence numerous aspects of HCMV pathogenesis in different cell types. Furthermore, their deregulation under conditions of non-productive viral growth may help understand the association of HCMV with complex inflammatory conditions, vascular disease, and cancer (Soderberg-Naucler, 2008; Soroceanu & Cobbs, 2011; Dziurzynski et al., 2012). Precisely how this exquisite control of host mRNA translation is achieved in HCMV-infected cells is now ripe for future investigation, and the relative contribution of mRNA structure, translation factors, and ribosomes to this complex, post-transcriptionally-regulated gene expression program induced upon virus infection can be evaluated (Xue & Barna, 2012; Lee et al., 2013).
EXPERIMENTAL PROCEDURES
Polysome Isolation and Microarray Analysis
Cell extracts (from 1 × 107 mock or HCMV-infected NHDFs (MOI=3); or uninfected NHDFs that express dox-inducible UL38 −/+ dox; McKinney et al., 2013) were sedimented through 15–50% linear sucrose gradients at 36,000 RPM (SW40 rotor) for 1.5 hours (4°C). Gradients were fraction ated while monitoring RNA absorbance at 254 nM. RNA was isolated from pooled polysome fractions by phenol-CHCl3 extraction and precipitated with isopropanol.
DNA array analysis was performed at the NYU Genome Technology Center. After analyzing RNA quantity (Nanodrop-2000) and quality (Agilent 2100 Bioanalyzer), biotinylated cRNA probes were prepared and hybridized to GeneChip HGU133A 2.0 arrays according to the manufacturer (Affymetrix). Raw data were normalized by Robust Multichip Average (RMA) involving a background adjustment, quantile normalization and summarization using GeneSpring (Agilent) software version GX11 (Irizarry et al., 2003). Differentially abundant mRNAs were identified by t-test with the p-value cut-off of 0.05 at alpha level. Individual differential abundance data obtained from independent, duplicate samples were extensively validated by qPCR and immunoblotting in lieu of applying corrections for multiple testing due to the investigative, rather than corroborative nature of the microarray experiment. Pavlidis Template Matching algorithm available in the open source TM4 analytical suite and fold-change thresholding were used to define specific mRNA abundance profile types across experimental conditions (Saeed et al., 2003). Microarray data was deposited to the NCBI Gene Expression Omnibus database (accession GSE50938).
Supplementary Material
HIGHLIGHTS.
HCMV manipulates cell mRNA translation without globally impairing protein synthesis
HCMV selectively controls which cellular mRNAs are translated
The landscape of translationally-regulated host mRNAs regulates HCMV replication
The HCMV imposed translational signature shares similarities with cancer cells
Acknowledgments
We thank the NYU Genome Technology Center (GTC) for assistance with the microarray experiments; D. Yu, R. Schneider, N. Landau and T. Shenk for helpful reagents; D. Walsh and H. Burgess for critically reviewing the manuscript; members of the Mohr lab and A. Wilson for many discussions. This work was supported by NIH Grants AI073898 and GM056927 (to I.M.). CM was supported by NIH training Grant T32 AI007647. The NYU GTC is supported in part by NIH grant P30 CA016087-30.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain
References
- Badura M, Braunstein S, Zavadil J, Schneider RJ. DNA damage and eIF4G1 in breast cancer cells reprogram translation for survival and DNA repair mRNAs. Proc Natl Acad Sci USA. 2012;109:18767–18772. doi: 10.1073/pnas.1203853109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilanges B, Argonza-Barrett R, Kolesnichenko M, Skinner C, Nair M, Chen M, Stokoe D. Tuberous sclerosis complex proteins 1 and 2 control serum-dependent translation in a TOP-dependent and -independent manner. Mol Cell Biol. 2007;27:5746–5764. doi: 10.1128/MCB.02136-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziurzynski K, Chang SM, Heimberger AB, Kalejta RF, McGregor Dallas SR, Smit M, Soroceanu L, Cobbs CS. Consensus on the role of human cytomegalovirus in glioblastoma. Neuro Oncol. 2012;14:246–255. doi: 10.1093/neuonc/nor227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakki M, Geballe AP. Cellular serine/threonine phosphatase activity during human cytomegalovirus infection. Virology. 2008;380:255–263. doi: 10.1016/j.virol.2008.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh AC, Liu Y, Edlind MP, Ingolia NT, Janes MR, Sher A, Shi EY, Stumpf CR, Christensen C, Bonham MJ, et al. The translational landscape of mTOR signaling steers cancer initiation and metastasis. Nature. 2012;485:55–61. doi: 10.1038/nature10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Shermab BT, Lempicki RA. Systemic and integrative analysis of large lists using DAVID bioinformatics resources. Nat Protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucl Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M, Wilson AC, Chao MV, Mohr I. Control of viral latency in neurons by axonal mTOR signaling and the 4E-BP translation repressor. Genes Dev. 2012;14:1527–1532. doi: 10.1101/gad.190157.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudchodkar SB, Yu Y, Maguire TG, Alwine JC. Human cytomegalovirus infection induces rapamycin-insensitive phosphorylation of downstream effectors of mTOR kinase. J Virol. 2004;78:11030–11039. doi: 10.1128/JVI.78.20.11030-11039.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson O, Morita M, Topisirovic I, Alain T, Blouin MJ, Pollak M, Sonenberg N. Distinct perturbation of the translatome by the antidiabetic drug metformin. Proc Natl Acad Sci USA. 2012;109:8977–8982. doi: 10.1073/pnas.1201689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ASY, Burdinick-Kerr R, Whelan SPJ. A ribosome-specialized translation initiation pathway is required for cap-dependent translation of vesicular stomatitis virus mRNAs. Proc Natl Acad Sci USA. 2013;110:324–329. doi: 10.1073/pnas.1216454109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loreni F, Mancino M, Biffo S. Translation factors and ribosomal proteins control tumor onset and progression: how? Oncogene. 2013 doi: 10.1038/onc.2013.153. [DOI] [PubMed] [Google Scholar]
- Mamane Y, Petroulakis E, Martineau Y, Sato TA, Larsson O, Rajasekhar VK, Sonenberg N. Epigenetic activation of a subset of mRNAs by eIF4E explains its effects on cell proliferation. PLoS One. 2007;2:e242. doi: 10.1371/journal.pone.0000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney C, Perez C, Mohr I. Poly(A) binding protein abundance regulates eukaryotic translation initiation factor 4F assembly in human cytomegalovirus-infected cells. Proc Natl Acad Sci USA. 2012;109:5627–5632. doi: 10.1073/pnas.1202829109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney C, Yu D, Mohr I. A new role for the cellular PABP repressor Paip2 as an innate restriction factor capable of limiting productive cytomegalovirus replication. Genes Dev. 2013;27:1809–1820. doi: 10.1101/gad.221341.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyuhas O. Synthesis of the translational apparatus is regulated at the translational level. Eur J Biochem. 2000;267:6321–6330. doi: 10.1046/j.1432-1327.2000.01719.x. [DOI] [PubMed] [Google Scholar]
- Mohr I, Sonenberg N. Host translation at the nexus of infection and immunity. Cell Host Microbe. 2012;12:470–483. doi: 10.1016/j.chom.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorman NJ, Cristea IM, Terhune SS, Rout MP, Chait BT, Shenk T. Human cytomegalovirus protein UL38 inhibits host cell stress responses by antagonizing the tuberous sclerosis protein complex. Cell Host Microbe. 2008;3:253–262. doi: 10.1016/j.chom.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park EH, Zhang F, Warringer J, Sunnerhagen P, Hinnebusch AG. Depletion of eIF4G from yeast cells narrows the range of translational efficiencies genome-wide. BMC Genomics. 2011;12:68. doi: 10.1186/1471-2164-12-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsyan A, Svitkin Y, Shahbazian D, Gkogkas C, Lasko P, Merrick WC, Sonenberg N. mRNA helicases: the tacticians of translational control. Nat Rev Mol Cell Biol. 2011;12:235–245. doi: 10.1038/nrm3083. [DOI] [PubMed] [Google Scholar]
- Perez C, McKinney C, Chuluunbaatar U, Mohr I. Translational control of cytoplasmic poly A binding protein (PABP) abundance in HCMV-infected cells. J Virol. 2011;85:156–164. doi: 10.1128/JVI.01778-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Z, Xuan B, Gualberto N, Yu D. The human cytomegalovirus protein pUL38 suppresses endoplasmic reticulum stress-mediated cell death independently of its ability to induce mTORC1 activation. J Virol. 2011;85:9103–9113. doi: 10.1128/JVI.00572-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Silvera D, Formenti SC, Schneider RJ. Translational control and cancer. Nat Rev Cancer. 2010;10:254–266. doi: 10.1038/nrc2824. [DOI] [PubMed] [Google Scholar]
- Soderberg-Naucler C. HCMV microinfections in inflammatory diseases and cancer. J Clin Virol. 2008;41:218–223. doi: 10.1016/j.jcv.2007.11.009. [DOI] [PubMed] [Google Scholar]
- Soroceanu L, Cobbs C. Is HCMV a tumor promoter? Virus Res. 2011;157:93–203. doi: 10.1016/j.virusres.2010.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton RJ, McSharry BP, Rickards CR, Wang EC, Tomasec P, Wilkinson GW. Cytomegalovirus destruction of focal adhesions revealed in a high-throughput Western blot analysis of cellular protein expression. J Virol. 2007;81:7860–7872. doi: 10.1128/JVI.02247-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern-Ginossar N, Weisburd B, Michalski A, Le VT, Hein MY, Huang SX, Ma M, Shen B, Qian SB, Hengel H, et al. Decoding human cytomegalovirus. Science. 2012;338:1088–1093. doi: 10.1126/science.1227919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinski MF. Synthesis of proteins and glycoproteins in cells infected with human cytomegalovirus. J Virol. 1977;23:751–767. doi: 10.1128/jvi.23.3.751-767.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Furukawa T, Plotkin SA. Human cytomegalovirus stimulates host cell RNA synthesis. J Virol. 1975;15:297–304. doi: 10.1128/jvi.15.2.297-304.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terhune S, Torigoi E, Moorman N, Silva M, Qian Z, Shenk T, Yu D. Human cytomegalovirus UL38 protein blocks apoptosis. J Virol. 2007;81:3109–3123. doi: 10.1128/JVI.02124-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485:109–113. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topisirovic I, Sonenberg N. mRNA translation and energy metabolism in cancer: the role of MAPK and mTORC1 pathways. Cold Spring Harb Symp Quant Biol. 2011;76:355–367. doi: 10.1101/sqb.2011.76.010785. [DOI] [PubMed] [Google Scholar]
- Virshup DM, Shenolikar S. From promiscuity to precision: protein phosphatases get a makeover. Mol Cell. 2009;33:537–545. doi: 10.1016/j.molcel.2009.02.015. [DOI] [PubMed] [Google Scholar]
- Walsh D, Mohr I. Viral subversion of the host cell protein synthesis machinery. Nat Rev Micro. 2011;9:860–875. doi: 10.1038/nrmicro2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh D, Perez C, Notary J, Mohr I. Regulation of the translation initiation factor eIF4F by multiple mechanisms in human cytomegalovirus-infected cells. J Virol. 2005;79:8057–8064. doi: 10.1128/JVI.79.13.8057-8064.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue S, Barna M. Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat Rev Mol Cell Biol. 2012;13:355–369. doi: 10.1038/nrm3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.