Among the mammalian adenylyl cyclases, the soluble type 10 cyclase is unique in its activation by bicarbonate and calcium and its lack of a transmembrane region. Here, the recombinant production and crystallization of the catalytic core of the human type 10 adenylyl cyclase are described.
Keywords: soluble adenylyl cyclase, type 10, mammalian, catalytic core
Abstract
The second messenger cAMP is synthesized in mammals by ten differently regulated adenylyl cyclases (AC1–10). These ACs are grouped into nucleotidyl cyclase class III based on homologies in their catalytic domains. The catalytic domain of AC10 is unique, however, in being activated through direct interaction with calcium and bicarbonate. Here, the production, crystallization and X-ray diffraction analysis of the catalytic domain of human AC10 are described as a basis for structural studies of regulator binding sites and mechanisms. The recombinant protein had high specific AC activity, and crystals of AC10 in space group P63 diffracted to ∼2.0 Å resolution on a synchrotron beamline. A complete diffraction data set revealed unit-cell parameters a = b = 99.65, c = 98.04 Å, indicating one AC10 catalytic domain per asymmetric unit, and confirmed that the obtained crystals are suitable for structure solution and mechanistic studies.
1. Introduction
cAMP is a ubiquitous second messenger that regulates various physiological processes, such as gene expression and cellular metabolism (Hanoune & Defer, 2001 ▶). In mammals, cAMP is synthesized by ten differentially regulated adenylyl cyclases (ACs), which all belong to the nucleotidyl cyclase class III that is defined by homologous catalytic domains (Hanoune & Defer, 2001 ▶; Kamenetsky et al., 2006 ▶). Nine of the ten mammalian enzymes are transmembrane ACs (tmACs) localized in the cell membrane. The single soluble mammalian AC10, also called soluble AC (sAC), can be found in different splice variants and in various intracellular compartments (Kamenetsky et al., 2006 ▶). Nuclear AC10 contributes to transcription regulation via cAMP-responsive element binding protein (CREB; Zippin et al., 2004 ▶), and in mitochondria AC10 forms a regulatory system with phosphodiesterase 2 (PDE2) controlling the activity of the respiratory chain (Acin-Perez et al., 2011 ▶).
Class III comprises ACs from many organisms, and also all mammalian ACs and guanylyl cyclases (GCs). Crystal structures of the conserved class III catalytic core (Sinha & Sprang, 2006 ▶; Rauch et al., 2008 ▶) revealed a symmetric or pseudo-symmetric arrangement of two identical or two highly similar catalytic subunits (C1 and C2). In mammalian AC, C1 and C2 reside on one polypeptide chain. Fused to the catalytic domains, tmACs harbour transmembrane segments and sometimes regulatory regions (Linder, 2006 ▶; Kamenetsky et al., 2006 ▶). C-terminal to its catalytic domains, AC10 comprises ∼1100 residues with little understood function and architecture (Linder, 2006 ▶; Kamenetsky et al., 2006 ▶) except for the known presence of a haem-binding domain (Middelhaufe et al., 2012 ▶) and a small autoinhibitory motif (Chaloupka et al., 2006 ▶). The best characterized AC1–10 regulators, however, directly target the catalytic domains and show isoform-specific effects despite the homology. The AC10 catalytic core is specifically activated by bicarbonate and calcium, while it is insensitive to the major tmAC regulators calmodulin and heterotrimeric G proteins (Kamenetsky et al., 2006 ▶; Sunahara & Taussig, 2002 ▶). AC10 acts as a sensor for bicarbonate in a variety of tissues and functions, such as sperm capacitation and mitochondrial energy metabolism (Acin-Perez et al., 2011 ▶; Hess et al., 2005 ▶). Bicarbonate-regulated AC10-like enzymes also exist in lower organisms and contribute, for example, to CO2 sensing in fungi (Hall et al., 2010 ▶). Crystallographic studies on the AC10 homologue CyaC from Spirulina platensis indicated that bicarbonate induces an active-site closure of the enzyme–substrate complex (Steegborn, Litvin, Levin et al., 2005 ▶), but the bicarbonate binding site and the mechanism initiating this conformational change remained to be revealed. Type 10 ACs also differ from tmACs in their pharmacological regulation. They are relatively insensitive to the adenosine-derived so-called ‘p-site’ tmAC inhibitors and to the tmAC activator forskolin (Kamenetsky et al., 2006 ▶). Instead, type 10 ACs are inhibited by BCC2 and BCC8 (Schlicker et al., 2008 ▶), two isoform-specific ligands for the binding site of the nonspecific AC inhibitor catechol oestrogen (Steegborn, Litvin, Hess et al., 2005 ▶), and by the high-throughput screen-derived compound KH7 (Hess et al., 2005 ▶), whose binding site and inhibition mechanism remain to be revealed. We describe here the recombinant production, crystallization and preliminary diffraction analysis of human AC10 catalytic domains as a basis for structural studies on binding sites and modulation mechanisms of AC10 regulators.
2. Materials and methods
2.1. Cloning, expression and purification
The gene fragment coding for the catalytic domain comprising residues 1–469 of human AC10 (AC10-cat; UniGene Hs.320892) was PCR-amplified, introducing a C-terminal His tag and two stop codons through the 3′-primer, to generate an expression construct similar to those described previously (Chen et al., 2000 ▶). The PCR product was cloned into the pVL1392 transfer vector using PstI and BamHI, and a recombinant baculovirus for AC10-cat expression was generated by Diarect AG (Freiburg, Germany) through co-transfection with linearized baculovirus DNA and plaque selection of recombinant virus. AC10-cat baculovirus was amplified in Sf21 cells. BIIC (baculovirus infected insect cells) were stored in the vapour phase in a liquid-nitrogen storage tank as the virus source for production experiments as described in Wasilko et al. (2009 ▶). For expression, Hi5 insect cells were infected with recombinant AC10-cat baculovirus by adding 1 ml BIIC stock to 1 l of Hi5 cells grown to 0.8 × 106 cells ml−1. The infection process was continued until an increase in cell diameter of 2–3 µm was reached. Cultures were diluted to keep the cell concentration below 2 × 106 cells ml−1. Cells were harvested 48 h after proliferation stop, snap-frozen and stored at 193 K.
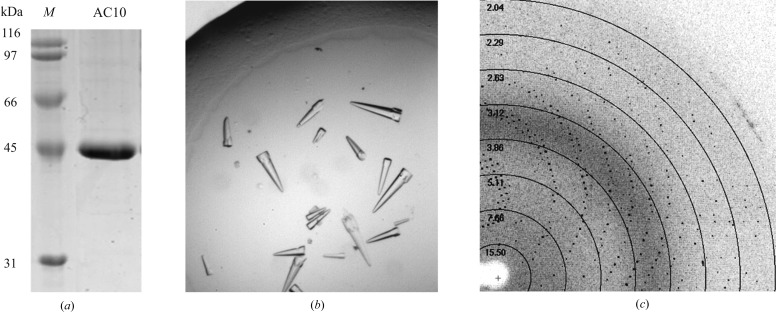
For purification of the recombinant His-tagged AC10-cat protein (calculated mass 54 119 Da), we modified the protocols described in Chen et al. (2000 ▶) and Litvin (2004 ▶) and performed all steps at 277 K. Insect cell pellets were thawed, resuspended in buffer A [50 mM Tris–HCl pH 7.5, 300 mM NaCl, 10%(v/v) glycerol, 20 mM imidazole, 2 mM β-mercaptoethanol and protease inhibitor mix (Roche)] and disrupted with a microfluidizer. After centrifugation (48 000g for 1 h at 277 K) the supernatant was loaded onto a HisTrap Ni Sepharose column (GE Healthcare). After washing with 20 column volumes (CV) of buffer A, the protein was eluted using a 20 CV gradient from 20 to 500 mM imidazole in buffer A. The buffer was changed to buffer B [50 mM Tris pH 7.5, 50 mM NaCl, 10%(v/v) glycerol, 2 mM β-mercaptoethanol] using an NAP column (GE Healthcare) and the protein loaded onto a Mono Q anion-exchange column (GE Healthcare). The protein was eluted with a 20 CV gradient from buffer B to 30% buffer B + 1 M NaCl. The protein was then subjected to gel filtration on a Superdex 75 column (GE Healthcare) in 50 mM Tris–HCl pH 7.5, 350 mM NaCl, 10%(v/v) glycerol, 5 mM β-mercaptoethanol. The purity and identity of the protein were confirmed by 14% SDS–PAGE (Fig. 1 ▶ a) and by mass spectrometry (MS). For MS analysis, 10 µg AC10-cat was digested with trypsin for 18 h, followed by electron spray ionization (ESI) MS analysis of the resulting peptides on an LTQ Velos (Thermo Fisher Scientific) as described in Gertz et al. (2008 ▶). The protein was concentrated to 6.8 mg ml−1, flash-cooled in liquid nitrogen and stored at 193 K. Protein concentrations were determined by absorbance spectroscopy using the calculated extinction coefficient ∊280 = 51 340 M −1 cm−1.
Figure 1.
Purification, crystallization and preliminary diffraction analysis of the human type 10 AC catalytic domains. (a) SDS–PAGE analysis of purified recombinant human AC10-cat protein. Lane M, molecular-mass marker. (b) Rocket-shaped crystals of human AC10-cat obtained in hanging-drop vapour-diffusion experiments at 277 K. (c) Diffraction pattern of human AC10-cat crystals on BESSY beamline MX 14.1.
2.2. Crystallization
For AC10-cat crystallization, we initially screened protein with and without an ATP analogue (7.5 mM α,β-methylene-ATP, 7.5 mM MgCl2, 7.5 mM CaCl2) against the Qiagen JCSG+ factorial screen in 0.2 µl drops (0.1 µl protein solution + 0.1 µl reservoir solution) set up using a Phoenix nanodispenser (Art Robbins Instruments). Crystals appeared in drops both with and without ATP analogue under the same condition [0.2 M tripotassium citrate, 20%(w/v) PEG 3350] but showed cleaner morphology and grew to a much larger size in the absence of ATP analogue. We therefore refined this condition with AC10-cat without ATP analogue to 0.1 M sodium acetate pH 4.8, 0.2 M trisodium citrate, 15%(w/v) PEG 4000, 10%(v/v) glycerol. Crystals were grown at 277 K by using the hanging-drop vapour-diffusion method with equal volumes of crystallization solution and protein stock (1 µl:1 µl) in the drop and a reservoir volume of 500 µl. Diffraction-quality crystals appeared reproducibly within 24 h.
2.3. Crystallographic studies
For cryoprotection, 20%(v/v) glycerol was added to the reservoir solution and the PEG 4000 concentration was increased to 18%(w/v), and all diffraction measurements were performed with crystals at 100 K. AC10-cat crystals diffracted to beyond 2 Å resolution on beamline MX 14.1 operated by the Helmholtz-Zentrum Berlin (HZB) at the BESSY II electron-storage ring (Berlin-Adlershof, Germany; Mueller et al., 2012 ▶). Indexing revealed the point group to be hexagonal, and a diffraction data set covering 75° (150 frames of 0.5° each) was collected at 100 K on BESSY II beamline MX 14.1 (Mueller et al., 2012 ▶). The crystal space group was identified as P63, and diffraction data were indexed, integrated and merged using XDS (Kabsch, 2010 ▶), resulting in a 99.8% complete data set (Table 1 ▶).
Table 1. Diffraction data statistics for a crystal of human AC10-cat.
Values in parentheses refer to the outermost shell.
| Space group | P63 |
| Unit-cell parameters (Å) | a = b = 99.65, c = 98.04 |
| Beamline | MX 14.1 |
| Wavelength (Å) | 0.9171 |
| Resolution range (Å) | 86.30–1.95 (2.05–1.95) |
| No. of reflections | 186918 (20026) |
| No. of unique reflections | 40209 (5487) |
| Completeness (%) | 99.8 (98.7) |
| Multiplicity | 4.6 (3.7) |
| R meas (%) | 9.5 (51.3) |
| Average I/σ(I) | 14.8 (3.1) |
| Temperature (K) | 100 |
| Solvent content (%) | 52.7 |
| Matthews coefficient (Å3 Da−1) | 2.6 |
3. Results
Our initial trials to express AC10 constructs comprising both catalytic domains in soluble form in Escherichia coli failed. Expression of the two sAC catalytic domains individually, an approach that had enabled purification and crystallization of an active heterodimeric tmAC catalytic core (Tesmer et al., 1997 ▶), yielded only C2 protein, which could be purified as a homodimer. The AC10-C2 homodimer was inactive, as expected for a dimer lacking the catalytic C1 residues (Kamenetsky et al., 2006 ▶), and the protein did not form crystals. We therefore expressed a C-terminally His-tagged construct comprising C1 and C2 of human AC10 (AC10-cat) in insect cells. The protein could be purified to apparent homogeneity through affinity, anion-exchange and size-exclusion chromatography (Fig. 1 ▶ a), yielding about 200 µg AC10-cat from 3 l cultured cells. The elution volume of AC10-cat in the size-exclusion chromatography was consistent with the expected monomeric state, the protein showed high specific AC activity and its identity as AC10-cat was confirmed through peptide mass spectrometry after tryptic digest.
The recombinant AC10-cat protein could be crystallized in vapour-diffusion experiments. Crystals mounted from a primary screen drop showed diffraction up to 3 Å resolution on a synchrotron beamline, and optimized crystals from refinement of this condition diffracted to beyond 2 Å resolution. Analysis of a complete diffraction data set collected at 1.95 Å resolution revealed that the crystal belonged to space group P63, with unit-cell parameters a = b = 99.65, c = 98.04 Å. A Matthews coefficient of 2.6 Å3 Da−1 indicates one AC catalytic domain per asymmetric unit and a solvent content of 52.7% (Matthews, 1968 ▶). The processing statistics for the diffraction data set (Table 1 ▶) show that our AC10-cat protein can form crystals that are suitable for structure solution and mechanistic studies on AC10 regulation.
Acknowledgments
We thank Dr Frank Fischer for support with mass-spectrometric analysis, Thomas Simon (Diarect AG, Freiburg, Germany) for help with baculovirus generation, the Helmholtz Protein Sample Production Facility (PSPF) for help with large-scale protein expression in insect cells and the BESSY beamline staff for support with diffraction data collection.
References
- Acin-Perez, R., Russwurm, M., Günnewig, K., Gertz, M., Zoidl, G., Ramos, L., Buck, J., Levin, L. R., Rassow, J., Manfredi, G. & Steegborn, C. (2011). J. Biol. Chem. 286, 30423–30432. [DOI] [PMC free article] [PubMed]
- Chaloupka, J. A., Bullock, S. A., Iourgenko, V., Levin, L. R. & Buck, J. (2006). Mol. Reprod. Dev. 73, 361–368. [DOI] [PMC free article] [PubMed]
- Chen, Y., Cann, M. J., Litvin, T. N., Iourgenko, V., Sinclair, M. L., Levin, L. R. & Buck, J. (2000). Science, 289, 625–628. [DOI] [PubMed]
- Gertz, M., Fischer, F., Wolters, D. & Steegborn, C. (2008). Proc. Natl Acad. Sci. USA, 105, 5705–5709. [DOI] [PMC free article] [PubMed]
- Hall, R. A., De Sordi, L., Maccallum, D. M., Topal, H., Eaton, R., Bloor, J. W., Robinson, G. K., Levin, L. R., Buck, J., Wang, Y., Gow, N. A., Steegborn, C. & Mühlschlegel, F. A. (2010). PLoS Pathog. 6, e1001193. [DOI] [PMC free article] [PubMed]
- Hanoune, J. & Defer, N. (2001). Annu. Rev. Pharmacol. Toxicol. 41, 145–174. [DOI] [PubMed]
- Hess, K. C., Jones, B. H., Marquez, B., Chen, Y., Ord, T. S., Kamenetsky, M., Miyamoto, C., Zippin, J. H., Kopf, G. S., Suarez, S. S., Levin, L. R., Williams, C. J., Buck, J. & Moss, S. B. (2005). Dev. Cell, 9, 249–259. [DOI] [PMC free article] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Kamenetsky, M., Middelhaufe, S., Bank, E. M., Levin, L. R., Buck, J. & Steegborn, C. (2006). J. Mol. Biol. 362, 623–639. [DOI] [PMC free article] [PubMed]
- Linder, J. U. (2006). Cell. Mol. Life Sci. 63, 1736–1751. [DOI] [PMC free article] [PubMed]
- Litvin, T. N. (2004). PhD thesis, Cornell Medical College, New York City, USA.
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Middelhaufe, S., Leipelt, M., Levin, L. R., Buck, J. & Steegborn, C. (2012). Biosci. Rep. 32, 491–499. [DOI] [PMC free article] [PubMed]
- Mueller, U., Darowski, N., Fuchs, M. R., Förster, R., Hellmig, M., Paithankar, K. S., Pühringer, S., Steffien, M., Zocher, G. & Weiss, M. S. (2012). J. Synchrotron Rad. 19, 442–449. [DOI] [PMC free article] [PubMed]
- Rauch, A., Leipelt, M., Russwurm, M. & Steegborn, C. (2008). Proc. Natl Acad. Sci. USA, 105, 15720–15725. [DOI] [PMC free article] [PubMed]
- Schlicker, C., Rauch, A., Hess, K. C., Kachholz, B., Levin, L. R., Buck, J. & Steegborn, C. (2008). J. Med. Chem. 51, 4456–4464. [DOI] [PMC free article] [PubMed]
- Sinha, S. C. & Sprang, S. R. (2006). Rev. Physiol. Biochem. Pharmacol. 157, 105–140. [DOI] [PubMed]
- Steegborn, C., Litvin, T. N., Hess, K. C., Capper, A. B., Taussig, R., Buck, J., Levin, L. R. & Wu, H. (2005). J. Biol. Chem. 280, 31754–31759. [DOI] [PMC free article] [PubMed]
- Steegborn, C., Litvin, T. N., Levin, L. R., Buck, J. & Wu, H. (2005). Nature Struct. Mol. Biol. 12, 32–37. [DOI] [PMC free article] [PubMed]
- Sunahara, R. K. & Taussig, R. (2002). Mol. Interv. 2, 168–184. [DOI] [PubMed]
- Tesmer, J. J., Sunahara, R. K., Gilman, A. G. & Sprang, S. R. (1997). Science, 278, 1907–1916. [DOI] [PubMed]
- Wasilko, D. J., Lee, S. E., Stutzman-Engwall, K. J., Reitz, B. A., Emmons, T. L., Mathis, K. J., Bienkowski, M. J., Tomasselli, A. G. & Fischer, H. D. (2009). Protein Expr. Purif. 65, 122–132. [DOI] [PubMed]
- Zippin, J. H., Farrell, J., Huron, D., Kamenetsky, M., Hess, K. C., Fischman, D. A., Levin, L. R. & Buck, J. (2004). J. Cell Biol. 164, 527–534. [DOI] [PMC free article] [PubMed]