The recombinant α-glucosidase HaG from Halomonas sp. Strain H11 was overexpressed, purified and crystallized to 2.15 Å, and its structure has been determined by molecular replacement.
Keywords: HaG, Halomonas sp. strain H11
Abstract
The α-glucosidase HaG from the halophilic bacterium Halomonas sp. strain H11 catalyzes the hydrolysis of the glucosidic linkage at the nonreducing end of α-glucosides, such as maltose and sucrose, to release α-glucose. Based on its amino-acid sequence, this enzyme is classified as a member of glycoside hydrolase family 13. HaG has three unique characteristics: (i) a very narrow substrate specificity, almost exclusively hydrolyzing disaccharides; (ii) activation by monovalent cations, such as K+, Rb+, Cs+ and NH4 +; and (iii) high transfer activity of the glucose moiety to the OH group of low-molecular-weight compounds, including glycerol and 6-gingerol. Crystallographic studies have been performed in order to understand these special features. An expression vector was constructed and recombinant HaG protein was overexpressed, purified and crystallized. A data set to 2.15 Å resolution was collected and processed. The crystal belonged to space group P212121, with unit-cell parameters a = 60.2, b = 119.2, c = 177.2 Å. The structure has been determined by molecular replacement using the isomaltulose synthase PalI as the search model (PDB entry 1m53).
1. Introduction
α-Glucosidases are enzymes that catalyze the exohydrolysis of α-1,4-glucosidic linkages, with the release of α-glucose, at the nonreducing end of substrates (EC 3.2.1.20). They play critical roles in biology ranging from digestion and decomposition of polysaccharides to biosynthesis of glycoproteins (Lovering et al., 2004 ▶). Transglucosylation catalyzed by α-glucosidase has also been utilized to improve the physicochemical properties of biologically useful substances (Murase et al., 1997 ▶; Kurimoto et al., 1997 ▶; Takenaka & Uchiyama, 2000 ▶). According to the CAZy (Carbohydrate-active Enzymes) glycoside hydrolase (GH) classification, α-glucosidases are classified into two major families, GH13 and GH31, and three minor families, GH4, GH97 and GH63 (Henrissat & Davies, 1997 ▶). The GH13 family contains a wide range of glycoside-processing enzymes, including α-amylases and glycosyltransferases including cyclodextrin glucanotransferase (CGTase), branching enzymes and amylosucrases. All of the enzymes included in GH13 catalyze the reaction through a two-step double-displacement mechanism. Two highly conserved catalytic residues access the substrate to form a β-d-glycosyl enzyme intermediate in the first step, which can then be hydrolyzed in the second step or can transfer a molecule of glucose to an acceptor if a free hydroxyl group attacks the intermediate instead of water (McCarter & Withers, 1996 ▶).
HaG, an α-glucosidase belonging to the GH13 family, was found in the halophilic bacterium Halomonas sp. strain H11, which was isolated from the surface of corals (Ojima, Saburi et al., 2012 ▶). The optimum conditions of the enzymological reaction of HaG towards maltose are pH 6.5 and a temperature of 303 K, and this enzyme shows 50% of the maximum activity even at 277 K. In comparison with the other enzymes that have similar functions, HaG has three unique characteristics: (i) a very narrow substrate specificity, hydrolyzing almost exclusively disaccharides, which is notable because bacterial α-glucosidases are generally capable of hydrolyzing longer maltooligosaccharides; (ii) activation by monovalent cations, such as K+, Rb+, Cs+ and NH4 +, while most known α-glucosidases in the GH13 family are independent of monovalent cations; and (iii) high transglucosylation activity towards OH groups of low-molecular-weight compounds.
Although there have been many reports on enzymes in the GH13 family, contributing to the biological importance of these enzymes (Henrissat & Davies, 1997 ▶; Zhang et al., 2003 ▶; Møller et al., 2012 ▶; Ravaud et al., 2007 ▶; Watanabe et al., 1997 ▶), the structure of GSJ (PDB entry 2ze0), an α-glucosidase from Geobacillus sp. strain HTA-462, is the only published structure of an α-glucosidase from this family (Shirai et al., 2008 ▶). However, multiple alignment using the Clustal Omega program (Goujon et al., 2010 ▶; Sievers et al., 2011 ▶) indicated that GSJ shares only 36% sequence identity with HaG. For transglucosylation, GSJ is able to use various nonsugar molecules as acceptors, and even rather large molecules such as curcumin (molecular weight 368.39), owing to its wide open catalytic pocket. Similarly, HaG also has high transglucosylation activity towards glycerol and 6-gingerol, generating α-d-glucosylglycerol (αGG) and 5-α-d-glucosylgingerol, respectively. The product αGG has already been applied in cosmetics as a moisturizing agent (Ojima, Saburi et al., 2012 ▶; Yoshida et al., 2007 ▶), and no biocatalytic route for the synthesis of 5-α-d-glucosylgingerol has previously been described (Ojima, Aizawa et al., 2012 ▶). However, it is notable that HaG specifically transferred the glucosyl moiety to the alkyl OH rather than the phenolic OH.
To understand the structural elements responsible for the special properties of HaG, we produced recombinant HaG in Escherichia coli. This recombinant enzyme was purified and crystals were obtained which diffracted to 2 Å resolution. This will allow detailed analysis of the structure, which will contribute to a better understanding of the reaction mechanism of α-glucosidase acting as a glucosyltransferase.
2. Materials and methods
2.1. Construction and expression of α-glucosidase HaG
The HaG gene (GenBank accession No. AB663683) was cloned into the EcoRI and BglII sites of the pFLAG-CTS vector (Sigma, St Louis, Missouri, USA). The primers 5′-AAAAGAATTCCAAGACAACATGATGTGGT-3′ (sense, EcoRI site in bold) and 5′-AAAAAAGATCTTAGGCAACCTGCATAAAG-3′ (antisense, BglII site in bold), genomic DNA of Halomonas sp. strain H11 and KOD DNA polymerase (Toyobo, Osaka, Japan) were used for amplification of the DNA fragment. The expression vector was transformed into E. coli BL21 (DE3) cells to express recombinant HaG. The cells were grown at 310 K to an OD600 of 0.5 in 3 l LB medium containing 100 µg ml−1 ampicillin. HaG production was induced by adding isopropyl β-d-1-thiogalactopyranoside (IPTG) to a final concentration of 0.1 mM and cultivation was continued for 21 h at 291 K. Cells were harvested by centrifugation at 5000g for 10 min at 293 K and stored at 193 K.
2.2. Purification of HaG
The frozen cells were suspended in osmotic shock buffer (500 mM sucrose, 30 mM Tris–HCl pH 8.0, 2 mM EDTA) and centrifuged at 3500g for 10 min at 293 K. The pellet was resuspended in 240 ml ice-cold water and centrifuged again at 3500g for 10 min at 277 K. The supernatant was collected as the starting sample for purification.
The proteins in the supernatant were fractionated by ion-exchange chromatography with a Toyopearl DEAE-650M column (Toyobo). After column equilibration with 10 mM HEPES buffer pH 7.0, the proteins were eluted with a linear NaCl gradient (0–0.3 M) in the same buffer. The purified sample is shown in Fig. 1 ▶. Samples of active fractions were collected and dialyzed against 4 l 10 mM HEPES buffer pH 7.0 for 5 h. After dialysis three times, the proteins were concentrated to 10 mg ml−1 by ultrafiltration with Vivaspin 20 (Vivascience, Hanover, Germany) and stored at 193 K.
Figure 1.
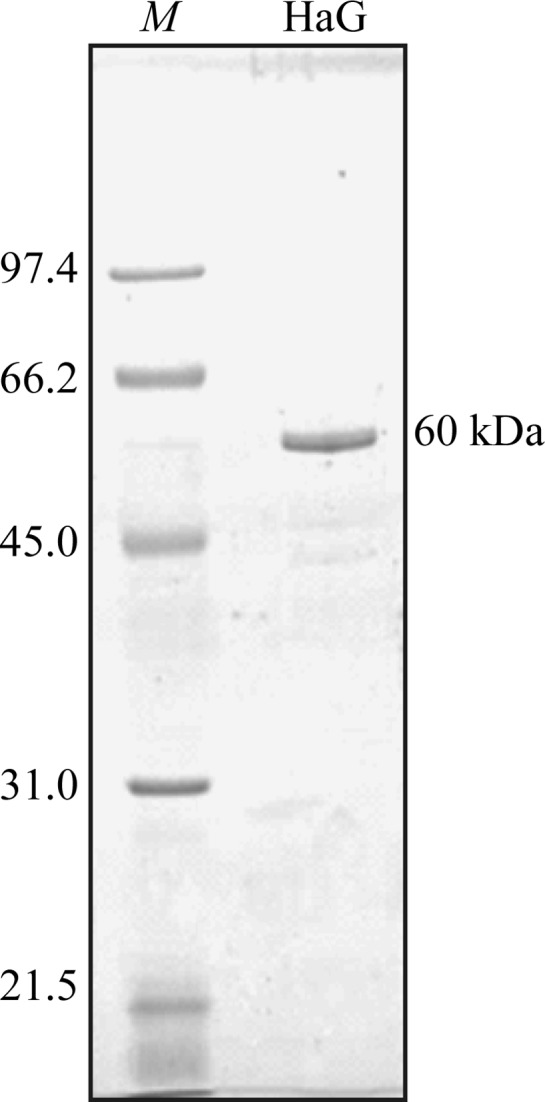
SDS–PAGE of the recombinant HaG produced in E. coli after purification. Purified protein (0.5 µg) was analyzed on 10% SDS–PAGE. Lane M contains molecular-mass marker (labelled in kDa). The gel was visualized by Coomassie Brilliant Blue staining.
2.3. Crystallization
Crystallization was carried out by the sitting-drop vapour-diffusion method in 96-well plates at 293 K. Drops consisting of 0.75 µl protein solution and 0.75 µl reservoir solution were equilibrated against 75 µl reservoir solution. Initial crystallization screening was performed using nine kits (Qiagen NeXtal Tube Suites; Qiagen, Hilden, Germany). Crystals were obtained under several conditions in The JCSG+, PEGs, PEGs II and Classics Suites, and the best crystals, which diffracted to high resolution, were obtained with The JCSG+ Suite. After optimizing the pH from 6.6 to 7.5 and the precipitant concentration from 15% to 25%, the best conditions were 0.1 M HEPES pH 7.0, 0.02 M magnesium chloride, 20% polyacrylic acid 5100 sodium salt. The large stick-shaped crystals (0.5 × 0.05 × 0.03 mm) were finally harvested within 10 d at 293 K (Fig. 2 ▶).
Figure 2.
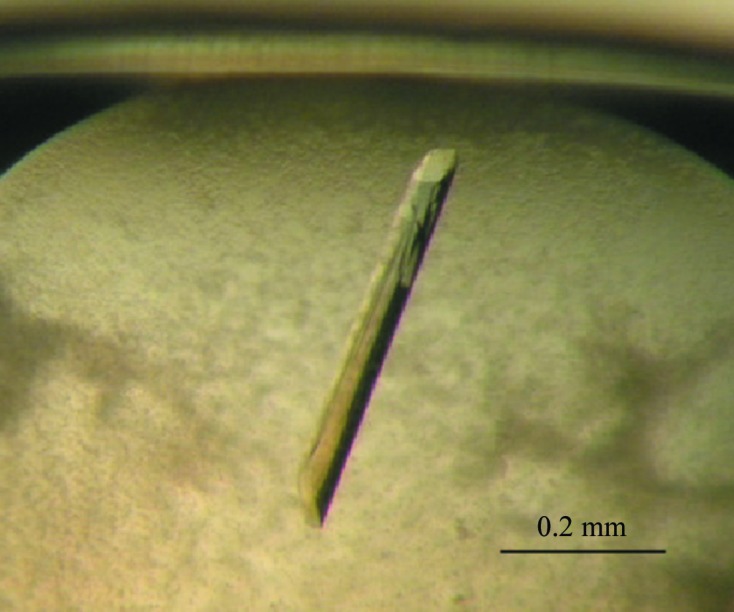
Crystal of α-glucosidase HaG from Halomonas sp. strain H11. The longest dimension is approximately 0.5 mm.
2.4. Data collection and processing
Crystals were soaked in cryoprotectant solution containing reservoir buffer with a final concentration of 15% glycerol for several seconds in one step before flash-cooling under a stream of nitrogen gas. X-ray diffraction data were collected from the crystal at a wavelength of 1.0000 Å using an ADSC Quantum 315r CCD detector on beamline BL-5A of the Photon Factory (PF), Tsukuba, Japan. A total of 600 images were collected with an oscillation angle of 0.3° and an exposure time of 2 s per image. The diffraction data were indexed, integrated, scaled and merged using the XDS and CCP4 packages (Kabsch, 2010 ▶; Winn et al., 2011 ▶). The data-processing statistics are summarized in Table 1 ▶.
Table 1. Statistics of data collection and refinement.
Values in parentheses are for the outermost resolution shell.
| Beamline | BL-5A, PF |
| Detector | ADSC Quantum 315r |
| Crystal-to-detector distance (mm) | 249.6 |
| Wavelength (Å) | 1.0000 |
| No. of images | 600 |
| Oscillation range (°) | 0–180 |
| Resolution range (Å) | 50.0–2.15 (2.28–2.15) |
| No. of total reflections | 518379 (82634) |
| No. of unique reflections | 71192 (11231) |
| Space group | P212121 |
| Unit-cell parameters (Å) | a = 60.2, b = 119.2, c = 177.2 |
| Completeness (%) | 99.7 (98.7) |
| R meas (%) | 8.9 (52.6) |
| 〈I/σ(I)〉 | 20.81 (5.75) |
| Crystal mosaicity (°) | 0.15 |
| Wilson B factor (Å2) | 35.27 |
3. Results
The expression vector pFLAG-CTS and host E. coli BL21 (DE3) cells were used to express recombinant HaG protein. After purification by anion-exchange column chromatography and dialysis, a total of 10 mg purified HaG protein was obtained from 3 l of culture medium.
A crystal diffracting to a resolution limit of 2.15 Å was finally obtained. The data-collection statistics are given in Table 1 ▶. Analysis of the diffraction pattern showed that the crystal belonged to space group P212121, with unit-cell parameters a = 60.2, b = 119.2, c = 177.2 Å. Assuming a molecular mass of 58 kDa, a chain length of 538 amino-acid residues and two molecules in the asymmetric unit gave a solvent content of 55.7% and a V M value of 2.78 Å3 Da−1 (Matthews, 1968 ▶).
Structure determination was performed using the molecular-replacement method with Phaser (McCoy et al., 2007 ▶). Many models were used as search models; finally, we solved the structure using the monomer structure of the isomaltulose synthase PalI from Klebsiella sp. LX3 (PDB entry 1m53), which shows 36% identity to HaG (Zhang et al., 2003 ▶). Rotation and translation functions were calculated using data in the 50–3 Å resolution range. Two molecules were revealed in the asymmetric unit, with TFZ scores of 12.8 and 12.5. Initial rigid-body refinement was performed with REFMAC (Murshudov et al., 2011 ▶). The model was then improved by AutoBuild (Terwilliger et al., 2008 ▶) and morph_model from PHENIX (Adams et al., 2010 ▶). Further model modification and refinement are currently in progress.
Acknowledgments
We are grateful to the staff at BL-5A, PF for their help in data collection. XS was supported by the CSC (China Scholarship Council).
References
- Adams, P. D. et al (2010). Acta Cryst. D66, 213–221.
- Goujon, M., McWilliam, H., Li, W., Valentin, F., Squizzato, S., Paern, J. & Lopez, R. (2010). Nucleic Acids Res. 38, W695–W699. [DOI] [PMC free article] [PubMed]
- Henrissat, B. & Davies, G. (1997). Curr. Opin. Struct. Biol. 7, 637–644. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Kurimoto, M., Nishimoto, T., Nakada, T., Chaen, H., Fukuda, S. & Tsujisaka, Y. (1997). Biosci. Biotechnol. Biochem. 61, 699–703. [DOI] [PubMed]
- Lovering, A. L., Lee, S. S., Kim, Y.-W., Withers, S. G. & Strynadka, N. C. (2004). J. Biol. Chem. 280, 2105–2115. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- McCarter, J. D. & Withers, S. G. (1996). J. Biol. Chem. 271, 6889–6894. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Møller, M. S., Fredslund, F., Majumder, A., Nakai, H., Poulsen, J. C., Lo Leggio, L., Svensson, B. & Abou Hachem, M. (2012). J. Bacteriol. 194, 4249–4259. [DOI] [PMC free article] [PubMed]
- Murase, H., Yamauchi, R., Kato, K., Kunieda, T. & Terao, J. (1997). Lipids, 32, 73–78. [DOI] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Ojima, T., Aizawa, K., Saburi, W. & Yamamoto, T. (2012). Carbohydr. Res. 354, 59–64. [DOI] [PubMed]
- Ojima, T., Saburi, W., Yamamoto, T. & Kudo, T. (2012). Appl. Environ. Microbiol. 78, 1836–1845. [DOI] [PMC free article] [PubMed]
- Ravaud, S., Robert, X., Watzlawick, H., Haser, R., Mattes, R. & Aghajari, N. (2007). J. Biol. Chem. 282, 28126–28136. [DOI] [PubMed]
- Shirai, T., Hung, V. S., Morinaka, K., Kobayashi, T. & Ito, S. (2008). Proteins, 73, 126–133. [DOI] [PubMed]
- Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., Lopez, R., McWilliam, H., Remmert, M., Söding, J., Thompson, J. D. & Higgins, D. G. (2011). Mol. Syst. Biol. 7, 539. [DOI] [PMC free article] [PubMed]
- Takenaka, F. & Uchiyama, H. (2000). Biosci. Biotechnol. Biochem. 64, 1821–1826. [DOI] [PubMed]
- Terwilliger, T. C., Grosse-Kunstleve, R. W., Afonine, P. V., Moriarty, N. W., Zwart, P. H., Hung, L.-W., Read, R. J. & Adams, P. D. (2008). Acta Cryst. D64, 61–69. [DOI] [PMC free article] [PubMed]
- Watanabe, K., Hata, Y., Kizaki, H., Katsube, Y. & Suzuki, Y. (1997). J. Mol. Biol. 269, 142–153. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Yoshida, K. T. F., Nitta, A. & Iki, M. (2007). Japan Patent 2007-137862.
- Zhang, D., Li, N., Lok, S. M., Zhang, L. H. & Swaminathan, K. (2003). J. Biol. Chem. 278, 35428–35434. [DOI] [PubMed]


